Abstract
Industrial strains of Rhizopus oryzae is known for its strong ability to produce L-( +)-lactic acid, ethanol, and fumaric acid at high yields. To better understand the underlying mechanism behind the physiology of R. oryzae, we conducted the proteome changes between two different morphologies using two-dimensional polyacrylamide gel electrophoresis and mass spectrometry. R. oryzae exhibited pellet morphology and filamentous morphology when the initial pH of the culture medium was 3.0 and 5.0, respectively. The concentration of lactic acid reached 63.5 g L−1 in the samples containing the pellet morphology, compared to 41.5 g L−1 produced by filamentous R. oryzae. Proteomic analysis indicated that expression levels of 128 proteins changed significantly. Of these, 17 protein spots were successfully identified by mass spectrometry and were deemed to be mainly involved in carbohydrate metabolism, genetic information processing, chitin metabolism, protein catabolism, protein folding, and antioxidative pathway. L-lactate dehydrogenase (RO3G_06188), enolase (RO3G_05466) and 2, 3-bisphosphoglycerate-independent phosphoglycerate mutase (RO3G_02462) were found to be upregulated, while isocitrate dehydrogenase (RO3G_13820) was downregulated in the samples with pellet morphology compared to the filamentous hyphae. These results suggested that more carbon flow was directed towards lactic acid biosynthesis in R. oryzae hyphae with pellet morphology.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The morphology of filamentous fungi has received attention in terms of both pure and applied science.
The filamentous mycelia of Rhizopus species can grow in different morphological forms such as clumps, filaments, and pellets, depending on the growth conditions (Kossen 2000). The particular morphology is determined not only by the genetic makeup of the fungal species but also the nature of inoculum as well as the chemical (medium constituents) and physical (pH, temperature, mechanical forces) culture conditions (Kossen 2000). Moreover, the morphology of filamentous fungi during fermentation is critical for maximum performance. Although the optimal morphology for the production of various chemicals differs among fungi, pellets are often the preferable morphological forms in industrial fermentation processes. For example, pellet morphology is necessary for maximum production of fumaric acid by R. oryzae 1526 (Das and Brar 2014), L-( +)-lactic acid by R. oryzae NRRL 395 (Liu et al. 2006) and heterologous proteins by Aspergillus niger (Xu et al. 2000). Our study also showed the effects of pellet characteristics on L-lactic acid fermentation by R. oryzae (Fu et al. 2014). However, it has also been reported that filamentous morphology is favorable for fumaric acid production by Rhizopus arrhizus (Byrne and Ward 1989), but the underlying molecular mechanism remains unknown.
R. oryzae is a filamentous fungus from the family Zygomycetes with a high capacity to produce L-( +)-lactic acid, ethanol, and fumaric acid using sugars derived from biomass as the substrate (Meussen et al. 2012). In this study, a lactic acid-producing strain of R. oryzae was chosen as the model organism. Many studies investigated the morphological changes of R. oryzae in response to different fermentation conditions. For example, pellets of R. oryzae were formed in shake flasks by controlling the culture conditions such as inoculum size, heavy metal and nitrogen concentrations (Liao et al. 2007; Zhou et al. 2000). A multi-stage preculture strategy was developed to control the morphology of R. oryzae by controlling the pH of the preculture medium (Fu et al. 2009). Pellets of R. oryzae were formed by the facile manipulation of the initial spore concentration when R. oryzae was cultivated in air-lift reactors (Miura et al. 2003) and bubble column reactors (Yin et al. 1997). Although different morphologies can be induced using available models and practices based almost entirely on the influence of culture condition on morphology, the mechanism involved in the regulation of morphology in R. oryzae is poorly understood. Moreover, knowledge of the genes and enzymes involved in the determination of fungal morphology is necessary for the application of genetic engineering to control the morphology. The genome of R. oryzae strain 99–880, with a size of 45.3 Mbp and 13,895 predicted protein-coding genes, was published previously (Ma et al. 2009). This strain was the first organism in the polyphyletic basal lineage of Zygomycetes to be sequenced, which made a great contribution to this research field and provided new insights for molecular techniques.
Proteomics is a powerful tool for understanding the global protein expression differences between cells grown under different conditions. In this study, the specific changes in the protein expression pattern of R. oryzae displaying filamentous and pellet morphology were determined. Proteins were separated by two-dimensional electrophoresis (2-DE), quantified densitometrically by digital imaging, and identified by MALDI-TOF mass spectrometry based on the peptide mass fingerprint. This work helps explain the mechanisms controlling the induction of pellet morphology and resulting overproduction of target compounds, which lays a basis for controlling the morphology of R. oryzae hyphae by genetic engineering.
Materials and methods
Microorganism and culture condition
R. oryzae LA-UN-1 (Yin et al. 2013), a mutant of R. oryzae NRRL 395, was used throughout this study. The fungus was grown on potato-dextrose agar slants at 30 °C for 6 days. For the experiments, the fungal spores in the slants were suspended in sterilized water maintained at 4 °C. The culture medium contained 10 g L−1 soluble starch, 5.0 g L−1 peptone, 5.0 g L−1 yeast extract, 0.2 g L−1 KH2PO4, 0.2 g L−1 MgSO4⋅7H2O. The pH of culture media was adjusted by adding 1 M H2SO4. The fermentation medium consisted of 90 g L−1 glucose, 3.02 g L−1 NH4SO4, 0.25 g L−1 MgSO4⋅7H2O, 0.15 g L−1 KH2PO4 and 0.04 g L−1 ZnSO4⋅7H2O. The media were autoclaved at 121 °C for 20 min.
To obtain the different morphological form of R. oryzae, a multi-stage culture method with small modification was used (Fu et al. 2009). For the induction of filamentous and pellet morphology, the initial pH of culture medium was adjusted to 5.0 and 3.0 using 2 M H2SO4, respectively. Firstly, the cultures inoculated with spores (a final concentration of 107 spores/ml) were grown in a 250 ml shake flask containing 50 ml of culture medium at 200 rpm and 30 °C for 16 h. In the second stage, a 300 ml inoculum of the preculture was transferred to a 5 L stirred-tank bioreactor (NBS, New Brunswick, USA) containing 3 L of culture medium and grown at 200 rpm and 30 °C for 16 h.
Measurements of lactic acid and glucose concentrations
To determine the glucose concentration, samples were collected by centrifugation and the supernatants were used. To determine the lactic acid concentration, samples were diluted with distilled water and heated at 80 °C until the culture liquid was clear. Then, the diluted suspensions were centrifuged and the supernatants were used. The concentrations of glucose and lactic acid were analyzed using an SBA-40D biosensor analyzer (Institute of Biology, Shandong Academy of Sciences) with glucose oxidase and lactate oxidase electrodes, respectively.
Electron microscopy
The morphology of R. oryzae was observed via scanning electron microscopy (XL-30 ESEM, Philips) according to a published method (Pera and Callieri 1997).
Protein extraction and quantification
Total proteins were collected using phenol extraction coupled with ammonium acetate precipitation (Isaacson et al. 2006). Deep-frozen cells were ground into a fine powder in liquid nitrogen with a pre-cooled mortar and pestle, after which the powder was suspended and fully lysed in 5 ml phenol saturated with Tris, pH 7.8. Then, 5 ml of cold phenol extraction buffer [0.7 M sucrose; 0.1 M KCl; 0.5 M Tris–HCl, pH 7.5 and 50 mM EDTA, 1% w/v DTT, pH 7.5; complete protease inhibitor cocktail (Roche Applied Science)] were added and centrifuged at 5000 g for 20 min at 4 °C. The upper phenolic phase was collected and the process repeat with the aqueous phase. Then, five volumes of cold 0.1 M ammonium acetate in methanol were added to the collected phenol phase and stored at − 20 °C overnight. The sample was centrifuged for 30 min at 5000 g and 4 °C, after which the supernatant was carefully removed. Next, the pellet was washed twice with 5 ml of ice-cold methanol and centrifuged for 10 min at 5000 g and 4 °C. Then, the pellet was washed twice with 5 ml of ice-cold acetone instead of methanol. The resulting clear precipitates were stored in aliquots at − 80 °C until analysis.
Two-dimensional electrophoresis
Two-dimensional electrophoresis (2-DE) was performed according to the manual of the gel kit (GE Healthcare Life Sciences, USA). The first dimension separates proteins according to their isoelectric points. For the isoelectric focusing electrophoresis, 120 µg of protein was diluted with rehydration solution containing 7 M urea, 2 M thiourea, 4% (w/v) CHAPS, 1% (w/v) DTT, 1% (v/v) IPG buffer pH 4–7 (GE, USA) and 0.002% bromophenol blue to 460 µl. Immobiline DryStrip gels (pH 4–7, 24 cm; Bio-Rad) were rehydrated with 460 µl mixture solutions in 24 cm strip holders and electro-focused using the Ettan IPGphor III Isoelectric Focusing System (GE Healthcare Life Sciences, USA). The focusing protocol was as follows: 50 mA per strip at 20 °C; (1) rehydration with 30 V for 12 h; (2) 300 V for 30 min (step and hold); (3) 700 V for 30 min (step and hold); (4) 1500 V for 1.5 h (step and hold); (5) 9000 V for 3 h (gradient); (6) 9000 V for 5 h (step and hold).
After isoelectric focusing, the strips were equilibrated twice in SDS buffer with gentle shaking. The first step was performed in an equilibration solution containing 6 M urea, 30% glycerol, 2% (w/v) SDS, 1% (w/v) DTT, 50 mM Tris–HCl buffer (pH 8.8), and 0.002% (w/v) bromphenol blue) for 15 min. The second step was carried out in an equilibration solution in which the DTT was substituted with 2.5% (w/v) iodoacetamide for 15 min. The second separation was performed using an Ettan DALT System (Amersham Bioscience, USA). When the equilibration was finished, the strips were loaded onto vertical SDS–polyacrylamide gels (12.5% T constant). The gels were run at 2 W per gel for the first 40 min and followed by 17 W per gel for 5 h until the dye reached the bottom of the gel. Three independent biological replicates were included for each condition.
Silver staining, image acquisition and data analysis
The resolved protein spots in the 2-D gels were visualized by silver staining. Stained gels were scanned using a PowerLook 1100 scanner (UMAX Technologies, USA), and the images were analyzed with ImageMaster 2D Platinum software version 5.0 (GE Healthcare Life Science). The protein spots were initially detected, background subtracted, total spot volume normalized and matched by manual assistance. The protein spots separated on the 2-D gels were quantitated in terms of their relative volume (spot volume/total spot volume). For comparison, the value of each spot was divided by that of each corresponding spot volume from the filamentous R. oryzae. Statistical significance of the differences in expression profiles was evaluated using Student’s t test with the significance threshold set at p < 0.05. Only those spots with a twofold or greater change in expression levels were selected for spot picking, trypsin digestion, and mass spectrometry analysis.
In-gel trypsin digestion
The 2-D gels of samples intended for mass spectrometry were loaded with 1.2 mg protein and stained overnight with Coomassie Blue R-250 (Bio-Rad, USA). The target protein spots were excised from the stained gels and transferred into Eppendorf tubes. The gel pieces were washed twice with deionized water (Millipore) and de-stained by washing with a 1:1 solution of 30 mM potassium ferricyanide and 100 mM sodium thiosulfate. Then, the gels were washed twice with deionized water (Millipore), dehydrated in acetonitrile, and dried in a SpeedVac (Thermo Savant, USA) for 20 min. Subsequently, the gel in each tube was rehydrated in 10 μl of proteomics grade trypsin (Sigma) solution (20 ng/μl in 40 mM NH4HCO3) and incubated at 37 °C for 16 h. Peptides were extracted twice by adding 30 μl of a solution containing 50% ACN and 5% trifluoroacetic acid (TFA). The extracted solutions were concentrated to 5 μl in a lyophilizer (Virtis, Gardiner, NY, USA). A protein-free gel piece was similarly processed and used as the control to identify autoproteolysis products derived from trypsin.
Mass spectrometry analysis and database search
Trypsin-digested peptide mixtures were combined with an equal volume of 10 mg/ml CHCA saturated with 50% ACN in 0.05% TFA and analyzed on an Autoflex MALDI-TOF-TOF mass spectrometry platform (Bruker Daltonics, Germany). The working mode was set to positive ion reflection mode, with a UV wavelength of 355 nm, laser frequency of 50 Hz and an accelerating voltage of 20 kV. The mass spectra ranging from 700 to 3200 Da were acquired with laser shots at 1000/spectrum. The matrix and autolytic peaks of trypsin were used for internal calibration. The acquired data were processed using flexAnalysis software (Bruker Daltonics). The processed mass spectra were submitted to Mascot 2.0 using BioTools software (Bruker Daltonics) for peptide search against the NCBI non-redundant database (updated on August 9, 2019). The main search parameters were as follows: (1) trypsin set as digestion enzyme with one missed cleavage site allowed; (2) peptide size of 800–4000 Da; (3) peptide mass tolerance ± 50 ppm, and fragment mass tolerance ± 0.5 Da; (4) taxonomy was set to fungi and peptide charge state to 1 + ; (5) oxidation of methionine and carbamidomethylation of cysteine were set as a variable and fixed modification, respectively. Only significant hits (p < 0.05) in each MASCOT search were retained in the list of identifications.
Quantitative real-time PCR (qRT-PCR)
Total RNA isolation and the synthesis of cDNA were performed as described previously (Xu et al. 2016). The RNA concentration was determined by measuring the A260/A280 ratio using a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies Inc., USA). The real-time PCR reaction was performed using the Mx3000P with a real-time PCR Detection System (Stratagene, USA). All primers are listed in Table 1.
Results and discussion
Analysis of lactic acid production and glucose consumption
In this study, R. oryzae exhibited a significant difference in lactic acid production when the spore suspension was added to the culture broth at pH 5.0 and 3.0 (Fig. 1). The glucose was completely consumed and the concentration of lactic acid reached 63.5 g L−1 after 54 h of cultivation when the initial pH of the culture medium was set to 3.0. Contrastively, 19.5 g L−1 glucose remained in the medium and the concentration of lactic acid reached only 41.5 g L−1 after 50 h of cultivation when the initial pH of the culture medium was 5.0 (Fig. 1).
Observation of morphology
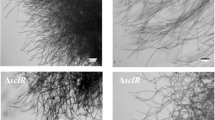
Previous studies had demonstrated that the optimal morphology for the production of various chemicals differed among fungi, whereby the pellet morphology was often preferable in industrial fermentation processes (Das and Brar 2014; Liu et al. 2006, 2012). Furthermore, the morphology of R. oryzae in this study was determined with scanning electron microscopy. As shown in Fig. 2, R. oryzae exhibited pellet morphology when the initial pH of the culture medium was 3.0, and filamentous morphology when the initial pH of the culture medium was 5.0.

taken from a 12 cm Petri-dish at the end of the cultivations by digital camera showed the different morphologies of R. oryzae
Scanning electron microscopy images revealing the morphology of the pellet (cultivation with an initial pH 3.0) and the filamentous (cultivation with the initial pH 5.0) R. oryzae, obtained at 500 µm (a) and 100 µm (b), respectively. Figures 2c and d
Based on these results, it was inferred that pellet morphology was also preferable for lactic acid fermentation by R. oryzae. Although the different morphologies could be induced by controlling the initial pH of culture medium in this study, the mechanism underlying the regulation of morphology in R. oryzae is still poorly understood. Information on proteins that are differentially expressed during fungal morphology determination may enable the construction of high-yield strains by rational genetic engineering.
Comparative proteomic analysis of R. oryzae hyphae displaying filamentous and pellet morphology
Proteomic analysis was applied to identify the protein profile with different morphologies. The total proteins were resolved into more than 1000 spots (ranging 1029–1355) in each SDS–polyacrylamide gel. The 2-D protein pattern was highly reproducible and similar results were obtained in triplicate cell extracts. Figure 3a and b show the silver-stained 2-DE gels of total extracted proteins from pellet and filamentous R. oryzae. Compared with filamentous R. oryzae, the 2-D pattern significantly changed in the pellet samples (Fig. 3). Overall, 55 upregulated protein spots (A01-A55, ratio > 1.5) were identified in the pellet samples (Fig. 3a), while 73 protein spots were upregulated (B01-B73, ratio > 1.5) in the filamentous samples (Fig. 3b).
Representative 2-DE images visualized by silver-staining. Proteins were separated by 2-DE and visualized by silver-staining as described in Sects. 2.3–2.6. The first dimension, IPG 4–7, linear; separation distance, 24 cm. The second dimension, vertical SDS-PAGE (12.5% T constant). One of the best images is presented and differentially accumulated proteins are indicated by green sashes. a pellet R. oryzae; b filamentous R. oryzae. Fifty-five upregulated protein spots (A01-A55, ratio > 1.5) in pellet R. oryzae are indicated in map (a). Seventy-three upregulated protein spots (B01-B73, ratio > 1.5) in filamentous R. oryzae are indicated in map (b)
Identification of differentially expressed protein by MALDI-TOF mass spectrometry
Protein spots with significant changes were cut out and subjected to trypsin digestion, followed by MALDI-TOF-TOF and a database search. Tables 2 and 3 list the protein spots which were successfully identified by MS based on PMF. Among them, 10 protein spots were upregulated in the pellet samples compared with the filamentous samples, whereas 7 spots were downregulated (Tables 2 and 3) (Fig. 3). Moreover, 17 identified proteins were classified according to their biological functions based on annotations from the KEGG database (Table 4; Figs. 4, 5). As shown in Table 4, ten of the identified proteins found to be involved in different metabolic pathways, including carbohydrate metabolism, chitin metabolism, protein catabolism, protein folding, and antioxidative pathways. Additionally, two of the identified proteins were related to genetic information processing, while the remaining five were proteins of unknown function.
Five functional proteins involved in carbohydrate metabolism were identified, including 2, 3-bisphosphoglycerate-independent phosphoglycerate mutase (A13), enolase (A16), aldehyde dehydrogenase (A19), l-lactate dehydrogenase (A31) and isocitrate dehydrogenase (B16). Compared with the filamentous sample, the expression levels of 2,3-bisphosphoglycerate independent phosphoglycerate mutase (A13), enolase (A16), aldehyde dehydrogenase (A19), and l-lactate dehydrogenase (A31) showed respective 4.70-, 1.87-, 7.73-, and 6.89-fold upregulation in the pellet samples, while isocitrate dehydrogenase (B16), an important enzyme of the TCA cycle, was downregulated 4.19-fold (Tables 2, 3). As a key enzyme of glycolysis, 2, 3-bisphosphoglycerate independent phosphoglycerate mutase catalyzes the interconversion between 3-phosphoglycerate and 2-phosphoglycerate, while enolase catalyzes the conversion of 2-phosphoglycerate into phosphoenolpyruvate in the ninth and penultimate step of glycolysis. Moreover, l-lactate dehydrogenase is a vital enzyme in lactic acid synthesis. Metabolomic studies confirmed that both the upregulation of enzymes related to glycolysis and the downregulation of enzymes related to the TCA cycle in R. oryzae hyphae with pellet morphology increase the carbon flux toward organic acid biosynthesis (Xu et al. 2016). These results are in agreement with the observation that R. oryzae with pellet morphology produces more lactic acid that the filamentous form in fermentation processes.
Chitin is a structurally important component of the fugal cell wall that can be enzymatically deacetylated to chitosan by chitin deacetylases. Chitinases, which belong to the glycoside hydrolase family, are responsible for the hydrolysis of β-1, 4 linkages in chitin (Cantarel et al. 2009). Chitinases play important roles in cell wall remodeling, influencing cell wall integrity, cell separation and stress resistance (Langner and Gohre 2016). In this study, two proteins related to chitin catabolism were found to be differentially expressed in pellets and filamentous hyphae. The protein levels of chitinase (RO3G_13635) and chitin deacetylase 2 (RO3G_01408) were increased 3.37- and 16.65-fold in pellet R. oryzae compared with filamentous R. oryzae, respectively (Table 2). As has been previously reported, the chitin deacetylase encoding gene RO3G_16202 and chitinase encoding gene RO3G_16170 were found to be upregulated in R. oryzae pellets compared with the filamentous R. oryzae at the transcriptional level (Xu et al. 2016). Additionally, there was less chitin in R. oryzae hyphae with pellet morphology than in filamentous hyphae (Xu et al. 2016). Therefore, it was speculated that upregulating the expression of proteins related to chitin catabolism is beneficial for pellet formation.
Moreover, peroxiredoxin-1 (A12) and hsp70-like protein (A33) were upregulated 8.66- and 4.93-fold in the pellet-type R. oryzae compared to the filamentous R. oryzae (Table 2). Peroxiredoxins play major roles in preventing oxidative damage, while heat shock proteins are known for their role in protecting cells from high temperature and other forms of stress by promoting correct protein folding and refolding of denatured proteins. This indicated that R. oryzae may be subjected to oxidative stress during the formation of pellet morphology in agreement with an earlier report that the reactive oxygen species (ROS) levels in pellet samples were higher than in filamentous samples (Xu et al. 2016).
Furthermore, the proteins downregulated in the pellet-type R. oryzae (Table 3) included eukaryotic translation initiation factor 6 (B48) and transcription factor MBP1 (B63). Similarly, the morphology-regulating transcription factor gene COM1 was reported to play a critical role in maintaining the normal shape of Magnaporthe oryzae (Bhadauria et al. 2010; Yang et al. 2010). The transcription factor MBP1 (B63) may exert its effect by controlling the expression of other genes, but further studies are needed to clarify the mechanism.
Interestingly, rhizopuspepsin-1 (RO3G_12822) was upregulated in pellet-type R. oryzae. This is a secretory aspartic protease that is distinct from other Rhizopus pepsins (Chen et al. 2009). In addition, five hypothetical proteins were identified, one of which (A41) was upregulated and 4 of which (B32, B43, B49 and B71) were downregulated in pellet-type R. oryzae (Tables 2 and 3). Homology searches across the available genomic databases revealed no open reading frames with significant sequence similarity to these five unknown proteins, and their possible roles in controlling the morphology or metabolism of R. oryzae during lactic acid production merit further study.
Verification of gene expression by quantitative real-time PCR
To verify that the differentially abundant proteins identified by MS were differentially regulated on the genetic level, quantitative real-time PCR was used to analyze the gene expression at the mRNA level. In this study, five typical genes were chosen, with the gene encoding β-actin as the internal reference. The relative expression level of each gene was normalized according to the actin gene expression. As shown in Fig. 5, the expression of these genes at the mRNA level was positively correlated with the abundance of proteins. These results further confirmed that the proteins identified through the proteomic analysis are indeed subjected to differential genetic regulation. The fold-change of some genes was different from that observed at the protein level, which can be explained by various levels of regulation during protein synthesis, including post-transcriptional, translational, and post-translational regulation. Importantly, the change trends were consistent.
Conclusions
In this study, R. oryzae exhibited pellet morphology and filamentous morphology when the initial pH of the culture medium was set to 3.0 and 5.0, respectively. Moreover, the production of lactic acid was strongly correlated with the morphology and pellets were the preferable morphology in the lactic acid fermentation processes of R. oryzae. To better understand the underlying mechanism, proteomic analysis was applied to identify the protein profile associated with the different morphologies. Ten of the identified differentially expressed proteins were found to be involved in different metabolic pathways, including carbohydrate metabolism, chitin metabolism, protein catabolism, protein folding, and antioxidative pathways. Two of the identified proteins were found to be related to genetic information processing, while the remaining five were proteins with unknown function. The results indicate that the R. oryzae hyphae with pellet morphology had a stronger carbon flux toward lactic acid biosynthesis, and the up-regulation of proteins related to chitin catabolism may be beneficial for the formation of pellet morphology.
References
Bhadauria V, Wang LX, Peng YL (2010) Proteomic changes associated with deletion of the Magnaporthe oryzae conidial morphology-regulating gene COM1. Biol Direct 5:61. https://doi.org/10.1186/1745-6150-5-61
Byrne GS, Ward OP (1989) Effect of nutrition on pellet formation by Rhizopus arrhizus. Biotechnol Bioeng 33:912–914. https://doi.org/10.1002/bit.260330715
Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res 37:D233–238. https://doi.org/10.1093/nar/gkn663
Chen CC, Cho YC, Lai CC, Hsu WH (2009) Purification and characterization of a new Rhizopuspepsin from Rhizopus oryzae NBRC 4749. J Agric Food Chem 57:6742–6747. https://doi.org/10.1021/jf8040337
Das RK, Brar SK (2014) Enhanced fumaric acid production from brewery wastewater and insight into the morphology of Rhizopus oryzae 1526. Appl Biochem Biotechnol 172:2974–2988. https://doi.org/10.1007/s12010-014-0739-z
Fu YQ, Xu Q, Li S, Huang H, Chen Y (2009) A novel multi-stage preculture strategy of Rhizopus oryzae ME-F12 for fumaric acid production in a stirred-tank reactor. World J Microbiol Biotechnol 25:1871–1876. https://doi.org/10.1007/s11274-009-0076-5
Fu YQ, Yin LF, Zhu HY, Jiang R, Li S, Xu Q (2014) Effects of pellet characteristics on L-lactic acid fermentation by R. oryzae: pellet morphology, diameter, density, and interior structure. Appl Biochem Biotechnol 174:2019–2030. https://doi.org/10.1007/s12010-014-1146-1
Isaacson T, Damasceno CMB, Saravanan RS, He Y, Catalá C, Saladié M, Rose JKC (2006) Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nat Protoc 1:769–774. https://doi.org/10.1038/nprot.2006.102
Kossen NW (2000) The morphology of filamentous fungi. Adv Biochem Eng Biotechnol 70:1–33. https://doi.org/10.1007/3-540-44965-5_1
Langner T, Gohre V (2016) Fungal chitinases: function, regulation, and potential roles in plant/pathogen interactions. Curr Genet 62:243–254. https://doi.org/10.1007/s00294-015-0530-x
Liao W, Liu Y, Chen S (2007) Studying pellet formation of a filamentous fungus Rhizopus oryzae to enhance organic acid production. Appl Biochem Biotechnol 137–140:689–701. https://doi.org/10.1007/s12010-007-9089-4
Liu XH, Gao HM, Xu F, Lu JP, Devenish RJ, Lin FC (2012) Autophagy vitalizes the pathogenicity of pathogenic fungi. Autophagy 8:1415–1425. https://doi.org/10.4161/auto.21274
Liu Y, Liao W, Liu C, Chen S (2006) Optimization of L-(+)-lactic acid production using pelletized filamentous Rhizopus oryzae NRRL 395. Appl Biochem Biotechnol 131:844–853. https://doi.org/10.1385/ABAB:131:1-3:844
Ma LJ et al (2009) Genomic analysis of the basal lineage fungus Rhizopus oryzae reveals a whole-genome duplication. PLoS Genet 5:e1000549. https://doi.org/10.1371/journal.pgen.1000549
Meussen BJ, de Graaff LH, Sanders JP, Weusthuis RA (2012) Metabolic engineering of Rhizopus oryzae for the production of platform chemicals. Appl Microbiol Biotechnol 94:875–886. https://doi.org/10.1007/s00253-012-4033-0
Miura S, Arimura T, Hoshino M, Kojima M, Dwiarti L, Okabe M (2003) Optimization and scale-up of L-lactic acid fermentation by mutant strain Rhizopus sp. MK-96-1196 in airlift bioreactors. J Biosci Bioeng 96:65–69. https://doi.org/10.1016/s1389-1723(03)90098-3
Pera LM, Callieri DA (1997) Influence of calcium on fungal growth, hyphal morphology and citric acid production in Aspergillus niger. Folia Microbiol (Praha) 42:551–556. https://doi.org/10.1007/BF02815463
Xu J, Wang L, Ridgway D, Gu T, Moo-Young M (2000) Increased heterologous protein production in Aspergillus niger fermentation through extracellular proteases inhibition by pelleted growth. Biotechnol Prog 16:222–227. https://doi.org/10.1021/bp000006s
Xu Q, Liu Y, Li S, Jiang L, Huang H, Wen J (2016) Transcriptome analysis of Rhizopus oryzae in response to xylose during fumaric acid production. Bioprocess Biosyst Eng 39:1267–1280. https://doi.org/10.1007/s00449-016-1605-x
Yang J, Zhao X, Sun J, Kang Z, Ding S, Xu JR, Peng YL (2010) A novel protein Com1 is required for normal conidium morphology and full virulence in Magnaporthe oryzae. Mol Plant Microbe Interact 23:112–123. https://doi.org/10.1094/MPMI-23-1-0112
Yin LF, Ruan QC, Fu YQ (2013) Strain improvement of Rhizopus oryzae for over-production of lactic acid by random mutations. Afr J Microbiol Res 7:2970–2975. https://doi.org/10.5897/AJMR2013.5425
Yin P, Nishina N, Kosakai Y, Yahiro K, Pakr Y, Okabe M (1997) Enhanced production of l (+)-lactic acid from corn starch in a culture of Rhizopus oryzae using an air-lift bioreactor. J Ferment Bioeng 84:249–253. https://doi.org/10.1016/s0922-338x(97)82063-6
Zhou Y, Du J, Tsao GT (2000) Mycelial pellet formation by Rhizopus oryzae ATCC 20344. Appl Biochem Biotechnol 84–86:779–789. https://doi.org/10.1385/ABAB:84-86:1-9:779
Acknowledgements
This work was supported by a grant from the National Natural Science Foundation of China (21106091), a grant from Taizhou Science and Technology Plan Project (1701hb01) and a grant from Zhejiang Provincial Natural Science Foundation of China (LGG20B060001).
Author information
Authors and Affiliations
Contributions
LY and YF conceived and designed research. LY, XL, YZ, WY and WZ conducted research and collected data. LY was responsible for data analysis and wrote the manuscript. YF revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all the authors, the corresponding author states that there is no conflict of interest.
Rights and permissions
About this article
Cite this article
Yin, L., Luo, X., Zhang, Y. et al. Comparative proteomic analysis of Rhizopus oryzae hyphae displaying filamentous and pellet morphology. 3 Biotech 10, 469 (2020). https://doi.org/10.1007/s13205-020-02458-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-020-02458-0