Abstract
Mycoplasma genitalium (M. genitalium) has emerged as a sexually transmitted infection (STI) all over the world in the last three decades. It has been identified as a cause of male urethritis, and there is now evidence that it also causes cervicitis and pelvic inflammatory disease in women. However, the precise role of M. genitalium in diseases such as pelvic inflammatory disease, and infertility is unknown, and more research is required. It is a slow-growing organism, and with the advent of the nucleic acid amplification test (NAAT), more studies are being conducted and knowledge about the pathogenicity of this organism is being elucidated. The accumulation of data has improved our understanding of the pathogen and its role in disease transmission. Despite the widespread use of single-dose azithromycin in the sexual health field, M. genitalium is known to rapidly develop antibiotic resistance. As a result, the media frequently refer to this pathogen as the “new STI superbug.” Despite their rarity, antibiotics available today have serious side effects. As the cure rates for first-line antimicrobials have decreased, it is now a challenge to determine the effective antimicrobial therapy. In this review, we summarise recent M. genitalium research and investigate potential therapeutic targets for combating this pathogen.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
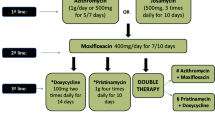
Mycoplasma genitalium (M. genitalium) is known to be a growing source of sexually transmitted infections (STIs) over the world, and it has been linked to both male and female urogenital infections, that include urethritis, cervicitis, pelvic inflammatory disease and preterm birth (Smolarczyk et al. 2021; Sethi et al. 2012; Jensen 2004; Lis et al. 2015; Lewis et al. 2020; Averbach et al. 2013). Non-gonococcal urethritis (NGU) cases were recorded continuously in about 30% of total STI cases (Manhart et al. 2013; Greydanus et al. 2022). M. genitalium co-infections with other human STIs such as the human immunodeficiency virus (HIV), chlamydia, and gonorrhoeae have also been reported in various studies (Napierala Mavedzenge et al. 2015; Fernández-Huerta and Espasa 2019; Harrison et al. 2019; Mahlangu et al. 2019). The Centers for Disease Control and Prevention (CDC) includes a guideline for M. genitalium infection diagnostics, management, and therapy in the Sexually Transmitted Diseases Treatment Guidelines, 2021, due to its clinical significance (Workowski et al. 2021). Despite these recommendations, treating M. genitalium infection remains difficult (Horner and Martin 2017; Sweeney et al. 2022b; Ross and Jensen 2006). It soon became evident that treatment was being restricted by antibiotic resistance (Raj et al. 2022). The degrees of resistance vary by location and amongst populations within a given area (Jensen et al. 2008; Workowski and Bolan 2015; Deguchi et al. 2017; Valentine-King et al. 2019). Another problem while treating M. genitalium is the high persistence of this bacterium in humans (Cina et al. 2019; Romano et al. 2019). This emphasizes the significance of conducting antibiotic susceptibility test prior to starting treatment. As this pathogen lacks a cell wall, it is immune to antibiotics that target cell wall production, such as beta-lactams (penicillins and cephalosporins) (Jensen and Bradshaw 2015; Iwuji et al. 2022). Antibiotics such as macrolides, tetracycline, and quinolones have been shown to be useful in treating this infection in various studies (Horner et al. 2016; Bradshaw et al. 2017; Sweeney et al. 2022a). The rapid rise in antimicrobial resistance to these drugs, on the other hand, is concerning. The recommended 1 g single dose of azithromycin for NGU treatment, for example, has not only been ineffective against M. genitalium but has also resulted in macrolide-resistant M. genitalium (Taylor-Robinson 2014; Jensen et al. 2016; Gnanadurai and Fifer 2020). Second-line antibiotics (such as moxifloxacin) are also effective against M. genitalium, however, resistance to these antibiotics has been reported in several countries (Manhart et al. 2015; Murray et al. 2017; Muller et al. 2019; Ke et al. 2020; Pitt and Fifer 2022). Researchers are exploring novel therapeutic targets in M. genitalium to develop new lead/drug molecules to cure this devastating infection because of the growing antibiotic resistance.
In this review, we have focused on the therapeutic targets of M. genitalium. A summary of all therapeutic targets is shown in Table 1. We hope that the reader will have a better understanding of these therapeutic targets that can aid in the current treatment of M. genitalium resistance development and the development of novel lead/drug molecules against these targets.
Drug target identification
Target identification in a biological pathway is one of the important steps in drug development process. In theory, when a pathogen's target is identified, it is critical that the putative target is either absent in the host or significantly different from the host homolog so that it may be used as a therapeutic target (Zhang et al. 2022). The evolution of antimicrobial resistance in Gram-positive bacteria has prompted researchers to look for new therapeutic targets that could help in the development of new antibiotics. Their cell organization differs greatly from that of mammalian cells, making it possible to identify pathogen-specific targets. The chosen target must be extremely important for the pathogen’s survival (Naorem et al. 2022). The biochemical properties of the target should also be considered. It must have a small molecule binding pocket so that specific inhibitors could be designed, and if the target protein is an enzyme, its inhibition should result in a loss of cell viability. It is critical that the target chosen be assayable (Uddin et al. 2021; Chandela et al. 2022). A general workflow for drug target identification is depicted in Fig. 1. The application of bioinformatics tools in comparative genomics is extremely beneficial in predicting novel therapeutic targets against a pathogen. The goal is to identify essential and non-homologous proteins in the pathogen with distinct metabolic pathways that could be used as novel drug targets. Based on this, a manual comparison of M. genitalium metabolic pathways with human host is performed in order to identify pathogen-specific metabolic pathways. Furthermore, the entire proteome of M. genitalium is analysed using various bioinformatics databases and tools to identify essential, non-homologous, and cytoplasmic proteins involved in pathogen-specific metabolic pathways. These proteins are then tested for druggability in the DrugBank database. Following further investigation, druggable proteins are chosen as novel putative therapeutic targets (Nogueira et al. 2021; Fatoba et al. 2021). For high-throughput screening of compounds against these targets, an affordable and specific assay system should be available (Fatoba et al. 2021).
Drug targets in Mycoplasma genitalium
Prior knowledge
M. genitalium is a common sexually transmitted bacterium that causes up to 25% of cases of nongonococcal urethritis and has been linked to cervicitis and pelvic inflammatory disease. The macrolide antibiotic azithromycin is the standard first-line treatment, however, an increase in treatment failure over the last five years shows the development of antibiotic resistance. In many M. genitalium populations, mutations causing macrolide resistance have been discovered in the 23S rRNA gene. When azithromycin became ineffective, the fluoroquinolone, moxifloxacin was deemed to be a viable option. However, several investigations confer the fluoroquinolone (second-line antibiotic) resistance due to mutations in the parC and gyrA genes. However, various quinolones, like moxifloxacin, show substantial antimicrobial activity against M. genitalium, and it is recommended as a second-line treatment for persistent or recurrent NGU (Jernberg et al. 2008). Quinolones shows their antimicrobial properties by inhibiting topoisomerases II, DNA gyrase (composed of two GyrA and two GyrB subunits), and topoisomerase IV (composed of two ParC and two ParE subunits). Quinolone resistance has been linked to alterations in the genes that determine quinolone resistance of the genes encoding DNA gyrase and/or topoisomerase IV in different species of bacteria, including mycoplasmas (Yoshida et al. 1990; Pan et al. 1996; Bebear et al. 1999; Gruson et al. 2005).
Current advances
Glycosyltransferase
Glycoglycerolipids (galactolipids) are membrane components linked with photosynthetic tissues found in plant chloroplasts as well as in cyanobacteria (Hölzl and Dörmann 2007; Benning 2008). Gram-positive bacteria create glycolipids with a wider structural diversity, in which the diacylglycerol headgroups contain glucosyl, galactosyl, or mannosyl units with a range of glycosidic linkages (Hölzl and Dörmann 2007). Their functions range from serving as membrane anchors for other biomolecules (such as lipoteichoic acids) to act as free components in the bilayer stability of membranes (Kiriukhin et al. 2001; Gründling and Schneewind 2007; Hölzl and Dörmann 2007). As part of the subsequent role, glycosyl residues are incorporated into monoglycosylidiacylglycerol to produce diglycosylidiacylglycerol that forms bilayers (Wieslander et al. 1980; Lindblom et al. 1986; Dahlqvist et al. 1995). Glycosyltransferases (GTs) function on a diacylglycerol substrate to start the synthesis of glycoglycerolipids. Glycolipid synthases are divided into GT families based on their sequence (carbohydrate-active enzyme classification). Despite the fact that only a few have been studied enzymatically, most have mono- or di synthase ability, with one enzyme for diacylglycerol glycosylation and another for further glycosylation to produce the diglycosyldiacylglycerol product (Cantarel et al. 2009). Monoglucosyldiacylglycerol (MGlcDAG) synthases belong to the GT2 family of enzymes in cyanobacteria (Awai et al. 2006), while digalactosyldiacylglycerol (DGalDAG) synthases are tentatively referred to the GT4 family (Sakurai et al. 2007). It has been shown that Mycoplasma pneumoniae contains a processive GT2 that produces mostly galactoglycerolipids (Klement et al. 2007). It is believed that GTs implicated in the biosynthesis of glycoglycerolipids could be viable therapeutic targets due to the critical significance of these components in membrane stability in Mycoplasma and the fact that these lipid structures are lacking in the animal host cells. A high-throughput screening test for GT based on fluorescence polarisation (FP-tag) was utilised to discover marine natural compounds as lead inhibitors (Gao et al. 2019). GT was also considered a suitable target in various other studies on Staphylococcus aureus, Citrobacter rodentium and Escherichia coli (E. coli) (El Qaidi et al. 2018; Kattke et al. 2019; Zhu et al. 2021). GT2 enzymes are found in the majority of Mycoplasma genomes (over 18 species) (Klement et al. 2007). They could be used to develop novel antibiotics.
Thymidylate kinase
Thymidylate kinase (TMK) has been proposed as a potential therapeutic targets by various researchers in M. genitalium (Kazakiewicz et al. 2015; Butt et al. 2012). It catalyses the phosphorylation of thymidine 5ʹ-monophosphate (dTMP) in the presence of adenosine triphosphate (ATP) and Mg2 + to generate thymidine 5'-diphosphate (dTDP), which is then transformed to thymidine 5ʹ-triphosphate (dTTP) by nucleoside-diphosphate kinase (NDK) (Reichard 1988; Huang et al. 1994; Lavie et al. 1997). dTTP is a crucial component of DNA synthesis, and its activity is carefully regulated throughout the cell cycle and during cell proliferation (Reichard 1988; Ke et al. 2005). TMK is also the last specific enzyme in the routes for the synthesis of dTTP, a critical component of DNA synthesis. To prevent the onset of such diseases interrupting dTTP metabolism could play as key role (Cui et al. 2013). TMKs are intensively investigated as prospective therapeutic targets in various other infectious diseases such as infections caused by Staphylococcus aureus and Mycobacterium tuberculosis (Martínez-Botella et al. 2013; Gul et al. 2020). In addition, Keating et al. reported piperidinylthymines (a group of synthetic molecules) as potential inhibitors of TMK in Staphylococcus aureus infection in mouse model (Keating et al. 2012).
Biosynthesis of secondary metabolites
Small bioactive compounds produced by bacteria in the stationary phase of growth are known as secondary metabolites (Ruiz et al. 2010). They are not needed for bacteria’s growth and survival, but they do give them a competitive advantage (Craney et al. 2013). Several studies have demonstrated the modulation of bacterial secondary metabolite pathways and their use in developing antibiotics, antiviral medications, and other pharmaceuticals (Weber et al. 2003; Ozcengiz and Demain 2013; Onaka 2017). Phosphomannomutase, Acyl carrier protein homolog, and Phosphate acyltransferase are all discovered in M. genitalium and are found to be involved in secondary metabolite biosynthesis (Fatoba et al. 2021). The conversion of mannose-6-phosphate to mannose-1-phosphate, the main function of phosphomannomutase (ManB) is involved in the biosynthesis of guanosine diphosphate (GDP)-mannose for a variety of processes in prokaryotes and eukaryotes, including the synthesis of structural carbohydrates, the production of alginates and ascorbic acid, and the post-translational modification of proteins (Yang et al. 2010).
Universal and highly conserved Acyl carrier protein (ACP) plays a major role in fatty acid biosynthesis as a carrier of acyl intermediate. The fatty acid synthase polyprotein (type 1 FAS) found in yeast and mammals contains ACP as a separate domain, whereas the type II FAS found in bacteria and plastids contains ACP as a small monomeric protein. A bacterial acyl carrier protein is involved in the synthesis of different products like endotoxin and acylated homoserine lactones as acyl donors. These products have an essential role in quorum sensing (Byers and Gong 2007). ACP-dependent enzymes are attractive antimicrobial drug targets because of their distinct and essential roles in growth and pathogenesis (Fatoba et al. 2021; Byers and Gong 2007). Furthermore, ACP homologs have a role in the production of secondary metabolites such as polyketides and nonribosomal peptides (Byers and Gong 2007).
The role of Phosphate acyltransferase is the reversible synthesis of acyl-phosphate (acyl-PO4) from acyl-[acyl-carrier-protein], which is catalysed by this enzyme. It uses acyl-ACP instead of acyl-CoA as a fatty acyl donor (Zhang and Rock 2008).
These three proteins involvement is invariable with observations from earlier studies showing the proteins implicated in this pathway are essential, non-homologous, and specific to various bacterial pathogens such as Staphylococcus saprophyticus and Vibrio parahaemolyticus (Hasan et al. 2020; Shahid et al. 2020).
Quorum sensing
Communication between microbial cells is known as quorum sensing (QS) and it relies on the detection and creation of signals called autoinducers (AIs) to access cell density and species complexity in populations. QS supports the coordination of collective behaviour among bacteria. AI ligands are quite specific to some QS receptors, while others are more promiscuous. QS is required for bacterial species to regulate crucial cellular processes that are required for surveillance, survival, and adaptation to changing environments. Bacteria can correlate with population density and species complexity changes in the vicinity and respond as a group when they monitor the accumulation of specific AIs (Hawver et al. 2016). The discovery of microbial quorum-sensing systems in recent years has sparked renewed interest in investigating drug resistance mechanisms as well as combating drug resistance. The findings reveal that the quorum-sensing system regulates a variety of cellular functions in microbes, including pathogenic gene expression, toxin production, and extracellular polysaccharide synthesis. It also plays a major regulatory role in drug efflux pumps and microbial biofilm development (Turan et al. 2017; Bäuerle et al. 2018; Wei and Zhao 2018). Antibiotic synthesis and sporulation are two further activities governed by quorum sensing (Abisado et al. 2018). In M. genitalium, the protein translocase subunit, which has been identified as a potential therapeutic target, is implicated in the quorum-sensing metabolic pathway (Fatoba et al. 2021). The SecYEG preprotein conducting channel interacts with a part of the Sec protein translocase complex, as shown in Fig. 2, serves as an ATP-driven molecular motor that drives the progressive translocation of polypeptide chains across the cell membrane, connecting ATP hydrolysis to protein transfer into and across the cell membrane (Veenendaal et al. 2004). Biofilm production contributes to antibiotic resistance in M. genitalium, according to a recent study by Daubenspeck and co-workers (Daubenspeck et al. 2020). Proteins from this metabolic pathway have been identified as possible therapeutic targets due to the substantial involvement of quorum sensing in biofilm development in most bacteria (Haque et al. 2018; Jiang et al. 2019; Daubenspeck et al. 2020). Furthermore, the protein translocase component was found to be involved in the metabolic process of the bacterial secretion system (Butt et al. 2012). In the development of an effective antibacterial treatment against M. genitalium, the bacterial secretion system was also proposed as a novel therapeutic target to consider. These systems have been considered as therapeutic targets in many bacterial species such as Staphylococcus aureus, Listeria monocytogenes, Vibrio cholerae, Klebsiella pneumoniae, and Yersinia enterocolitica (Cegelski et al. 2008; McShan and De Guzman 2015; Green and Mecsas 2016).
Schematic representation of the preprotein translocase of E. coli. The cartoon shows the different units of the preprotein translocase including the motor protein SecA which drives preproteins across the membrane via a channel made up of an oligomeric assembly of SecYEG complexes. The SecDFyajC is another heterotrimeric membrane protein complex that stimulates the preprotein translocation. The ribosome contacts the SecYEG complex and SecA protein to facilitates the co-translational insertion of membrane proteins (Veenendaal et al. 2004)
Bacterial secretion system
Typically bacterial secretion system allows proteins to be transported from their cytoplasm to their host environment or straight into their host cells. This results in serious damage to the host cell due to increased virulence and pathogenicity of the bacteria (Waksman 2012). Proteins involved in bacterial secretion system such as protein translocase subunit SecA could be a suitable therapeutic target in M. genitalium (Fatoba et al. 2021). SecA is required for bacterial pathogenicity and survival as it aids in the release of important proteins, toxins, and virulent factors. Furthermore, since SecA is a membrane protein in its translocation functional form, SecA inhibitors can directly access SecA without entering the cytoplasmic space (Chaudhary et al. 2015). Sodium azide, an inorganic compound, was the first described inhibitor of SecA in E. coli (Oliver et al. 1990). Later, various classes of small molecule SecA inhibitors were discovered which include Rose bengal, Thiouracil-pyrimidine and Triazole-pyrimidine analogues (Jin et al. 2016).
Two-component system
In response to environmental signals, bacteria commonly use a two-component system (TCS). Histidine kinase and a sensor regulator make up TCS. It is related to the expression of virulence and antibiotic resistance responses in pathogenic bacteria (Lingzhi et al. 2018). VicRK (a TCS), for example, has been shown to influence biofilm formation, lipid metabolism, and pathogenicity (Ng et al. 2005; Ahn and Burne 2007). Similarly, in enterococci, the VanS/VanR two-component system regulates vancomycin resistance (Gagnon et al. 2011). Chromosomal replication initiator protein (DnaA) is predicted as a novel drug target involved in the TCS metabolic pathway (Fatoba et al. 2021). It is essential for the beginning and control of chromosomal replication. It binds to the replication origin at a 9-bp consensus (DnaA box): 5ʹ-TTATC [CA]A[CA]A-3ʹ (Kitchen et al. 1999). In the literature, no TCS has been found in M. genitalium (Martinez et al. 2013). Signal transduction systems such as serine/threonine phosphatases (STP) and serine/threonine kinases (STK) have been shown to fulfill similar activities to TCS (Hoch 2000). Martinez and co-workers also discovered that STP plays a role in M. genitalium pathogenicity (Martinez et al. 2013). Other TCSs involved in bacterial pathogenicity and biofilm formation include DegS-DegU, Pho regulon (such as PhoR-PhoP, PhoR–PhoS, and PhoP-PhoQ), and QseC-QseB, which are found in Bacillus subtilis, Corynebacterium glutamicum, Shigella species, and enterohaemorrhagic Escherichia coli. As a result, a TCS can be used as an antibiotic target (Hirakawa et al. 2020).
Type I restriction-modification protein
Comparative genomics approaches have suggested that Type I restriction-modification protein could be a suitable therapeutic target against M. genitalium as well as in various other bacteria (Nogueira et al. 2021; Nammi et al. 2016). The restriction-modification system (RM system) is made up of big pentameric proteins known as type I restriction enzymes (REases) (Calisto et al. 2005). Restriction (R), methylation (M), and DNA sequence recognition (s) are all distinct subunits. Bacteria and other prokaryotic species have RM systems that defend them from foreign DNA such as bacteriophage DNA (Luria 1953). A restriction endonuclease and a methylase are the main components of this defence system. In foreign DNA such as viruses (phases) the restriction endonuclease cuts at a specific recognition site. The methylase enzyme adds a methyl group to a specific nucleotide shortly after replication, in the same target position as the restriction enzyme, to protect the bacterial DNA as illustrated in Fig. 3 (Vasu and Nagaraja 2013). Although Type I REases were the first to be found and purified, they have proven difficult to define. As genome analysis reveals their genes and methylome research reveals their recognition sequences, more information is getting available on these proteins for the researchers (Loenen et al. 2014; Nogueira et al. 2021).
Restriction-modification (R-M) system as defence mechanism. Incoming foreign DNA, such as phage genomes, is recognised by R-M system for its methylation status. Methylated sequences are recognised as self, but nonmethylated sequences on incoming DNA are recognised as nonself and cleaved by restriction endonuclease (REase). The cognate methyltransferase (MTase) of the R-M system maintains the methylation state at the genomic recognition sites (Vasu and Nagaraja 2013)
Hydroperoxide reductase
Hydroperoxide reductase has been found a potential therapeutic target against Mycobacterium tuberculosis and Candida albicans in various studies (Seneviratne et al. 2008; Dhiman and Singh 2018). It has also been identified as a suitable therapeutic target against M. genitalium in various comparative genomics studies (Nogueira et al. 2021). Hydroperoxide reductase catalyses the conversion of hydrogen peroxide and organic hydroperoxides to water and alcohols, respectively (Seaver and Imlay 2001). It is an essential enzyme for cell defence against oxidative stress by detoxifying peroxides and preventing bacterial damage as hydrogen peroxide is produced during aerobic metabolism and can cause significant damage to biomolecules (Jacobson et al. 1989).
Ribosome-binding factor A
Ribosome-binding factor A (RbfA) was discovered to interact with free 30S subunits but not with 70S or polysomes. RbfA binds to the 5ʹ terminal helix region of 16S rRNA. It is an assembly cofactor that is hypothesised to enhance assembly more quickly in-vivo in comparison to in-vitro. RbfA mutations impair 17 s rRNA maturation as well as the capacity of the 30S subunits to bind with the 50S (Maguire 2009; Shajani et al. 2011). It is one of the primary proteins involved in the late maturation of the functional core of the 30S subunit, and it also functions as a cold-shock adaptation protein in Escherichia coli. RbfA-deficient cells are unable to adapt to low temperatures and have slower growth rates as well as altered ribosome profiles (Bylund et al. 1998; Xia et al. 2003). It is required for successful pre-16S rRNA processing and belongs to a broad family of small proteins found in many bacterial species, making it a prime target for structural proteomics (Huang et al. 2003). This protein is identified as a potential therapeutic target for therapeutic applications against M. genitalium (Nogueira et al. 2021).
Hypothetical protein
Gene identification techniques anticipate hypothetical proteins during genome analysis, but there is no experimental proof that they are expressed in vivo (Prava et al. 2018). Bioinformatics tools are currently used to identify new genes during the annotation stages of genome assembly, and when it finds open reading frames (ORFs) in the genome that show less identity than required to a known, identified protein in the database, the sequence is labelled as “putative” or “hypothetical protein.” Structural and functional characterisation of such hypothetical proteins can be done by various in-silico and in-vitro approaches. The hypothetical proteins which have been found to play a vital role in the growth and pathogenesis of the organism can be considered potential therapeutic targets. These approaches are very crucial for developing therapeutic agents against drug-resistant infections caused by various organisms such as Plasmodium falciparum, Chlamydia trachomatis and Klebsiella pneumonia (Singh and Gupta 2022; Turab Naqvi et al. 2017; Pranavathiyani et al. 2020). Structural and functional characterisation of such hypothetical proteins present in M. genitalium provides novel therapeutic targets for screening to find new lead molecules against M. genitalium infection (Araújo et al. 2019; Nogueira et al. 2021).
DUF3217 domaincontaining protein
DUFs (domains of unknown function) are a huge collection of uncharacterized protein families that are assembled in the Pfam database, accounting for 20% of all known protein families (Bateman et al. 2010). The Pfam database is a database of protein families and domains that is frequently used to annotate sequenced genomes and assemblies (Finn et al. 2014). The DUF3217 (PF11506) domain is found in Mycoplasma proteins and appears to be confined to them. All proteins having this domain appear to belong to Mycoplasma, according to the Pfam database. While certain members of this protein family may be classified as MG376, this has yet to be proven. These proteins are suitable therapeutic targets for screening for a variety of disorders including M. genitalium infection (Nogueira et al. 2021).
Ribosomal proteins
Ribosomal proteins (L32 and S17) have been considered as therapeutic targets in M. genitalium by various researchers (Yang et al. 2020; Nogueira et al. 2021). The 50S ribosomal subunit contains the L32 protein that, along with L17 and L22, forms a cluster that holds all of the 23S rRNA domains together (Walleczek et al. 1989). The 50S ribosomal protein L32 is present on the solvent side of the large subunit and belongs to the bacterial ribosomal protein bL32 family (Brauer and Röming 1979). The ribosomal protein S17 binds directly to 16S rRNA and assists in the nucleation of assembly of the platform and body of the 30S subunit by bringing together and stabilising connections between numerous distinct RNA helices. The shoulder and platform of the 30S subunit appear to be held together by a cluster of proteins derived from S8, S12, and S17 (Held et al. 1974; Yang et al. 2020).
Class Ib ribonucleosidediphosphate reductase assembly flavoprotein (NrdI)
Ribonucleotide reductases (RNRs) are enzymes that convert ribonucleoside diphosphates to deoxyribonucleoside diphosphates, which are required for DNA synthesis and repair (Reichard 1988). Till date, three classes of RNRs (I, II, and III) have been identified based on their diverse processes, quaternary structural differences, and cofactor differences. Class I RNRs are further subdivided into classes Ia and Ib (Jordan et al. 1997). A dimanganese-tyrosyl radical is found in Class Ib (NrdI) enzymes on a cysteine residue and it is involved in the formation of a transient thiyl (sulfanyl) radical. It was found in Salmonella that in presence of NrdI, NrdH and NrdEF have a stimulatory effect on their ribonucleotide reductase activities. In M. genitalium, protein NrdI has been discovered as a novel therapeutic drug target (Jordan et al. 1996; Nogueira et al. 2021).
Nucleotide adenylyltransferase
The Nucleotide adenylyltransferase is a crucial enzyme for nicotinamide adenine dinucleotide (NAD) production and plays an important role in a variety of redox processes. This protein is engaged in the first step of the deamido-NAD ( +) synthesis pathway from nicotinate D-ribonucleotide. This subpathway is a part of NAD ( +) biosynthesis route, which is part of the cofactor biosynthesis pathway (Rodionova et al. 2014). This protein has previously been identified as a therapeutic target in M. tuberculosis and has been linked to the development of new anti-mycobacterial drugs (Petrelli et al. 2011). Later, it was also suggested as a potential therapeutic target against M. genitalium by Yang et al. (Yang et al. 2020).
Phenylalanine–tRNA synthetase
The phenylalanine tRNA synthetase (PheRS), a member of the class II aminoacyl-tRNA synthetase family, catalyses the synthesis of phenylalanyl-tRNAPhe (Phe-tRNAPhe) (Mermershtain et al. 2011). As a result, PheRS, like other aminoacyl-tRNA synthetases, plays an important part in the cellular translation process. Its structural characterisation reveals remarkable architectural diversity ranging from monomer to hetero-oligomer. An editing domain in heterotetrameric PheRS prevents the inclusion of non-cognate amino acids (Banerjee and Chakraborty 2016). Monomeric PheRS, on the other hand, lacks editing activity. PheRS has also exhibited certain noncanonical capabilities including DNA binding and its involvement in complicated regulatory circuits. Engineered mutants of PheRS with relaxed substrate specificities could be a useful tool in chemical and synthetic biology (Banerjee and Chakraborty 2016). PheRS was considered a possible therapeutic target in drug development for the treatment of S. aureus multidrug resistance, as it has a natural substrate binding site (Uddin and Saeed 2014). Subsequently, PheRS was suggested as a potential therapeutic target against M. genitalium through comparative genomics approaches (Yang et al. 2020).
Phosphoglycerate mutase
Phosphoglycerate mutase (PGM) is an enzyme that involves in the interconversion of 3–phosphoglyceric acid (3PGA) and 2–phosphoglyceric acid (2PGA) in the glycolysis and gluconeogenesis pathways. There are two forms of PGMs: cofactor-dependent PGMs (dPGMs), which require 2, 3–diphosphoglyceric acid (DPG) for catalysis and use this molecule as a phosphate donor; and cofactor-independent PGMs (iPGMs), which do not use DPG. In vertebrates (including humans), certain invertebrates, fungi, and some bacteria, notably Gram-negative bacteria like Escherichia coli, the dPGMs are the most abundant and often the only PGMs present. The phosphoglycerate mutase enzyme in Geobacillus stearothermophilus is a well-studied drug target (Jedrzejas 2000). As a novel drug target phosphoglycerate mutase was considered to be a potential drug target in drug development in Mycoplasma genitalium (Yang et al. 2020).
Phosphate acetyltransferase
Phosphate acetyltransferase has been explored as a potential drug target against various organisms such as Bacillus subtilis and Staphylococcus aureus (Yang et al. 2020; Morya et al. 2012). It has been identified as a suitable target against M. genitalium in comparative genomics (Yang et al. 2020). This enzyme is engaged in step 2 of the acetate-to-acetyl-CoA conversion process from acetate. This process is part of the acetyl-CoA biosynthesis pathway, which is part of the metabolic intermediate biosynthesis pathway (Shin et al. 1999). In a computer simulation of M. genitalium, phosphate acetyltransferase, which is involved in acetyl-CoA production, was considered as a potential therapeutic target. Acetylphosphate inhibits phosphate acetyltransferase, a therapeutic target in Bacillus subtilis (Yang et al. 2020).
Conclusion
M. genitalium is a sexually transmitted pathogen that causes various human diseases such as acute and chronic urethritis, cervicitis, pelvic inflammatory disease, and possibly female infertility. It has emerged as a superbug, with rising resistance in this bacterium and only a few treatment options available. Future research should focus on finding new potential therapeutic targets and the development of novel antimicrobials. As monotherapy is no more effective, combination therapy along with antimicrobial resistance testing is essential. Because of the misuse of antibiotics as part of syndromic management, etiology-based treatment could help against this emerging antimicrobial resistance. In this article potential therapeutic targets of M. genitalium have been discussed. Inhibiting or modulating these targets could be a rational strategy for developing novel antimicrobial drug molecules against various M. genitalium infections which require extensive further investigations.
Data availability statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Abisado RG, Benomar S, Klaus JR et al (2018) Bacterial quorum sensing and microbial community interactions. Mbio 9:e02331-e2417
Ahn S-J, Burne RA (2007) Effects of oxygen on biofilm formation and the AtlA autolysin of Streptococcus mutans. J Bacteriol 189:6293–6302
Araújo CL, Alves J, Nogueira W et al (2019) Prediction of new vaccine targets in the core genome of Corynebacterium pseudotuberculosis through omics approaches and reverse vaccinology. Gene 702:36–45
Averbach SH, Hacker MR, Yiu T et al (2013) Mycoplasma genitalium and preterm delivery at an urban community health center. Int J Gynecol Obstet 123:54–57
Awai K, Kakimoto T, Awai C et al (2006) Comparative genomic analysis revealed a gene for monoglucosyldiacylglycerol synthase, an enzyme for photosynthetic membrane lipid synthesis in cyanobacteria. Plant Physiol 141:1120–1127
Banerjee R, Chakraborty S (2016) Phenylalanyl-tRNA synthetase. Res Rep Biochem 6:25
Bateman A, Coggill P, Finn RD (2010) DUFs: families in search of function. Acta Crystallogr, Sect f: Struct Biol Cryst Commun 66:1148–1152
Bäuerle T, Fischer A, Speck T, Bechinger C (2018) Self-organization of active particles by quorum sensing rules. Nat Commun 9:1–8
Bebear CM, Renaudin J, Charron A et al (1999) Mutations in the gyrA, parC, and parE genes associated with fluoroquinolone resistance in clinical isolates of Mycoplasma hominis. Antimicrob Agents Chemother 43:954–956
Benning C (2008) A role for lipid trafficking in chloroplast biogenesis. Prog Lipid Res 47:381–389
Bradshaw CS, Jensen JS, Waites KB (2017) New horizons in Mycoplasma genitalium treatment. J Infect Dis 216:S412–S419
Brauer D, Röming R (1979) The primary structure of protein S3 from the small ribosomal subunit of Escherichia coli. FEBS Lett 106:352–357
Butt AM, Tahir S, Nasrullah I et al (2012) Mycoplasma genitalium: a comparative genomics study of metabolic pathways for the identification of drug and vaccine targets. Infect Genet Evol 12:53–62
Byers DM, Gong H (2007) Acyl carrier protein: structure–function relationships in a conserved multifunctional protein family. Biochem Cell Biol 85:649–662
Bylund GO, Wipemo LC, Lundberg LC, Wikström PM (1998) RimM and RbfA are essential for efficient processing of 16S rRNA in Escherichia coli. J Bacteriol 180:73–82
Calisto BM, Pich OQ, Piñol J et al (2005) Crystal structure of a putative type I restriction–modification S subunit from Mycoplasma genitalium. J Mol Biol 351:749–762
Cantarel BL, Coutinho PM, Rancurel C et al (2009) The carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233–D238
Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ (2008) The biology and future prospects of antivirulence therapies. Nat Rev Microbiol 6:17–27
Chandela R, Jade D, Mohan S et al (2022) Identification of therapeutic drug target of Stenotrophomonas maltophilia through subtractive genomic approach and in-silico screening based on 2D similarity filtration and molecular dynamic simulation. Comb Chem High Throughput Screen 25:123–138
Chaudhary AS, Chen W, Jin J et al (2015) SecA: a potential antimicrobial target. Future Med Chem 7:989–1007
Cina M, Baumann L, Egli-Gany D et al (2019) Mycoplasma genitalium incidence, persistence, concordance between partners and progression: systematic review and meta-analysis. Sex Transm Infect 95:328–335
Craney A, Ahmed S, Nodwell J (2013) Towards a new science of secondary metabolism. J Antibiot 66:387–400
Cui Q, Shin S, W, Luo Y, et al (2013) Thymidylate kinase: an old topic brings new perspectives. Curr Med Chem 20:1286–1305
Dahlqvist A, Nordstroem S, Karlsson OP et al (1995) Efficient modulation of glucolipid enzyme activities in membranes of Acholeplasma laidlawii by the type of lipids in the bilayer matrix. Biochemistry 34:13381–13389
Daubenspeck JM, Totten AH, Needham J et al (2020) Mycoplasma genitalium biofilms contain poly-GlcNAc and contribute to antibiotic resistance. Front Microbiol. https://doi.org/10.3389/fmicb.2020.585524
Deguchi T, Ito S, Yasuda M et al (2017) Emergence of Mycoplasma genitalium with clinically significant fluoroquinolone resistance conferred by amino acid changes both in GyrA and ParC in Japan. J Infect Chemother 23:648–650
Dhiman R, Singh R (2018) Recent advances for identification of new scaffolds and drug targets for Mycobacterium tuberculosis. IUBMB Life 70:905–916
El Qaidi S, Zhu C, McDonald P et al (2018) High-throughput screening for bacterial glycosyltransferase inhibitors. Front Cell Infect Microbiol 8:435
Fatoba AJ, Okpeku M, Adeleke MA (2021) Subtractive genomics approach for identification of novel therapeutic drug targets in Mycoplasma genitalium. Pathogens 10:921
Fernández-Huerta M, Espasa M (2019) Mycoplasma genitalium co-infection with Chlamydia trachomatis and Neisseria gonorrhoeae among asymptomatic patients: the silent wick for macrolide resistance spread. Sex Transm Infect 95:391
Finn RD, Bateman A, Clements J et al (2014) Pfam: the protein families database. Nucleic Acids Res 42:D222–D230
Gagnon S, Lévesque S, Lefebvre B et al (2011) vanA-containing Enterococcus faecium susceptible to vancomycin and teicoplanin because of major nucleotide deletions in Tn 1546. J Antimicrob Chemother 66:2758–2762
Gao Z, Ovchinnikova OG, Huang B-S et al (2019) High-throughput “FP-Tag” assay for the identification of glycosyltransferase inhibitors. J Am Chem Soc 141:2201–2204
Gnanadurai R, Fifer H (2020) Mycoplasma genitalium: a review. Microbiology 166:21–29
Green ER, Mecsas J (2016) Bacterial secretion systems: an overview. Microbiol Spectr 4:4–1
Greydanus DE, Cabral MD, Patel DR (2022) Pelvic inflammatory disease in the adolescent and young adult: an update. Dis Mon 68:101287
Gründling A, Schneewind O (2007) Synthesis of glycerol phosphate lipoteichoic acid in Staphylococcus aureus. Proc Natl Acad Sci 104:8478–8483
Gruson D, Pereyre S, Renaudin H et al (2005) In vitro development of resistance to six and four fluoroquinolones in Mycoplasma pneumoniae and Mycoplasma hominis, respectively. Antimicrob Agents Chemother 49:1190–1193
Gul S, Khalil R, Ul-Haq Z, Mubarak MS (2020) Computational overview of mycobacterial thymidine monophosphate kinase. Curr Pharm Des 26:1676–1681
Haque S, Ahmad F, Dar SA et al (2018) Developments in strategies for quorum sensing virulence factor inhibition to combat bacterial drug resistance. Microb Pathog 121:293–302
Harrison SA, Olson KM, Ratliff AE et al (2019) Mycoplasma genitalium co-infection in women with Chlamydia trachomatis Infection. Sex Transm Dis 46:e101
Hasan M, Azim KF, Imran MAS et al (2020) Comprehensive genome based analysis of Vibrio parahaemolyticus for identifying novel drug and vaccine molecules: subtractive proteomics and vaccinomics approach. PLoS ONE 15:e0237181
Hawver LA, Jung SA, Ng W-L (2016) Specificity and complexity in bacterial quorum-sensing systems. FEMS Microbiol Rev 40:738–752
Held WA, Ballou B, Mizushima S, Nomura M (1974) Assembly mapping of 30 S ribosomal proteins from Escherichia coli: further studies. J Biol Chem 249:3103–3111
Hirakawa H, Kurushima J, Hashimoto Y, Tomita H (2020) Progress overview of bacterial two-component regulatory systems as potential targets for antimicrobial chemotherapy. Antibiotics 9:635
Hoch JA (2000) Two-component and phosphorelay signal transduction. Curr Opin Microbiol 3:165–170
Hölzl G, Dörmann P (2007) Structure and function of glycoglycerolipids in plants and bacteria. Prog Lipid Res 46:225–243
Horner PJ, Martin DH (2017) Mycoplasma genitalium infection in men. J Infect Dis 216:S396–S405
Horner PJ, Blee K, Falk L et al (2016) 2016 European guideline on the management of non-gonococcal urethritis. Int J STD AIDS 27:928–937
Huang SH, Tang A, Drisco B et al (1994) Human dTMP kinase: gene expression and enzymatic activity coinciding with cell cycle progression and cell growth. DNA Cell Biol 13:461–471
Huang YJ, Swapna GVT, Rajan PK et al (2003) Solution NMR structure of ribosome-binding factor A (RbfA), a cold-shock adaptation protein from Escherichia coli. J Mol Biol 327:521–536
Iwuji C, Pillay D, Shamu P et al (2022) A systematic review of antimicrobial resistance in Neisseria gonorrhoeae and Mycoplasma genitalium in sub-Saharan Africa. J Antimicrob Chemother 77:2074–2093
Jacobson FS, Morgan RW, Christman MF, Ames BN (1989) An alkyl hydroperoxide reductase from Salmonella typhimurium involved in the defense of DNA against oxidative damage: purification and properties. J Biol Chem 264:1488–1496
Jedrzejas MJ (2000) Structure, function, and evolution of phosphoglycerate mutases: comparison with fructose-2, 6-bisphosphatase, acid phosphatase, and alkaline phosphatase. Prog Biophys Mol Biol 73:263–287
Jensen JS (2004) Mycoplasma genitalium: the aetiological agent of urethritis and other sexually transmitted diseases. J Eur Acad Dermatol Venereol 18:1–11
Jensen JS, Bradshaw C (2015) Management of Mycoplasma genitalium infections–can we hit a moving target? BMC Infect Dis 15:1–9
Jensen JS, Bradshaw CS, Tabrizi SN et al (2008) Azithromycin treatment failure in Mycoplasma genitalium–positive patients with nongonococcal urethritis is associated with induced macrolide resistance. Clin Infect Dis 47:1546–1553
Jensen JS, Cusini M, Gomberg M, Moi H (2016) 2016 European guideline on Mycoplasma genitalium infections. J Eur Acad Dermatol Venereol 30:1650–1656
Jernberg E, Moghaddam A, Moi H (2008) Azithromycin and moxifloxacin for microbiological cure of Mycoplasma genitalium infection: an open study. Int J STD AIDS 19:676–679
Jiang Q, Chen J, Yang C et al (2019) Quorum sensing: a prospective therapeutic target for bacterial diseases. BioMed Res Int. https://doi.org/10.1155/2019/2015978
Jin J, Hsieh YH, Cui J et al (2016) Using chemical probes to assess the feasibility of targeting seca for developing antimicrobial agents against gram-negative bacteria. ChemMedChem 11(22):2511–2521
Jordan A, Aragall E, Gibert I, Barbé J (1996) Promoter identification and expression analysis of Salmonella typhimurium and Escherichia coli nrdEF operons encoding one of two class I ribonucleotide reductases present in both bacteria. Mol Microbiol 19:777–790
Jordan A, Åslund F, Pontis E et al (1997) Characterization of Escherichia coli NrdH: a glutaredoxin-like protein with a thioredoxin-like activity profile. J Biol Chem 272:18044–18050
Kattke MD, Gosschalk JE, Martinez OE et al (2019) Structure and mechanism of TagA, a novel membrane-associated glycosyltransferase that produces wall teichoic acids in pathogenic bacteria. PLoS Pathog 15:e1007723
Kazakiewicz D, Karr JR, Langner KM, Plewczynski D (2015) A combined systems and structural modeling approach repositions antibiotics for Mycoplasma genitalium. Comput Biol Chem 59:91–97
Ke P-Y, Kuo Y-Y, Hu C-M, Chang Z-F (2005) Control of dTTP pool size by anaphase promoting complex/cyclosome is essential for the maintenance of genetic stability. Genes Dev 19:1920–1933
Ke W, Li D, Tso LS et al (2020) Macrolide and fluoroquinolone associated mutations in Mycoplasma genitalium in a retrospective study of male and female patients seeking care at a STI Clinic in Guangzhou, China, 2016–2018. BMC Infect Dis 20:1–8
Keating TA, Newman JV, Olivier NB et al (2012) In vivo validation of thymidylate kinase (TMK) with a rationally designed, selective antibacterial compound. ACS Chem Biol 7:1866–1872
Kiriukhin MY, Debabov DV, Shinabarger DL, Neuhaus FC (2001) Biosynthesis of the glycolipid anchor in lipoteichoic acid of Staphylococcus aureus RN4220: role of YpfP, the diglucosyldiacylglycerol synthase. J Bacteriol 183:3506–3514
Kitchen JL, Li Z, Crooke E (1999) Electrostatic interactions during acidic phospholipid reactivation of DnaA protein, the Escherichia coli initiator of chromosomal replication. Biochemistry 38:6213–6221
Klement MLR, Öjemyr L, Tagscherer KE et al (2007) A processive lipid glycosyltransferase in the small human pathogen Mycoplasma pneumoniae: involvement in host immune response. Mol Microbiol 65:1444–1457
Lavie A, Vetter IR, Konrad M et al (1997) Structure of thymidylate kinase reveals the cause behind the limiting step in AZT activation. Nat Struct Biol 4:601–604
Lewis J, Horner PJ, White PJ (2020) Incidence of pelvic inflammatory disease associated with Mycoplasma genitalium infection: evidence synthesis of cohort study data. Clin Infect Dis 71:2719–2722
Lindblom G, Wieslander A, Sjoelund M et al (1986) Phase equilibria of membrane lipids for Acholeplasma laidlawii: importance of a single lipid forming nonlamellar phases. Biochemistry 25:7502–7510
Lingzhi L, Haojie G, Dan G et al (2018) The role of two-component regulatory system in β-lactam antibiotics resistance. Microbiol Res 215:126–129
Lis R, Rowhani-Rahbar A, Manhart LE (2015) Mycoplasma genitalium infection and female reproductive tract disease: a meta-analysis. Clin Infect Dis 61:418–426
Loenen WA, Dryden DT, Raleigh EA, Wilson GG (2014) Type I restriction enzymes and their relatives. Nucleic Acids Res 42:20–44
Luria SE (1953) Cold Spring Harbor Symp Quant Biol 18:237
Maguire BA (2009) Inhibition of bacterial ribosome assembly: a suitable drug target? Microbiol Mol Biol Rev 73:22–35
Mahlangu MP, Müller EE, Venter JM et al (2019) The prevalence of Mycoplasma genitalium and association with human immunodeficiency virus infection in symptomatic patients, Johannesburg, South Africa, 2007–2014. Sex Transm Dis 46:395
Manhart LE, Gillespie CW, Lowens MS et al (2013) Standard treatment regimens for nongonococcal urethritis have similar but declining cure rates: a randomized controlled trial. Clin Infect Dis 56:934–942
Manhart LE, Jensen JS, Bradshaw CS et al (2015) Efficacy of antimicrobial therapy for Mycoplasma genitalium infections. Clin Infect Dis 61:S802–S817
Martinez MA, Das K, Saikolappan S et al (2013) A serine/threonine phosphatase encoded by MG_207 of Mycoplasma genitalium is critical for its virulence. BMC Microbiol 13:1–14
Martínez-Botella G, Loch JT, Green OM et al (2013) Sulfonylpiperidines as novel, antibacterial inhibitors of gram-positive thymidylate kinase (TMK). Bioorg Med Chem Lett 23:169–173
McShan AC, De Guzman RN (2015) The bacterial type III secretion system as a target for developing new antibiotics. Chem Biol Drug Des 85:30–42
Mermershtain I, Finarov I, Klipcan L et al (2011) Idiosyncrasy and identity in the prokaryotic phe-system: crystal structure of E. coli phenylalanyl-tRNA synthetase complexed with phenylalanine and AMP. Protein Sci 20:160–167
Morya VK, Dewaker V, Kim EK (2012) In silico study and validation of phosphotransacetylase (PTA) as a putative drug target for Staphylococcus aureus by homology-based modelling and virtual screening. Appl Biochem Biotechnol 168(7):1792–1805
Muller EE, Mahlangu MP, Lewis DA, Kularatne RS (2019) Macrolide and fluoroquinolone resistance-associated mutations in Mycoplasma genitalium in Johannesburg, South Africa, 2007–2014. BMC Infect Dis 19:1–8
Murray GL, Bradshaw CS, Bissessor M et al (2017) Increasing macrolide and fluoroquinolone resistance in Mycoplasma genitalium. Emerg Infect Dis 23:809
Nammi D, Srimath-Tirumala-Peddinti RC, Neelapu NR (2016) Identification of drug targets in helicobacter pylori by in silico analysis : possible therapeutic implications for gastric cancer. Curr Cancer Drug Targets 16(1):79–98
Naorem RS, Pangabam BD, Bora SS et al (2022) Identification of putative vaccine and drug targets against the methicillin-resistant Staphylococcus aureus by reverse vaccinology and subtractive genomics approaches. Molecules 27:2083
Napierala Mavedzenge S, Müller EE, Lewis DA et al (2015) Mycoplasma genitalium is associated with increased genital HIV type 1 RNA in Zimbabwean women. J Infect Dis 211:1388–1398
Ng W-L, Tsui H-CT, Winkler ME (2005) Regulation of the pspA virulence factor and essential pcsB murein biosynthetic genes by the phosphorylated VicR (YycF) response regulator in Streptococcus pneumoniae. J Bacteriol 187:7444–7459
Nogueira WG, Jaiswal AK, Tiwari S et al (2021) Computational identification of putative common genomic drug and vaccine targets in Mycoplasma genitalium. Genomics 113:2730–2743
Oliver DB, Cabelli RJ, Dolan KM, Jarosik GP (1990) Azide-resistant mutants of Escherichia coli alter the SecA protein, an azide-sensitive component of the protein export machinery. Proc Natl Acad Sci U S A 87:8227–8231
Onaka H (2017) Novel antibiotic screening methods to awaken silent or cryptic secondary metabolic pathways in actinomycetes. J Antibiot 70:865–870
Ozcengiz G, Demain AL (2013) Recent advances in the biosynthesis of penicillins, cephalosporins and clavams and its regulation. Biotechnol Adv 31:287–311
Pan X-S, Ambler J, Mehtar S, Fisher LM (1996) Involvement of topoisomerase IV and DNA gyrase as ciprofloxacin targets in Streptococcus pneumoniae. Antimicrob Agents Chemother 40:2321–2326
Petrelli R, Felczak K, Cappellacci L (2011) NMN/NaMN adenylyltransferase (NMNAT) and NAD kinase (NADK) inhibitors: chemistry and potential therapeutic applications. Curr Med Chem 18:1973–1992
Pitt R, Fifer H (2022) Antimicrobial resistance in bacterial sexually transmitted infections. Medicine 50:277–279
Pranavathiyani G, Prava J, Rajeev AC et al (2020) Novel target exploration from hypothetical proteins of Klebsiella pneumoniae MGH 78578 reveals a protein involved in host-pathogen interaction. Front Cell Infect Microbiol 10:109
Prava J, Pranavathiyani G, Pan A (2018) Functional assignment for essential hypothetical proteins of Staphylococcus aureus N315. Int J Biol Macromol 108:765–774
Raj JS, Rawre J, Dhawan N et al (2022) Mycoplasma genitalium: a new superbug. Indian J Sex Transm Dis AIDS 43:1
Reichard P (1988) Interactions between deoxyribonucleotide and DNA synthesis. Annu Rev Biochem 57:349–374
Rodionova IA, Schuster BM, Guinn KM et al (2014) Metabolic and bactericidal effects of targeted suppression of NadD and NadE enzymes in mycobacteria. Mbio 5:e00747-e813
Romano SS, Jensen JS, Lowens MS et al (2019) Long duration of asymptomatic Mycoplasma genitalium infection after syndromic treatment for nongonococcal urethritis. Clin Infect Dis 69:113–120
Ross JDC, Jensen JS (2006) Mycoplasma genitalium as a sexually transmitted infection: implications for screening, testing, and treatment. Sex Transm Infect 82:269–271
Ruiz B, Chávez A, Forero A et al (2010) Production of microbial secondary metabolites: regulation by the carbon source. Crit Rev Microbiol 36:146–167
Sakurai I, Mizusawa N, Wada H, Sato N (2007) Digalactosyldiacylglycerol is required for stabilization of the oxygen-evolving complex in photosystem II. Plant Physiol 145:1361–1370
Seaver LC, Imlay JA (2001) Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J Bacteriol 183:7173–7181
Seneviratne CJ, Wang Y, Jin L et al (2008) Candida albicans biofilm formation is associated with increased anti-oxidative capacities. Proteomics 8:2936–2947
Sethi S, Singh G, Samanta P, Sharma M (2012) Mycoplasma genitalium: an emerging sexually transmitted pathogen. Indian J Med Res 136:942
Shahid F, Ashfaq UA, Saeed S et al (2020) In silico subtractive proteomics approach for identification of potential drug targets in Staphylococcus saprophyticus. Int J Environ Res Public Health 17:3644
Shajani Z, Sykes MT, Williamson JR (2011) Assembly of bacterial ribosomes. Annu Rev Biochem 80:501–526
Shin B-S, Choi S-K, Park S-H (1999) Regulation of the Bacillus subtilis phosphotransacetylase gene. J Biochem 126:333–339
Singh G, Gupta D (2022) In-Silico functional annotation of Plasmodium falciparum hypothetical proteins to identify novel drug targets. Front Genet 13:821516
Smolarczyk K, Mlynarczyk-Bonikowska B, Rudnicka E et al (2021) The impact of selected bacterial sexually transmitted diseases on pregnancy and female fertility. Int J Mol Sci 22:2170
Sweeney EL, Bradshaw CS, Murray GL, Whiley DM (2022a) Individualised treatment of Mycoplasma genitalium infection—incorporation of fluoroquinolone resistance testing into clinical care. Lancet Infect Dis 22:e267–e270
Sweeney EL, Whiley DM, Murray GL et al (2022b) Mycoplasma genitalium: enhanced management using expanded resistance-guided treatment strategies. Sex Health 19(4):248–254
Taylor-Robinson D (2014) Diagnosis and antimicrobial treatment of Mycoplasma genitalium infection: sobering thoughts. Expert Rev Anti Infect Ther 12:715–722
Turab Naqvi AA, RahmanRubi S et al (2017) Genome analysis of Chlamydia trachomatis for functional characterization of hypothetical proteins to discover novel drug targets. Int J Biol Macromol 96:234–240
Turan NB, Chormey DS, Büyükpınar Ç et al (2017) Quorum sensing: little talks for an effective bacterial coordination. TrAC, Trends Anal Chem 91:1–11
Uddin R, Saeed K (2014) Identification and characterization of potential drug targets by subtractive genome analyses of methicillin resistant Staphylococcus aureus. Comput Biol Chem 48:55–63
Uddin R, Arif A, Zahra N-U-A, Sufian M (2021) Comparative proteome-wide study for in-silico identification and characterization of indispensable hypothetical proteins of food borne-pathogen Campylobacter jejuni (CJJ) by subtractive genomics approach. Pak J Pharm Sci 34:1359–1367
Valentine-King MA, Cisneros K, James MO et al (2019) Turning the tide against antibiotic resistance by evaluating novel, halogenated phenazine, quinoline, and NH125 compounds against ureaplasma species clinical isolates and Mycoplasma type strains. Antimicrob Agents Chemother 63:e02265-e2318
Vasu K, Nagaraja V (2013) Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol Mol Biol Rev 77:53–72
Veenendaal AK, van der Does C, Driessen AJ (2004) The protein-conducting channel SecYEG. Biochimica et Biophysica Acta (BBA)-Mol Cell Res 1694:81–95
Waksman G (2012) Bacterial secretion comes of age. Philos Trans R Soc b: Biolog Sci 367:1014–1015
Walleczek J, Martin T, Redl B et al (1989) Comparative cross-linking study on the 50S ribosomal subunit from Escherichia coli. Biochemistry 28:4099–4105
Weber T, Welzel K, Pelzer S et al (2003) Exploiting the genetic potential of polyketide producing streptomycetes. J Biotechnol 106:221–232
Wei C, Zhao X (2018) Induction of viable but nonculturable Escherichia coli O157: H7 by low temperature and its resuscitation. Front Microbiol. https://doi.org/10.3389/fmicb.2018.02728
Wieslander A, Christiansson A, Rilfors L, Lindblom G (1980) Lipid bilayer stability in membranes. Regulation of lipid composition in Acholeplasma laidlawii as governed by molecular shape. Biochemistry 19:3650–3655
Workowski KA, Bachmann LH, Chan PA et al (2021) (2021) Sexually transmitted infections treatment guidelines. Morb Mortal Wkly Rep 70(4):1–187
Xia B, Ke H, Shinde U, Inouye M (2003) The role of RbfA in 16 S rRNA processing and cell growth at low temperature in Escherichia coli. J Mol Biol 332:575–584
Yang Y-H, Song E, Park S-H et al (2010) Loss of phosphomannomutase activity enhances actinorhodin production in Streptomyces coelicolor. Appl Microbiol Biotechnol 86:1485–1492
Yang Z, Hou J, Mu M, Wu SY (2020) Subtractive proteomics and systems biology analysis revealed novel drug targets in Mycoplasma genitalium strain G37. Microb Pathog 145:104231
Yoshida H, Bogaki M, Nakamura M, Nakamura S (1990) Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother 34:1271–1272
Zhang Y-M, Rock CO (2008) Thematic review series: glycerolipids. Acyltransferases in bacterial glycerophospholipid synthesis. J Lipid Res 49:1867–1874
Zhang X, Wu F, Yang N et al (2022) In silico methods for identification of potential therapeutic targets. Interdiscip Sci, Comput Life Sci 14(2):285–310
Zhu C, El Qaidi S, McDonald P et al (2021) YM155 inhibits NleB and SseK arginine glycosyltransferase activity. Pathogens 10:253
Acknowledgements
Authors thank Central University of South Bihar (CUSB), India for infrastructural support.
Funding
The work presented here is not covered by any funding agency.
Author information
Authors and Affiliations
Contributions
KB: data collection, compilation, drafting and art work of the manuscript. PKA: editing the manuscript. AKS: manuscript moderation. AK: primary investigator of the work presented.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no competing conflict of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Barik, K., Arya, P.K., Singh, A.K. et al. Potential therapeutic targets for combating Mycoplasma genitalium. 3 Biotech 13, 9 (2023). https://doi.org/10.1007/s13205-022-03423-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-022-03423-9