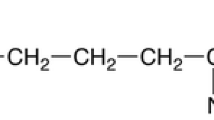Abstract
To enhance the ε-poly-l-lysine (ε-PL) productivity of Streptomyces sp. FEEL-1, a novel breeding strategy of atmospheric and room temperature plasma (ARTP) mutagenesis with streptomycin resistance screening was developed. After three rounds of recursive breeding, one strain designated AS3-14 was selected with a ε-PL yield of 2.91 g/L in shake-flask fermentation, which was elevated by 66.3 % compared to that of the parent strain FEEL-1. Subsequent batch and fed-batch fermentation was performed in a 5 L fermenter, and 8.2 g/L and 41.2 g/L ε-PL productions were achieved, which were 49.1 % and 68.1 % higher, respectively, than those of the parent strain. Analysis of enzyme activities indicated that key enzymes, e.g., hexokinase, pyruvate kinase, citrate synthase and aspartate kinase, were more active in the mutant AS3-14 than in FEEL-1, which might explain the enhanced ε-PL productivity. The results of amplified fragment length polymorphism analysis indicated that the genomic DNA of AS3-14 became more variable after ARTP mutagenesis. Therefore, ARTP combined with streptomycin resistance is an efficient breeding approach for the rapid evolution of ε-PL-producing Streptomyces strains.
Similar content being viewed by others
Introduction
ε-Poly-l-lysine (ε-PL)—a natural homopolymer produced mostly by Streptomyces—usually consists of 25–35 l-lysine residues linked by α-carboxyl and ε-amino groups (Shima and Sakai 1977, 1981a, b). ε-PL is well known for its antimicrobial activity towards bacteria, fungi, and some viruses, and therefore has been used widely as a biological preservative in Japan, Korea, and the United States (Hiraki 2000). Besides, ε-PL is water soluble, biodegradable, edible, and non-toxic towards humans and the environment. Thus, the fields of application of ε-PL are expected to broaden in future (Shih et al. 2006; Ren et al. 2014).
However, wild ε-PL-producing strains have low ε-PL biosynthesis capability so it is difficult to meet industrial needs. For example, production of ε-PL by Streptomyces albulus Z-18, Streptomyces noursei NRRL 5126, and Streptomyces griseofuscus H1 in shake flask fermentation yielded only 0.24, 0.42, and 0.7 g/L, respectively (Zhang et al. 2006; Bankar and Singhal 2010). Despite fermentation of Streptomyces noursei NRRL 5126 in optimized medium in shake-flask culture, ε-PL production was still lower than 1 g/L (Li et al. 2011). Thus, it is necessary to improve the ε-PL productivities of wild strains. Until now, several traditional breeding methods have been undertaken for ε-PL production enhancement. The most successful example was screening for mutants with S-(2-aminoethyl)-l-cysteine (AEC) plus glycine resistance after treatment of nitrosoguanidine (NTG) and ultraviolet (UV) mutagenesis. After 20 years of continuous work for, this breeding strategy had succeeded in isolating a mutant, Streptomyces albulus 11011A, with a maximum ε-PL production of 2.11 g/L, nearly 10-fold higher than that of the wild strain (Hiraki et al. 1998). With this strain, ε-PL production as high as 48.3 g/L was obtained in a 5-L jar fermenter using a two-stage pH control strategy (Kahar et al. 2001), and a ~1,000 t/a industrial scale was accomplished in Chisso Corporation in Japan in 2001. Recently, Zong et al. (2012) reported that they had improved the ε-PL yield of Streptomyces albulus A-29 from an initial 0.4 g/L to 1.59 g/L through atmospheric and room temperature plasma (ARTP) mutagenesis combined with AEC and glycine resistance screening. However, although an efficient selective marker, AEC is very expensive at about US $180 per gram. Wang et al. (2012) proposed a combined resistance screening method with sulfaguanidine + glycine + l-lysine + dl-3-hydroxynorvaline as selective markers after ARTP mutagenesis. Finally, an improved mutant Streptomyces diastatochromogenes L9 yielding ε-PL production of 0.77 g/L was obtained, which was 15 % higher than that of original strain. It can be seen that the primary principle of these latter approaches were generally based on deleting or weakening the feedback inhibition of target end products such as amino acids; however, such strategies are laborious, time-consuming and expensive.
It is well known that drug-resistance can significantly enhance the production of secondary metabolites in Streptomyces mutants. For example, a streptomycin resistant mutant Streptomyces chattanoogensis can produce fredericamycin in amounts 5-fold greater than the parent strain (Hu and Ochi 2001). Moreover, the mutants gained not only streptomycin resistance, but also could grow well under higher concentrations of other antibiotics (Hosoya et al. 1998; Ochi et al. 2004). ε-PL—a secondary metabolite in Streptomyces—may be enhanced through increasing the tolerance to antibiotics of the producing strains. Furthermore, antibiotics could also be an effective selective marker for the screening of high-producing mutants.
In our previous studies (Li et al. 2012), genome shuffling was adopted to enhance ε-PL production of wild-type strains through improving tolerance to high concentrations of carbon sources such as glucose, sulfaguanidine and succinic acid. Although this approach proved effective, the ε-PL titer in the shuffled strains remained low. Thus, a further breeding strategy was applied, i.e., interspecific hybridization involving stochastic protoplast fusion within the shuffled strains. As a result, a hybrid designated Streptomyces sp. FEEL-1 was constructed, which had a higher ε-PL production of 1.12 g/L in shake-flask fermentation (Li et al. 2013). However, genome shuffling and protoplast fusion are time-consuming and laborious due to the lack of an effective selective marker, thus a new breeding method that is brief and effective is urgently needed. In this study, a breeding method combining ARTP mutagenesis with streptomycin resistance has been developed to improve the ε-PL production of Streptomyces sp. FEEL-1. The highest ε-PL production mutant found was then evaluated in batch and fed-batch fermentations. Finally, differences in key enzymes and genomic DNA polymorphism between parent and mutant strains were investigated.
Materials and methods
Strain and medium
Streptomyces sp. FEEL-1, with a ε-PL yield of 1.75 g/L in shake-flask fermentation, is a hybrid obtained by interspecific hybridization among Streptomyces in our previous study (Li et al. 2013). It was unable to grow on agar plates containing more than 3 μg/mL streptomycin. BTN medium was used in solid culture, and M3G was employed as seed medium (Li et al. 2012; Hiraki et al. 1998). Fermentation medium was composed of glycerol 60 g, yeast extract 8 g, (NH4)2SO4 5 g, MgSO4·7H2O 2 g, KH2PO4 2 g, ZnSO4 0.03 g and FeSO4 0.04 g per liter; and its pH was adjusted to 6.8 using 2 mol/L NaOH before sterilization at 121 °C for 20 min.
Strain mutagenesis by ARTP
In each ARTP mutagenesis, a loop of starting strain was inoculated in a BTN agar plate and incubated for 7 days. Spores were washed with 10 mL sterilized phosphate buffer (2 M) and diluted to 106–107 CFU/mL. Then, 10 μL spore suspension was poured into a sterilized sample plate and exposed to the plasma jet downstream of the plasma torch nozzle exit at 2 mm distance. The operating parameters were as follows: radio-frequency power input of 120 W, gas flow at 8 L/min (Hua et al. 2010), pretreatment time of 400 s with 87.3 % lethality rate. The detailed ARTP mutagenesis procedure is shown in Fig. 1a.
Successive mutagenesis and mutant screening
After ARTP mutagenesis, the treated spores were spread on BTN agar plates containing 3 μg/mL streptomycin and cultivated for 7 days at 30 °C. A library of mutants with rapid growth was constructed, and the corresponding round of mutagenesis was described as AS1. Spores of each colony in AS1 were picked out and mixed together, being used as starters for the next round of ARTP mutagenesis (AS2). Four successive rounds of ARTP mutagenesis were performed by the same method, and in rounds AS2, AS3, and AS4, the concentrations of streptomycin on BTN agar plates were raised to 6, 12, and 24 μg/mL streptomycin, respectively. The detailed procedures of rounds AS1–AS4 are illustrated in Fig. 1b. No colonies appeared on the plates of AS4, which indicated the end of the mutagenesis. After each round of ARTP treatment, 100 mutants were isolated randomly to carry out fermentation tests in shake-flask culture, with the purposes of (1) examining the relationship between ε-PL production and streptomycin resistance (Table 2) and (2) calculating the mutation rate and positive mutation rate (Table 3).
Two control assays were carried out, control A (A1-3) and control B (S1-2); the detailed procedure of the two controls is illustrated in Fig. 1b. In control A, the spore suspension of FEEL-1 was treated by three successive rounds of ARTP mutagenesis. After each round, it was spread on BTN agar plates without streptomycin to determine whether the improved ε-PL production could be achieved without the addition of streptomycin. In control B, the spore suspension of FEEL-1 was spread on agar plates containing different concentrations of streptomycin, from 3 μg/mL to 12 μg/mL, without ARTP mutagenesis. No colonies appeared on medium containing streptomycin beyond 6 μg/mL, indicating the end of the assay. The ε-PL production of the potential strains in this study allowed 5 % random error, i.e., only those strains with more than a 5 % increase in ε-PL yield were considered as positive mutants.
Evaluation efficiency of mutagenesis to Streptomyces sp. FEEL-1 by ARTP
The positive mutation rate and the mutation rate were calculated based on the following equations:
Where R M and R P are the mutation rate and the positive mutation rate, respectively. M is the total CFU of the mutants, and P is the CFU of the mutants whose yield of ε-PL increased at least 5 % over than that of the starting strains selected from the previous round. The corresponding data are shown in Table 3.
Shake-flask and 5-L fermenter fermentation conditions
For the primary screening, three loops of aerial spores were transferred into a 250-mL shake flask containing 40 mL fermentation medium and incubated at 30 °C, 200 rpm on a rotating shaker (HYL-C, Qiang Le Laboratory Equipment Co., Taicang, China) for a 96-h cultivation. For secondary screening, the mutants with the highest ε-PL production in each round were selected out through subcultivation and incubated in three independent shake flasks; the results were expressed as mean values ± standard deviation (Table 2).
To evaluate the potential advantage of mutant AS3-14 in ε-PL production, batch and fed-batch fermentations were performed in a 5-L jar-fermenter (Baoxing, Shanghai, China) and compared with the parent FEEL-1. The 5-L fermenter containing 3.2 L sterilized fermentation medium was inoculated with 320 mL 24-h seed culture from a 500-mL shake flask cultivation. The initial pH of the culture broth was adjusted to 6.8 using NH3·H2O solution (12.5 % v/v). When the initial pH was decreased to the optimal pH of 3.8, NH3·H2O solution was added automatically to maintain this pH until fermentation finished. The pH decrease was due to (NH4)2SO4 consumption and some uncertain organic acid production. Dissolved oxygen was maintained at about 30 % of air saturation, and adjusted by varying the agitation speed from 200 to 800 rpm. The residual glycerol during fermentation was maintained at about 10 g/L by supplementing the broth with sterile glycerol via automatic pulses using a peristaltic pump. Similarly, the concentration of NH3-N was maintained at about 1 g/L in the same way as the feeding of glycerol by adding a sterile 600 g/L (NH4)2SO4 solution.
Determination of hexokinase, pyruvate kinase, citrate synthase and aspartokinase activities
Spores of strains were inoculated into fermentation medium at 30 °C, 200 rpm on a rotating shaker. After 30 h incubation, mycelium of each strain in 150 mL culture broth was prepared for analysis of cell-free crude enzymes (Chen and Mao 2013). Enzyme activities of hexokinase (HK), pyruvate kinase (PK), citrate synthase (CS) and aspartokinase (AsK) were determined according to methods previously published by Teichgraber et al. (1972), Morris et al. (1984), Zeng et al. (2014) and Chen and Mao (2013), respectively. All assays of enzyme activities in two strains were performed in triplicate and the results were expressed as mean values ± standard deviation (Table 4). Protein concentrations were measured as described by Bradford (1976). All chemicals, including substrates, coenzymes and buffers were obtained from Sigma-Aldrich (St. Louis, MO).
Amplified fragment length polymorphism analysis
Amplified fragment length polymorphism (AFLP) analysis was conducted as described by Jin et al. (2008) with a little modification. The genomic DNA was obtained by 20-mg/mL lysozyme treatment and extraction using a nucleic acid purification kit (Dongsheng Biotech Co., Guangzhou, China), and then 0.5 μg genomic DNA was digested with Pst I/Sac II (Takara Biotechnology Co., Dalian, China). The restriction sites in the digested DNA were ligated to Pst I- and Sac II-adapters (Table 1) by adding T4 DNA ligase for 4 h at 16 °C. Next, the ligated DNA sample was used as a template for preamplification polymerase chain reaction (PCR). The preamplification PCR was conducted by adding Taq polymerase, Pst I and Sac II preamplification primers in a Mastercycler pro PCR thermal cycler (Eppendorf, Hamburg, Germany) for 20 cycles of 30 s at 94 °C, 1 min at 56 °C and 1 min at 72 °C. After the preamplification PCR, the product obtained was diluted 50-fold and used as the template for the following selective amplification PCR. As each of the four selective primers consists of one selective nucleotide (Table 1), the restriction enzyme combination of PstI/SacII has 16 unique primer pairs for each strain in the assay. The selective amplification PCR was conducted by adding Taq polymerase, Pst I and Sac II selective primers in a Mastercycler pro PCR thermal cycler with the following program: (1) 1 cycle of 1 min at 94 °C, (2) 12 cycles of denaturation for 30 s at 94 °C, annealing with a decreasing rate at [65 − (n − 0.5)]°C for 30 s (here n is the cycle number), and extension for 60 s at 72 °C, and (3) 22 cycles of denaturation for 30 s at 94 °C, annealing for 30 s at 56 °C, and extension at 72 °C for 60 s. The processes of electrophoresis and silver staining were conducted as described by Jin et al. (2008).
Analysis of dry cell weight, ε-PL concentration and glycerol concentration
The fermentation broth was sampled every 6 h and centrifuged at 4,500 g for 10 min. The resulting precipitate was washed twice with sterile water and dried to a constant weight at 105 °C to measure the dry cell weight (DCW) of the culture, and the supernatant was used to determine the concentration of ε-PL and glycerol. ε-PL concentration in various solutions was determined according to the method described by Itzhaki (1972). The concentration of glycerol was detected by an HPLC system (Dionex U-3000, Thermo Fisher Scientific, Sunnyvale, CA) with an ion exchange column (Aminex HPX-87H, Bio-Rad, Hercules, CA) and a refraction index detector (Shodex RI-101, Tokyo, Japan). The elution of the ion exchange column was performed at 60 °C with 5 mM H2SO4, and a flow rate of 0.6 mL/min.
Results and discussion
Mutagenesis and selection of mutants with improved ε-PL productivity
After three rounds of streptomycin-aided ARTP mutagenesis, strain AS3-14 was obtained, which has a ε-PL yield of 2.91 g/L—66.3 % higher than the original strain FEEL-1 (Table 2). Statistical analysis on round AS1-3 revealed high mutation rates were obtained with 26.1–33.0 %, while the rates of UV mutagenesis and chemical mutagenesis in our previous work were only 7.6 % and 15.0 %, respectively (data not shown). One of the possible reasons for the high mutation rate in Table 3 is that the ARTP system produces atmospheric-pressure helium plasma, which consists mainly of different chemically active species upon irradiated of the stainless steel plate during mutagenesis (Zhang et al. 2014). The plasma jet temperature was kept below 40 °C to make sure that the damage to the spores was caused by active species rather than by high temperature. According to recent studies, variation in the plasma dosage in ARTP mutagenesis may contribute to a diverse pattern of breakage of plasmid DNA and oligonucleotides, and the mechanism of action differs significantly from UV and chemical mutagenesis as analyzed by the umu test (Oda et al. 1985). Besides, another key parameter, positive mutation rate, was 37 %–44.4 %, which, although expected, was a very large value. Therefore, selection of streptomycin-resistance mutants after ARTP treatment proved to be an effective approach for breeding of higher ε-PL-producing strains.
The result of control A showed that the highest ε-PL production of A3-29 was only 2.24 g/L, which means ARTP mutagenesis without addition of streptomycin can also enhance the ε-PL production. In recent publications, ARTP mutagenesis has been reported to improve the production of avermectin in Streptomyces avermitilis, and cellulase in Trichoderma viride. Avermectin production in Streptomyces avermitilis increased from 6,100 mg/L to 8,540 mg/L, and cellulase production in Trichoderma viride showed a 2.61-fold improvement compared to the parent strain (Wang et al. 2010; Xu et al. 2011). According to the data in Table 2, although the average R M of A1-3 was a little higher than that of AS1-3, the average R p was much lower than that of AS1-3. Therefore, ARTP seemed to increase the mutation rate a great deal, whereas monitoring streptomycin-resistance resulted in the efficient screening of high-yield strains. In control B, the result showed that no colonies appeared on medium containing streptomycin beyond 6 μg/mL, indicating that enhanced streptomycin-resistance could be obtained efficiently only by successive rounds of ARTP mutagenesis. Furthermore, the highest production of strain S2-2 was 2.15 g/L, which means resistance to streptomycin can also lead to the enhancement of ε-PL production. As stated above, although the sole use of ARTP mutagenesis or streptomycin-resistance can improve the yield to some extent, the difference in ε-PL production between the control and AS3-14 was still large. This study demonstrated that the combination of ARTP and streptomycin-resistance contributed to the improvement of ε-PL production; however, ARTP mutagenesis or streptomycin resistance alone has no significant effect.
During three rounds of ARTP mutagenesis, more than 900 colonies were observed in culture. Growth rate and morphology was seen to vary among the colonies. The morphology of mutants with the highest ε-PL production in each round of mutagenesis is illustrated in Fig. 2: the parent strain FEEL-1 appeared hoary, flat and glabrate with a few filamentous mycelia. However, mutants obtained from AS1–AS3 were rugous and rich in filamentous mycelia. In addition, the color of the spores of some mutants from AS1 and AS2 changed to green after cultivating for 4 days, whereas spores of FEEL-1 are gray. Therefore, the various morphologies observed in AS1 to AS3 proved that ARTP mutagenesis can change mutant morphology. The variety of morphology in the actinomycetes can reflect gene diversity and alternative products, thus morphological change is usually considered an indicator for gene mutation of a strain. Some explanation of the underlying reasons can be found in a report on Streptomyces avermitilis, whose entire genome has been reported, which states that many key regulatory steps involved in morphological differentiation and secondary metabolism of this organism share common genes (Wang et al. 2010). Mutants with different morphologies produced ε-PL at different levels. The production of stain F, which looks similar to FEEL-1, proved to be the same as FEEL-1. Thus the diversified morphology reduced the difficulties in screening for hyper-yield strains.
Typical images of colonies with the highest ε-PL production at each round of mutagenesis. FEEL-1 Parent strain, F a strain morphologically similar to FEEL-1 in the assays; S1-4, S2-2 mutants obtained in streptomycin mutagenesis (control B); A1-33, A2-57, A3-29 mutants developed from treated spores in corresponding rounds of ARTP mutagenesis (control A); AS1-24, AS2-68, AS3-14 mutants selected in corresponding rounds of ASn (n = 1–3); All pictures were obtained after the plate had been cultivated for 5 days at 30 °C
Performance of mutant AS3-14 in batch and fed-batch bioreactor fermentation
After three rounds mutagenesis, a mutant designated AS3-14 was selected with the highest ε-PL production of 2.91 g/L in shake-flask fermentation. Subsequently, large-scale fermentation was carried out to verify whether advantage could be taken of its high productivity to meet industrial needs. The batch fermentation performance of AS3-14 was compared with that of the parent strain FEEL-1 in a 5-L fermenter under the same fermentation conditions. As shown in Fig. 3, time profiles of those two strains exhibited the same tendency in terms of pH decline, glycerol consumption, cell growth and ε-PL production. With respect to pH, both pHs dropped naturally within 16 h to pH 3.8–4.0, which is the optimal pH for ε-PL production (Fig. 3a). However, AS3-14 exhibited a more rapid glycerol consumption rate and consumed up 60 g/L in 50 h, i.e., 5 h shorter than required by FEEL-1 (Fig. 3b). In terms of cell growth, both strains maintained the same growth rate and achieved a maximum biomass of 19 ± 0.5 g/L (Fig. 3c). As anticipated, mutant AS3-14 accumulated a high ε-PL level of 8.2 g/L, which was 49.1 % higher than that of the parent FEEL-1, indicating that ε-PL biosynthesis ability of AS3-14 has been improved significantly by the mutagenesis strategy used (Fig. 3d). As the fed-batch fermentation approach is widely employed for ε-PL production (Kahar et al. 2001; Ren et al. 2014; Chen and Mao 2013), AS3-14 was further evaluated using this approach by feeding glycerol and ammonium sulfate (Fig. 4). After a 192-h fed-batch fermentation, ε-PL production reached 41.2 g/L, whereas 24.5 g/L of ε-PL was realized with the parent strain FEEL-1 under the same operational conditions in our previous study (Li et al. 2013). The above result showed that the mutant AS3-14 has acquired enhanced ε-PL synthetic ability through ARTP mutation combined with streptomycin resistance screening.
Analysis of key enzyme activities in mutant AS3-14 and the parent FEEL-1
To further explore the improved ε-PL synthetic ability of AS3-14, key enzyme activities (i.e., HK, PK, CS and AK) were investigated and compared with those of the parent strain FEEL-1. As shown in Table 4, AS3-14 possessed clearly higher activities of HK, with a 2.47-fold enhancement compared to FEEL-1, which suggested that the Embden-Meyerhof-Parnass (EMP) pathway in AS3-14 was more active than that in FEEL-1. Furthermore, the elevated CS enzyme activity (1.84-fold enhancement) may direct more metabolic flux to the tricarboxylic acid (TCA) cycle, which can then provide an abundance of oxaloacetic acid (OAA), the precursor of the diaminopimelate (DAP) pathway. The strengthened TCA cycle may also offer more ATP for ε-PL synthesis. This finding is similar to that of a previous report that adding citrate to the medium could facilitate the ε-PL production of strain USE-51 (Hirohara et al. 2006). In addition, the activity of AK–the first key enzyme in the DAP pathway—was 160 % higher in AS3-14 than that in FEEL-1. This improvement resulted in a large metabolic flux through the DAP pathway, which can provide more precursor lysine. The improved activities of these key enzymes may go some way to explaining the high ε-PL production of the mutant.
Assessment of genomic variation analysis between parent and mutant strains by AFLP analysis
AFLP analysis combined with restriction enzymes Pst I/Sac II is an efficient method with which to distinguish genomic DNA variation among parent strains and the mutants, the ε-PL productivities of which were 1.75, 1.99, 2.62, and 2.91 g/L, respectively. Sixteen primer pairs were tested to detect the most appropriate pairs of selective primers that yielded the most informative and polymorphic AFLP banding patterns. Results obtained with the six primer pairs selected are illustrated in Fig. 5. The number of fragments amplified and the polymorphism rate were calculated and are shown in Table 5. The data showed that the average polymorphism rates reached were 1.9 %, 6.5 %, and 13.4 % for parent FEEL-1, and mutants AS2-68 and AS3-14, respectively, indicating an obvious increasing polymorphism in mutants compared to the parent. Moreover, this result also showed a positive relationship between polymorphism rate and ε-PL yield. The results presented in Fig. 5 show that the mutant AS2-68 had more polymorphic bands than mutant A2-57, suggesting that genomic variation happened more easily and extensively when streptomycin was added in the course of induced mutagenesis. As a result, the strategy of ARTP mutagenesis combined with streptomycin-resistance resulted in higher genomic variation of mutants and enhanced ε-PL production.
Amplified fragment length polymorphism (AFLP) banding patterns of the mutants derived in the course of ARTP mutagenesis assays and the ancestor FEEL-1. The examples demonstrated here were generated with the selective primer combination Pst I-A/Sac II-G. 1, 2 and 4 polymorphic bands (arrows) could be distinguished, respectively, from the mutant A2-57 obtained from control A, and the mutants AS2-68 and AS3-14 from rounds AS2 and AS3, respectively, which were absent in the ancestor FEEL-1
Conclusion
In this study, a mutant Streptomyces sp. AS3-14 producing 2.91 g/L ε-PL was obtained through ARTP mutagenesis combined with streptomycin-resistance screening. After a 192-h fed-batch fermentation, ε-PL production of 41.2 g/L was realized by mutant AS3-14, suggesting that the strain has potential for industrial application. Further analyses showed that there are significant differences between the mutant AS3-14 and parent FEEL-1 strain in key enzymes and in genomic DNA polymorphism. The results demonstrate that the strategy of ARTP mutagenesis combined with streptomycin-resistance screening provides a powerful approach for obtaining enhanced ε-PL-producing strains.
References
Bankar SB, Singhal RS (2010) Optimization of poly-ε-lysine production by Streptomyces noursei NRRL 5126. Bioresour Technol 101:8370–8375. doi:10.1016/j.biortech.2010.06.004
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Chen XS, Mao ZG (2013) Comparison of glucose and glycerol as carbon sources for ε-poly-l-lysine production by Streptomyces sp. M-Z18. Appl Biochem Biotechnol 170:185–197. doi:10.1007/s12010-013-0167-5
Hiraki J (2000) ε-Polylysine, its development and utilization. Fine Chem 29:18–25
Hiraki J, Masakazu H, Hiroshi M, Yoshikazu I (1998) Improved ε-poly-l-lysine production of an S-(2-aminoethyl)-l-cysteine resistant mutant of Streptomyces albulus. Seibutsu Kogakkaishi 76:487–493
Hirohara H, Takehara M, Saimura M, Masayuki A, Miyamoto M (2006) Biosynthesis of poly(ε-l-lysine)s in two newly isolated strains of Streptomyces sp. Appl Microbiol Biotechnol 73:321–331
Hosoya Y, Okamoto S, Muramatsu H, Ochi K (1998) Acquisition of certain streptomycin-resistant (str)mutations enhances antibiotic production in bacteria. Antimicrob Agents Chemother 42:2041–2047
Hu H, Ochi K (2001) Novel approach for improving the productivity of antibiotic-producing strains by inducing combined resistant mutations. Appl Environ Microbiol 67:1885–1892. doi:10.1128/AEM. 67.4.1885-1892.2001
Hua XF, Wang J, Wu ZJ, Zhang HX, Li HP, Xing XH, Zheng L (2010) A salt tolerant Enterobacter cloacae mutant for bioaugmentation of petroleum- and salt-contaminated soil. Biochem Eng J 49:201–206. doi:10.1016/j.bej.2009.12.014
Itzhaki RF (1972) Colorimetric method for estimating poly-lysine and polyarginine. Anal Biochem 50:569–574
Jin QC, Jin ZH, Xu B, Wang Q, Lei YL, Yao SJ, Cen P (2008) Genomic variability among high pristinamycin-producing recombinants of Streptomyces pristinaespiralis revealed by amplified fragment length polymorphism. Biotechnol Lett 30:1423–1429
Kahar P, Iwata T, Hiraki J, Park EY, Okabe M (2001) Enhancement of ε-polylysine production by Streptomyces albulus strain 410 using pH control. J Biosci Bioeng 91:190–194
Li S, Tang L, Chen XS, Liao LJ, Li F, Mao ZG (2011) Isolation and characterization of a novel ε-poly-l-lysine producing strain: Streptomyces griseofuscus. J Ind Microbiol Biotechnol 38:557–563. doi:10.1007/s10295-010-0803-9
Li S, Li F, Chen XS, Wang L, Xu J, Tang L, Mao ZG (2012) Genome shuffling enhanced ε-poly-l-lysine production by improving glucose tolerance of Streptomyces graminearus. Appl Biochem Biotechnol 166:414–423. doi:10.1007/s12010-011-9437-2
Li S, Chen XS, Dong CL, Zhao FL, Tang L, Mao ZG (2013) Combining genome shuffling and interspecific hybridization among Streptomyces improved ε-poly-l-lysine production. Appl Biochem Biotechnol 169:338–350. doi:10.1007/s12010-012-9969-0
Morris CN, Ainsworth S, Kinderlerer J (1984) The regulatory properties of yeast pyruvate kinase. Biochem J 217:641–647
Ochi K, Okamoto S, Tozawa Y, Inaoka T, Hosaka T, Xu J, Kurosawa K (2004) Ribosome engineering and secondary metabolite production. Adv Appl Microbiol 56:155–184
Oda Y, Nakamura S, Oki I, Kato T, Shinagawa H (1985) Evaluation of the new system (umu-test) for the detection of environmental mutagens and carcinogens. Mutat Res 147:219–229
Ren XD, Chen XS, Tang L, Sun QX, Zeng X, Mao ZG (2014) Efficient production of ε-poly-l-lysine from agro-industrial by-products by Streptomyces sp. M-Z18. Ann Microbiol. doi:10.1007/s13213-014-0913-0
Shih IL, Shen MH, Van YT (2006) Microbial synthesis of poly (ε-lysine) and its various applications. Bioresour Technol 97:1148–1159. doi:10.1016/j.biortech.2004.08.012
Shima S, Sakai H (1977) Poly-l-lysine produced by Streptomyces. Agric Biol Chem 41:1807
Shima S, Sakai H (1981a) Poly-l-lysine produced by Streptomyces. Part II. Taxonomy and fermentation studies. Agric Biol Chem 45:2497–2502
Shima S, Sakai H (1981b) Poly-l-lysine produced by Streptomyces. Part III. Chemical studies. Agric Biol Chem 45:2503–2508
Teichgraber P, Biesold D, Pigereva ZD (1972) Subcellular localization of hexokinase in the rat cortex. Biokhimiya 37:748–756
Wang LY, Huang ZL, Li G, Zhao HX, Xing XH, Sun WT, Li HP, Gou ZX, Bao CY (2010) Novel mutation breeding method for Streptomyces avermitilis using an atmospheric pressure glow discharge plasma. J Appl Microbiol 108:851–858. doi:10.1111/j.1365-2672.2009.04483.x
Wang T, Jia SR, Tan ZL, Dai YJ, Song S, Wang G (2012) Mutagenesis and selective breeding of a high producing ɛ-poly-l-lysine strain. Front Chem Sci Eng 6:179–183
Xu F, Wang J, Chen S, Qin W, Yu Z, Zhao H, Xing XH, Li H (2011) Strain improvement for enhanced production of cellulase in Trichoderma viride. Appl Biochem Microbiol 47:53–58. doi:10.1134/s0003683811010212
Zeng X, Chen XS, Ren XD, Liu QR, Wang L, Sun QX, Tang L, Mao ZG (2014) Insights into the role of glucose and glycerol as a mixed carbon source in the improvement of ε-poly-l-lysine productivity. Appl Biochem Biotechnol 10:1007–1021
Zhang C, Zhang DR, He W, Duan ZY, Mao ZG (2006) A simple and sensitive method for screening ε-PL producing strains from soils. J Shandong Univ (China) 44:1103–1107
Zhang X, Zhang FX, Li HP, Wang LY, Zhang C, Xing XH (2014) Atmospheric and room temperature plasma (ARTP) as a new powerful mutagenesis tool. Appl Microbiol Biotechnol 98:5387–5396. doi:10.1007/s00253-014-5755-y
Zong H, Zhan Y, Li X, Peng LJ, Feng FQ, Li D (2012) A new mutation breeding method for Streptomyces albulus by an atmospheric and room temperature plasma. Afr J Microbiol Res 6:3154–3158. doi:10.5897/ajmr11.1251
Acknowledgments
This work was supported by the Project of Scientific and Technical Supporting Program of Jiangsu (BE2012616), the Cooperation Project of Jiangsu Province among Industries, Universities and Institutes (BY2013015-11), the Open Project Program of the Key Laboratory of Industrial Biotechnology, Ministry of Education, China (KLIBKF201302), the National Natural Science Foundation of China (21376106), and the Jiangsu Province “Collaborative Innovation Center for Advanced Industrial Fermentation” Industry Development Program.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Wang, L., Chen, X., Wu, G. et al. Improved ε-poly-l-lysine production of Streptomyces sp. FEEL-1 by atmospheric and room temperature plasma mutagenesis and streptomycin resistance screening. Ann Microbiol 65, 2009–2017 (2015). https://doi.org/10.1007/s13213-015-1039-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13213-015-1039-8