Abstract
A novel cost-effective and widely applicable pH indicator was developed using anthocyanins extracted from the purple subtype of Ocimum sanctum L. and common lab filter paper. This pH indicator was successfully tested to monitor the pH of a wide range of buffers, solutions, irrigation water, and soil solution. Upon testing, the indicator displayed specific colors at corresponding pH ranges. Sucrose showed a stabilizing effect for the color of the extracted anthocyanins. Further, molecular analysis indicated that the leaves from the purple subtypes showed higher transcripts abundance for chalcone synthase, chalcone isomerase, anthocyanidin synthase, and dihydroflavonol 4-reductase than that of the green subtype. Similarly, transcription factors HY5 and a bHLH putatively involved in the biosynthesis of anthocyanins showed up-regulation in the purple subtype of O. sanctum.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Anthocyanins are flavonoids and natural pigments accumulated in various plant species. These pigments are soluble in water and well known for their free radical scavenging activities showing strong antioxidant properties. More than 250 types of anthocyanins have been isolated from various plant species. Plant anthocyanins are accumulated in vacuoles of many organs of plants like flowers, leaves, fruits, and roots (Dixon et al. 2005). Anthocyanins undergo glycosylation, acetylation, and methylation, and depending on their cyanine glycoside forms display a range of colors (Jackman and Smith 1996). Purple, reddish violate, red color of foliage, fruits, roots, and the inflorescence is due to the accumulation of anthocyanins. Due to free radical scavenging properties, these have been used for suppressing various types of cancer cells and well known as anti-aging agents, blood lipid-lowering agents, and as hepatoprotectives (Kong et al. 2003; Tsuda 2012; Kamiloglu et al. 2015; Zhu 2018). This class of flavonoids in solutions shows different colors under different pH levels; hence recently, these molecules have been used for the development of pH indicators. For example, anthocyanins extracted from the husk of Vitis amurensis, roselle, purple potato, purple cabbage, black bean seeds, black carrot and black rice bran have been used as pH indicators (Chen and Gu 2013; Golasz et al. 2013; Shahid et al. 2013; Choi et al. 2017; Pourjavaher et al. 2017; Prietto et al. 2017; Wu et al. 2018; Moradia et al. 2019; Tirtashi et al. 2019). Extracted anthocyanins have been incorporated in various biofilms made of starch/agar or other organic materials. Ultimately, these biofilms have emerged as pH indicators.
In developing countries like India, Bangladesh, Pakistan, Nepal, Bhutan, Shri-Lanka, etc. such crop plants have not been cultivated on a large scale. In these countries, climatic conditions, limited farm size, and limited consumer demand restrict the use and farming of such commodities. Ocimum genus belonging to the Lamiaceae family is widely distributed in tropical and sub-tropical regions of the world including India. Several Ocimum species like O. sanctum, O. basilicum, O. gratissimum L., O. kilimandscharicum, and O. americanum have been found in India. Among these species, O. sanctum is well known as Holy basil or Queen of Herb or Tulsi and is very common in India. Tulsi is grown even in houses for worship. This herb has gained medicinal values for its valuable metabolites present in essential oils like eugenol, methyl eugenol, chavicol, linalool, ursolic acid, methyl cinnamate, etc. (Saran et al. 2017). Moreover, O. sanctum has been popularized as a source of rosmarinic acid. Various pharmaceutical products have been in trade, which use O. sanctum and well known for their cough, cold, antioxidant, and antimicrobial properties (Pattanayak et al. 2010). Nowadays, the area under O. sanctum cultivation has been increasing with an increase in market demand.
O. sanctum has two subtypes depending on the color of foliage viz., Green (Rama Tulsi), and Purple (Shyama or Krishna Tulsi). The purple-colored O. sanctum subtype accumulates anthocyanin in foliage, stem, and inflorescences. The essential oil of Tulsi is biosynthesized in glandular trichomes known as peltate glands, whereas anthocyanins are accumulated in the rest of the leaf. Overall, purple Tulsi is an easily available plant and a good source of anthocyanins as it is easy to propagate under varied climatic conditions. Various research groups (Rastogi et al. 2015; Upadhyay et al. 2015) have reported the genome and transcriptome sequences of Tulsi. The biosynthesis of metabolites found in essential oils and anthocyanins shares common precursor metabolites in Tulsi. Secondary metabolites of Tulsi mainly belong to phenylpropanoid and terpene classes. Two metabolic pathways, shikimate, and MEP pathways supply the precursor metabolites to the biosynthesis of economically important metabolites like eugenol, chavicol, etc. (Rastogi et al. 2015). Expression analysis of genes involved in the phenylpropanoid pathway is well studied in green and purple subtypes of Tulsi. The key genes involved in the biosynthesis of anthocyanins in the purple subtype of Tulsi and its regulations are yet to identified and analyzed. Overall, Tulsi plants the same as other industrially valuable species have been grown for herbage yield either for essential oil or for natural active pharmaceutical products concerning its antioxidant and antimicrobial properties (Theapparat et al. 2019; Duan et al. 2019).
In the present study, the anthocyanins from the purple subtype of Tulsi have been extracted, analyzed for their spectral characteristics under varied pH range, and ultimately extracted anthocyanins were used to develop a novel pH indicator strip. Acid/base free filter paper was used as a base material for the strip development. This pH indicator strip was successfully used to test the pH of a variety of solutions including buffers used in molecular biology, soil solution, and irrigation water at farmers’ fields. Development of strip was very cost-effective and easy as compared to biofilms developed earlier using costly reagents and anthocyanins extracted from costly food crops. To understand the metabolic mechanism behind the purple color of Tulsi, key genes involved in the biosynthesis of anthocyanins and its regulation were identified. The expression analysis of these genes in the purple and the green subtype showed varied expression profiles. In addition, such alternative use can add value to an Indigenous crop and enable farmers to get higher income using fellow land for the cultivation of Tulsi. The present study aimed to assess the feasibility of anthocyanins extracted from the purple subtype of O. sanctum for the development of a pH indicator.
Materials and methods
Sample collection
Plants of the green and the purple subtypes of O. sanctum L. were grown on the farm at our Institute. Leaves were collected from three different mature plants of each subtype (as biological replicates) and either immediately processed for anthocyanins isolation and RNA isolations or stored at – 80 °C for further use.
Extraction, quantitative estimation, and spectral analysis of anthocyanins extracted from O. sanctum
Total anthocyanins from the purple Tulsi were extracted by acidic water. Chopped leaves from the purple subtype of O. sanctum (1 g) were emerged in 10 ml of water with 0.1% HCl for anthocyanin extraction. The solution was incubated in the microwave for 2–4 min with intermittent ventilation for several times. After complete extraction, the solution was filtered through Whatman no 1 filter paper. Total anthocyanins were estimated using the AOAC method (Lee 2005). Two buffers with pH 1 (potassium chloride, 0.025 M) and pH 4.5 (sodium acetate, 0.4 M) were used to determine the total anthocyanin content. Test solutions were prepared to have 1 part of plant extract and 4 parts of buffers. Absorptions were taken at 520 nm and 700 nm for each pH system. Total anthocyanins were estimated using the following formula:
where A = (A520 nm − A700 nm) pH1.0 – (A520 nm – A700 nm) pH4.5; MW (molecular weight) = 449.2 g/mol of cyanidin-3-glucoside; DF = dilution factor; l = path length in cm and ε = 26,900 molar extinction coefficient.
For colorimetric analysis of the extracted anthocyanins, solutions of different pH range having anthocyanins were prepared using HCl and NaOH. The spectral analysis was carried out using a spectrophotometer set at a spectral wavelength range between 400 to 650 nm. Anthocyanins change their color in different pH levels so photographs were taken at each specific pH.
Effect of pH and sucrose on stability during storage, photostability and thermal stability of anthocyanins
Effect of pH, sucrose, and heat on stability of anthocyanins was determined as described earlier (Matsufuji et al. 2007). Residual anthocyanins content was determined by the λvis-max and degradation index (DI). The value of DI was calculated by the ratio of absorption at 420 nm and λvis-max. For the effect of pH on photostability, pH 3, 5, and 7 solutions were used. Absorption analysis was conducted for these three pH levels for several days of light exposers. For photostability analysis, 0.2% (w/v) extracts of different pH (3, 5, and 7) were taken into a glass vial with screw caps and exposed to fluorescent light with 5000 lx for 24 h/d light period under 25 °C for a different periods. To evaluate the effect of sucrose on anthocyanin stability 1, 10, and 20% sucrose solutions were used. Thermostability was estimated by incubating extract solutions of pH levels 3, 5, and 7 at temperature 95 °C maintained in a water bath for different periods. Each sample was cooled in ice after incubation and residual anthocyanin (anthocyanin content after experiment/anthocyanin content initially × 100) was calculated by the pH differential method described above using UV–Vis spectrophotometer.
Characterization of genes involved in the biosynthesis of anthocyanins and its regulation
Anthocyanins are synthesized by the flavonoid pathway, which derives metabolites from phenylpropanoid pathways. The first enzyme for the flavonoid biosynthesis is chalcone synthase (CHS), and downstream key enzymes are chalcone isomerase (CI), dihydroflavonol 4-reductase (DFR) and anthocyanidin synthase (ANS). CHS and CI are early biosynthesis enzymes, whereas DFR and ANS are known as late biosynthesis enzymes. Transcripts or genomic sequences of early and late biosynthesis enzymes were identified using query protein sequences (CHS: A0A2Z4N9C3; CI: A0A2Z4N9C5; DFR: A0A2Z4N9A3; and ANS: A0A2Z4N991) available at UniProt database using tBLASTn. We identified transcription factors putatively involved in anthocyanin biosynthesis using HY5 (UniProt id O24646) and TT8 bHLH (UniProt id Q9FT81) as query sequences. For genome-wide identification, O. sanctum genomic and transcriptome sequences were downloaded. The protein sequences collected from Uniprot were used as query sequences against genome and transcriptome data. The tBLASTn approach was used to identify genes. The genomic sequences obtained from blast search were used for CDS identification using the AUGUSTUS tool (Stanke et al. 2006). The redundancy of sequences was eliminated manually. Identified transcripts were analyzed for their annotation against the protein database using BLASTx tool and domain features were analyzed by PROSITE web server, NCBI CD search tool and Pfam. Once, annotated primers were synthesized for their comparative gene expression analysis in the green and purple subtypes of O. sanctum. Homologs of identified proteins were collected from the NCBI database, and multiple sequence alignments were performed using the Clustal-W tool. Phylogenetic trees were constructed using the Neighbor-Joining method of MEGA 6 tool with 1000 bootstrap trails (Tamura et al. 2013).
Differential analysis of genes involved in the biosynthesis of anthocyanins
Total RNA was isolated from green and purple subtypes of O. sanctum using GeneJET Plant RNA Purification Kit (ThermoFisher). Total RNA was used for cDNA preparation using RevertAid First Strand cDNA Synthesis Kit (ThermoFisher) using total RNA (treated with 1 μg of DNAse) in 25 μl reaction volume. The real-time PCR analysis was performed in the CFX96 real-time PCR cycler (BioRad) machine. The primers used for RT-PCR analysis are listed in Table 1. Real-time analysis was performed using SYBR green master mix (Maxima SYBR Green). The reactions were set up for 25 µl with 200 ng cDNA and 12.5 µl Maxima master mix in each reaction. Thermal cycling was performed for three steps cycling with an initial denaturation at 95 °C, followed by denaturation 95 °C for 15 s, annealing 60 °C for 30 s and elongation 72 °C for 30 s. Fold change in gene expression was calculated as 2−ΔΔCt using ΔCt values.
Development of the pH strip
Extracted anthocyanins were used for the development of pH strips. For the stability of anthocyanins, sucrose (10%) was added to the extracted anthocyanin solution. Acid/Base free filter papers were cut into 5 cm × 2 cm strips and kept in glass plates. Extracted anthocyanin solution having sucrose was poured in these plates and incubated with filter paper strips for 10 min at room temperature. After incubation, the strip was transferred to a fresh glass plate and excess solution was removed with tissue paper. The strips were dried in an oven at 55 °C until completely dry. These strips were tested for color change at different pH levels. For this, Tris buffer at pH 8.0, distilled water as neutral pH 7.0, HCl solution at pH 2, and NaOH solution at pH10 were used. These strips were also used to measure the pH level of soil solution, irrigation water collected from farms, and lemon juice.
Statistical analysis
All experiments were performed at least in triplicate, and results were expressed as mean values with standard deviations. Statistical significance of the results was evaluated by ANOVA and Tukey test at p < 0.05 using the GraphPad Prism software.
Results
Anthocyanins from the purple O. sanctum and UV–Vis spectrum analysis
Total anthocyanins were extracted from the purple subtype of O. sanctum. Anthocyanins are supposed to be stable at acidic pH as compared to neutral pH. The quantitative estimation of anthocyanins from the purple subtype of O. sanctum showed 47.50 ± 1.121 mg/100 g fresh sample anthocyanins in leaves. The color of anthocyanins at different pH levels is shown in Fig. 1a. The color of the solution at pH 2 is bright red and which turned pink when pH raised to 4. Color of solution at pH 5 turned to violet and at neutral pH 7 turned to light violet. At pH range higher than 7, the color became green (pH 8) and dark green (pH 9). Further increase in the pH changed the color of the solution to brown (at pH 10) and light brown (at pH 11). The spectral analysis of anthocyanins at different pH levels is shown in Fig. 1b. The maximum absorption peak for the anthocyanins from O. sanctum was observed around 525 nm at the pH range 1–2. The peak of wavelength for the anthocyanin solutions was shifted toward higher wavelength with an increase in pH level as shown in Fig. 1b. The peak was shifted from 525 nm to 550 nm with an increase in the pH up to 6. At pH 7, the peak of solution shifted to 600 nm and a further increase in pH resulted in shifting of the peak towards 610 nm with an increase in absorption. Overall, anthocyanins extracted from Tulsi showed a bathochromic shift with increasing pH. It is believed that four different molecular forms of anthocyanins exist in aqueous solutions depending upon the pH of solutions, for example red flavylium cation, purple-violet quinoidal-base, colorless carbinol-base, and yellow-colored chalcones.
Stability analysis of the extracted anthocyanins
It is important to determine the stability of the anthocyanins before their use in any product development. The stability of anthocyanins extracted from O. sanctum was determined under various pH ranges and storage times. The effect of pH on photostability and thermostability was also determined. To identify a stabilizing agent, sucrose was analyzed. Figure 2 shows the stability analysis of extracted anthocyanins under various conditions. With the degradation of the anthocyanins, the residual content was decreased and DI (Degradation Index) value was increased. Results showed that the photostability of anthocyanins from O. sanctum depends on the pH of extract. At acidic pH (pH 3 and 5), the residual anthocyanins content was 68% after exposure to light for 25 days (Fig. 2a). At neutral pH, the anthocyanins resulted in degradation and reduced to only 30% even after 2 days exposer. In addition, DI value represents the degradation of anthocyanins. DI values of anthocyanins extract at pH 7 increased rapidly when exposed to light (Fig. 2b). The residual anthocyanin content and DI values indicated that anthocyanins are more stable to light exposer in acidic pH and were degraded in neutral pH. During heating, the red color of extract changed to brown-yellow. The pattern of increase in DI values under heating was similar to photodegradation as shown in Fig. 2c and d. The heating was more destructive for anthocyanins as compared to light as DI value increase sharply in case of heating. Overall, anthocyanins from O. sanctum are more stable in acidic pH to heat exposer than neutral pH. Figure 2e and f shows the stabilizing effect of sucrose on anthocyanin stability.
Stability analysis of the anthocyanins at different pH levels and in the presence of sucrose. Residual anthocyanins (anthocyanin content after experiment/anthocyanin content initially × 100%) were calculated by pH differential method. In the figure, a and b represent changes in residual anthocyanin percentage and DI (Degradation index: the ratio of absorption at 420 nm and λvis-max), respectively, during different light exposer time at different pH levels. Thermal stability (at 95 °C) of anthocyanins at different pH levels is shown in c (residual anthocyanin %) and d (DI). Effect of sucrose on the stability of anthocyanins is shown in e and f. Where, e represents residual anthocyanin % under different sucrose levels and f represents the visual color of immediately extracted anthocyanins (A), the color of anthocyanins after 3 months of extraction without sucrose (B) and color of anthocyanins extract with 10% sucrose (C) after 3 months stored at 25 °C
Identification and characterization of various genes involved in the biosynthesis of anthocyanins and its regulation
The raw transcripts and gene sequences of two genes of early (OsCHS and OsCHI) and two genes of late (OsDFR and OsANS) biosynthesis of anthocyanin in O. sanctum were identified. Genes for two key transcriptional regulators (OsHY5 and OsbHLH) were also identified. Transcripts for the genes OsDFR and OsANS could not be identified from the TSA database. Thus, the corresponding genomic sequences were collected. Identified sequences were annotated, and corresponding gene names were assigned. Figure 3 shows the annotation and domain organization of these identified proteins. Phylogenetic analysis suggested conserved amino acid residues among aligned protein sequences (Fig. 4). The conserved nature of residues indicated similar functions of homolog proteins. Identified transcription factors OsHY5 and OsbHLH are supposed to be involved in the biosynthesis of anthocyanins. Phylogenetic analysis suggested that OsCHS, OsCHI, OsANS, and OsDFR grouped with corresponding annotated homologs from other Lamiaceae members such as Salvia miltiorrhiza, Plectranthus scutellarioides, and Perilla frutescens. Interestingly, OsHY5 was grouped with HY5 from Salvia splendens. HY5 from Solanum lycopersicum and Nicotiana tomentosiformis also grouped with OsHY5. OsbHLH showed close homology with the Myc-F3G1 gene encoding bHLHprotein from Perilla frutiscens. This was reported to be regulating anthocyanin biosynthesis in Perilla frutiscens (Yamazaki et al. 2003).
Conserved domains identified in OsCHS, OsCHI, OsANS, OsDFR, OsHY5, and OsbHLH. These domains were identified using NCBI CD-search tool (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi)
Phylogenetic analysis of early (OsCHS and OsCHI represented in a and b respectively) and late (OsANS and OsDFR represented in c and d, respectively) genes of the biosynthesis pathway of anthocyanin in O. sanctum. Phylogenetic analysis of transcription factors (OsHY5 and OsbHLH represented in e and f, respectively) putatively involved in the biosynthesis of anthocyanins
OsCHS showed the presence of N and C terminal domains of chalcone and Stilbene synthase family proteins. OsCHI showed the chalcone isomerase superfamily protein domain. OsANS showed two domains 2OG-Fe(II) oxygenase superfamily and a non-haemdioxygenase domain. These are characteristic domains of ANS family proteins. OsDFR showed the characteristic domain of the NAD-dependent epimerase/dehydratase family. OsHY5 showed the Basic leucine zipper (bZIP) domain of Plant Elongated/Long Hypocotyl5 (HY5)-like transcription factors. OsbHLH showed the presence of bHLH-MYC and R2R3-MYB transcription factors N-terminal domain and downstream bHLH domain. This domain organization is a characteristic feature observed in many R2R3-MYB transcription factors involved in plant secondary metabolite biosynthesis. Domain organization and phylogenetic analysis indicated the similar structures-function relationship of homolog proteins identified in O. sanctum.
Differential gene expression analysis between the green and the purple subtypes
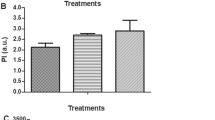
The purple subtype of Tulsi accumulates significantly higher anthocyanins in their foliage and inflorescences. The difference between green and purple subtypes is due to differentially expressed early and late genes involving in the biosynthesis of anthocyanins. Further, various transcriptional regulators regulate the expression of genes involved in anthocyanin biosynthesis. The main transcription factors for the biosynthesis of anthocyanin are HY5, TT8 (a bHLH transcription factor), and various MYB family proteins. HY5 generally involved in light regulatory pathways and anthocyanin biosynthesis. Whereas TT8 (bHLH class TF) acts as a part of multiprotein complex formation involved in the regulation of the biosynthesis of anthocyanins. The expression of OsCHS and OsCHI is higher in purple subtype than in the green subtype (Fig. 5), which indicates a higher carbon skeleton available for anthocyanin production. Also, the expression of OsANS and OsDFR was significantly higher in the purple subtype as compared to the green subtype (Fig. 5). These results correlated with the accumulation of anthocyanins in purple subtypes. The expression of OsHY5 and OsbHLH was also reported in green as well as purple subtypes. However, purple subtypes showed relatively higher expression for both OsHY5 and OsbHLH as indicated in Fig. 5.
Development of the pH strip using extracted anthocyanins
The pH strip developed using anthocyanins from O. sanctum and filter paper is shown in Fig. 6. The color of the strip changed when it was tested for different solutions having different pH levels. Different buffers and solutions were applied to strips by pipetting 10 μl volume. The strips showed immediate color change upon the application of buffers or solutions. The color of the strip becomes light violet when water at neutral pH was applied. The color of the strip became green when Tris buffer at pH 8 was applied. NaOH solution of higher pH like 10 changed strip color to brown. Application of HCl solution of pH 2 turned color of the strip to bright red (Fig. 6a). After the use of predetermined pH solutions, we tested strip for unknown pH solutions. Solutions of unknown pH (wet soil sample, irrigation water, and lemon juice) were used for their pH determination. Results showed that the pH of soil and irrigation water was slightly alkaline as the strip color changed to light green. While the application of lemon juice resulted in the pink color of the strip which indicated acidic pH of the lemon juice (Fig. 6b).
Discussion
Tulsi is known for its vast medicinal uses since ancient times. This plant species has been gaining commercial values due to its essential oil content. Our earlier study reported the germplasm of O. sanctum, which can be used as a source of eugenol (Saran et al. 2017). Apart from eugenol, the essential oil also has pharmaceutically important metabolites such as linalool, chavicol, methyl eugenol, methyl chavicol, cinnamate, citral, rosmarinic acid, etc. Nowadays, Tulsi has also been cultivated on farmer’s fields for herbage yield and its oil content to fulfill higher industrial demands. In crops like Tulsi, alternate use can add values and growers can get higher profits. In the present study, we tried to increase the utility of Tulsi beyond the use as food or therapeutic commodity. A pH strip was developed in which the anthocyanins isolated from the purple subtype of O. sanctum were used by absorbing on a filter paper. Before developing a strip, a detailed analysis was performed for anthocyanin extraction, spectral characterization, and stability of extracted anthocyanins. Simultaneously, key genes involved in the biosynthesis of anthocyanins and its regulation were identified and their relative gene expressions in the green and the purple subtypes were analyzed.
Total anthocyanins content showed that the purple Tulsi is a rich source of anthocyanins. The entire herbage part can be used as a source of anthocyanins. Thus, high herbage yield per unit area makes the Tulsi as a good source of anthocyanins. The extraction of anthocyanins can be achieved within a short duration by the use of the microwave. Our results indicated that microwaves could assist with the easy extraction of anthocyanins from Tulsi plants. Anthocyanins extracted from O. sanctum showed its characteristic absorption spectra. The anthocyanins change their color under various pH levels. Different molecular forms of anthocyanins are responsible for specific color at a specific pH. In acidic pH (2–5), anthocyanins of Tulsi showed red color due to flavylium cation form, which is converted to carbinol base at neutral pH (6–7), and color changed to light purple. Further increase in pH (8–12) anthocyanins of Tulsi changed to the yellow–brown quinoidal base form. The photostability and thermostability analysis indicated that anthocyanins from Tulsi are stable in acidic pH like anthocyanins extracted from different plant species. The results indicated that once isolated anthocyanins can be stored in acidic pH for further use. Similarly, anthocyanins isolated from red radish also showed thermostability and photostability under acidic pH (Matsufuji et al. 2007). Sucrose increased the stability of isolated anthocyanins. Sucrose may be used for the development of a cost-effective and harmless product using anthocyanins.
The purple subtype of Tulsi is more or less similar to all aspects like cultivation, climatic requirement, morphology, constituents of essential oil and pharmaceutical uses except the purple color of leaves. The molecular understanding of the regulation of biosynthesis of purple pigments in O. sanctum has not been reported earlier. Some preliminary studies have been reported for the differential expression of genes involved in the phenylpropanoid pathway and the shikimate pathway in the green and the purple subtypes (Maurya et al. 2019). Also, metabolic alterations have been reported in Ocimum species under different light and stress responses (Rastogi et al. 2019). The climatic conditions are also known to affect the metabolite distribution in Ocimum species. In O. basilicum, which shows similar gene content as O. sanctum, showed varied transcript abundance for genes involved in secondary metabolites between two Ocimum species (Rastogi et al. 2014). The present study reported that OsCHS, OsCHI, OsDFR, and OsANS were up-regulated in the purple morph than the green subtype indicating higher accumulation of starting metabolites for the biosynthesis of anthocyanins. However, some cultivars are also available which show intermediately accumulation of anthocyanins. Hence, only the purple subtype of Tulsi can be served as a source of anthocyanins due to the abundance of transcripts for OsCHS, OsCHI, OsANS, and OsDFR. Biosynthesis of anthocyanins in plants has been reported to be regulated by various transcription factors that form multiprotein complexes. Several regulatory mechanisms involving repressors, activators, and miRNAs operating in a sequence have been reported to regulate thebiosynthesis of anthocyanins. Protein trimeric MYB–bHLH–WDR (MBW) complex has been reported as a key activator for the transcription of DFR and ANS. The involvement of the HY5 factor in the biosynthesis of anthocyanins has also been shown in Arabidopsis, Tomato, Apple, etc. HY5 plays a key role by positively controlling MYB12 and negatively regulating MYB111. Also, HY5 showed regulation by mediating miRNAs (Shin et al. 2007; Zhang et al. 2011; An et al. 2017; Lim et al. 2017; Qiu et al. 2019). Overall, identification and characterization of HY5 and protein trimmer complex are necessary to understand the biosynthesis of anthocyanins plants. We identified a homolog of HY5 in O. sanctum, which was up-regulated in the purple subtype. Simultaneously, multiprotein complex regulating the late biosynthesis genes comprises a bHLH family factor. In Arabidopsis, TT8 was reported to act as bHLH protein involved in the multiprotein complex (Lim et al. 2017). We isolated homolog of Arabidopsis TT8 in O. sanctum and annotated as OsbHLH. Its expression was higher in purple subtype as compared to the green subtype. Overall, our study revealed various genes involved in the biosynthesis of anthocyanins in O. sanctum.
Anthocyanins extracted from Tulsi were successfully absorbed by filter paper. The results showed that the filter papers retained anthocyanins extracted in acidic solutions having sucrose even after drying for a longer time. Paper strip developed by the anthocyanins was very useful for estimating the pH of a wide range of buffers, solutions, and soil samples. When this pH strip was exposed to the solution of a particular pH, it immediately changed the color. Commercially available strips have been developed by using lichens. So far, no commercial strip was available utilizing any plant-based pigments. However, some research groups tried to develop plant anthocyanins-based biofilms for pH measurements. Recently, anthocyanins-based pH indicators have been proposed for specific purposes viz. for assessment of milk shelf life, chicken patties quality, etc. (Goodarzi et al. 2020; Talukder et al. 2020). Also, a pH indicator and antimicrobial cellulose nanofibres packaging film was developed using anthocyanin extracted from purple sweet potato and oregano essential oil (Chen et al. 2020). In a study, anthocyanins were used for pH biofilm development using cellulose nanofibres as a film matrix (Moradia et al. 2019). In another study, to monitor fish freshness, a film was developed with starch/polyvinyl alcohol as matrix and roselle anthocyanins (Zhai et al. 2017). Similarly, chitosan, methylcellulose, chitosan/corn starch blends, polyvinyl alcohol, chitosan/pectin polyelectrolyte complex and polyethylene terephthalate were used as solid support in anthocyanins-based pH indicators (Pacquit et al. 2007; Rukchon et al. 2014; Yoshida et al. 2014; Maciel et al. 2015; Pereira et al. 2015; Silva-Pereira et al. 2015). The cost involved in such matrix materials was comparatively higher than a strip developed with filter paper. In countries like India, where anthocyanin-rich food crops are costly, Tulsi can be used as a cheaper source for anthocyanins with wide applicability.
Conclusions
Tulsi is known for its pharmaceutical uses and has been cultivated for essential oil industries. We isolated total anthocyanins from the purple subtype of Tulsi. This purple subtype is different from the green subtype as the purple plants showed higher expression of genes involved in the biosynthesis of anthocyanins and its regulation. We reported transcription factors putatively regulating the biosynthesis of anthocyanins in O. sanctum. We performed spectral analysis of isolated anthocyanins and identified pH range and sucrose level at which anthocyanins from Tulsi can be stored for a longer time. Finally, a pH indicator strip was developed from anthocyanins isolated from purple Tulsi, which can be used to determine the pH of a wide range of commodities like soil solution, irrigation water, biological buffers, media, and other food products. Our study added values to Tulsi and can be utilized for the betterment of farmers as well as the use of anthocyanins.
Availability of data and material (data transparency)
Not applicable.
Code availability (software application or custom code).
Not applicable.
References
An J, Qu F, Yao J, Wang X, You C, Wang X, Hao Y (2017) The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in Apple. Hortic Res 4:1702
Chen X, Gu Z (2013) Absorption-type optical pH sensitive film based on immobilized purple cabbage pigment. Sensor Actuat B-Chem 178:207–211
Chen S, Wu M, Lu P, Gao L, Yan S, Wang S (2020) Development of pH indicator and antimicrobial cellulose nanofibre packaging film based on purple sweet potato anthocyanin and oregano essential oil. Int J BiolMacromol 149:271–280
Choi I, Lee JY, Lacroix M, Han J (2017) Intelligent pH indicator film composed of agar/potato starch and anthocyanin extracts from purple sweet potato. Food Chem 218:122–128
Dixon RA, Xie DY, Sharma SB (2005) Proanthocyanidins—a final frontier in flavonoid research. New Phyto 165:9–28
Duan C, Meng X, Meng J, Khan MIH, Dai L, Khan A, An X, Zhang J, Huq T, Ni Y (2019) Chitosan as a preservative for fruits and vegetables: a review on chemistry and antimicrobial properties. J BioresourBioproducts 4:11–21
Golasz LB, Silva JD, Silva SBD (2013) Film with anthocyanins as an indicator of chilled pork deterioration. Food SciTechnol 33:155–162
Goodarzi MM, Moradi M, Tajik H, Forough M, Ezati P, Kuswandi B (2020) Development of an easy-to-use colorimetric pH label with starch and carrot anthocyanins for milk shelf life assessment. Int J Biol Macromol 153:240–247
Jackman RL, Smith JL (1996) Anthocyanins and betalains. In: Hendry GAF, Houghton JD (eds) Natural food colorants, 2nd edn. Springer, US, pp 244–309
Kamiloglu S, Capanoglu E, Grootaert C, Van Camp J (2015) Anthocyanin absorption and metabolism by human intestinal Caco-2 cells—a review. Int J Mol Sci 16:21555–21574
Kong JM, Chia LS, Goh NK, Chia TF, Brouillard R (2003) Analysis and biological activities of anthocyanins. Phytochemistry 64:923–933
Lee J (2005) Determination of total monomeric anthocyanin pigment content of fruit juices, beverages, natural colorants, and wines by the pH differential method: collaborative study. J AOAC Int 88:1269–1278
Lim S, Kim D, Kim JK, Lee JY, Ha S (2017) A radish basic helix-loop-helix transcription factor, RsTT8 acts a positive regulator for anthocyanin biosynthesis. Front Plant Sci 8:1917
Maciel VBV, Yoshida CM, Franco TT (2015) Chitosan/pectin polyelectrolyte complex as a pH indicator. Carbohydr Polym 132:537–545
Matsufuji H, Kido H, Misawa H, Yaguchi J, Otsuki T, Chino M, Takeda M, Yamagata K (2007) Stability to light, heat, and hydrogen peroxide at different pH values and DPPH radical scavenging activity of acylated anthocyanins from red radish extract. J Agric Food Chem 55:3692–3701
Maurya S, Chandra M, Yadav RK, Narnoliya LK, Sangwan RS, Bansal S, Sandhu P, Singh U, Kumar D, Sangwan NS (2019) Interspecies comparative features of trichomes in Ocimum reveal insights for biosynthesis of specialized essential oil metabolites. Protoplasma 256(4):893–907
Moradia M, Tajik H, Almasic H, Forough M, Ezatib P (2019) A novel pH-sensing indicator based on bacterial cellulose nanofibers and black carrot anthocyanins for monitoring fish freshness. Carbohydr Poly 222:115030
Pacquit A, Frisby J, Diamond D, Lau KT, Farrell A, Quilty B, Diamond D (2007) Development of a smart packaging for the monitoring of fish spoilage. Food Chem 102(2):466–470
Pattanayak P, Behera P, Das D, Panda SK (2010) Ocimum sanctum Linn. A reservoir plant for therapeutic applications: an overview. Pharmacogn Rev 4:95–105
Pereira VA, de Arruda INQ, Stefani R (2015) Active chitosan/PVA films with anthocyanins from Brassica oleraceae (Red Cabbage) as time-temperature indicators for application in intelligent food packaging. Food Hydrocoll 43:180–188
Pourjavaher S, Almasi H, Meshkini S, Pirsa S, Parandi E (2017) Development of a colorimetric ph indicator based on bacterial cellulose nanofibers and red cabbage (Brassica oleraceae) extract. Carbohydr Polym 156:193–201
Prietto L, Mirapalhete TC, Pinto VZ, Hoffmann JF, Vanier NL, Lim L, Dias ARG, Zavareze E (2017) pH-sensitive films containing anthocyanins extracted from black bean seed coat and red cabbage. LWT-Food Sci Technol 80:492e500
Qiu Z, Wang H, Li D, Yu B, Hui Q, Yan S, Huang Z, Cui X, Cao B (2019) Identification of candidate HY5-dependent and -independent regulators of anthocyanin biosynthesis in Tomato. Plant Cell Physiol 60:643–656
Rastogi S, Meena S, Bhattacharya A, Ghosh S, Shukla RK, Sangwan NS, Lal RJ, Gupta MM, Lavania UC, Gupta V, Nagegowda DA, Shasany AK (2014) De Novo sequencing and comparative analysis of holy and sweet basil transcriptomes. BMC Genomics 15(1):588
Rastogi S, Kalra A, Gupta V, Khan F, Lal RK, Tripathi AK, Parameswaran S, Gopalakrishnan C, Ramaswamy G, Shasany AK (2015) Unravelling the genome of Holy basil: an “incomparable” “elixir of life” of traditional Indian medicine. BMC Genomics 16:413
Rastogi S, Shah S, Kumar R, Vashisth D, Akhtar MQ, Kumar A, Dwivedi UN, Shasany AK (2019) Ocimum metabolomics in response to abiotic stresses: cold, flood, drought and salinity. PLoS ONE 14(2):e0210903
Rukchon C, Nopwinyuwong A, Trevanich S, Jinkarn T, Suppakul P (2014) Development of a food spoilage indicator for monitoring freshness of skinless chicken breast. Talanta 130:547–554
Saran PL, Tripathy V, Saha A, Kalariya KA, Suthar MK, Kumar J (2017) Selection of superior Ocimum sanctum L. accessions for industrial application. Ind Crops Prod 108:700–707
Shahid M, Islam S-u, Mohammad F (2013) Recent advancements in natural dye applications: a review. J Clean Prod 53:310–331
Shin J, Park E, Choi G (2007) PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J 49:981–994
Silva-Pereira MC, Teixeira JA, Pereira-Júnior VA, Stefani R (2015) Chitosan/corn starch blend films with extract from Brassica oleraceae (red cabbage) as a visual indicator of fish deterioration. LWT Food Sci Technol 61(1):258–262
Stanke M, Schöffmann O, Morgenstern B, Waack B (2006) Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics 7:62
Talukder S, Mendiratta SK, Kumar RR, Agrawal RK, Soni A, Luke A, Chand S (2020) Jamun fruit (Syzgiumcumini) skin extract based indicator for monitoring chicken patties quality during storage. J Food SciTechnol 57(2):537–548
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 60. Mol Biol Evol 30:2725–2729
Theapparat Y, Khongthong S, Rodjan P, Lertwittayanon K, Faroongsarng D (2019) Physicochemical properties and in vitro antioxidant activities of pyroligneous acid prepared from brushwood biomass waste of Mangosteen, Durian, Rambutan, and Langsat. J For Res 30:1139–1148
Tirtashi FE, Moradi M, Tajik H, Forough M, Ezati P, Kuswandi B (2019) Cellulose/chitosan pH-responsive indicator incorporated with carrot anthocyanins for intelligent food packaging. Int J Biol Macromol 136:920–926
Tsuda T (2012) Dietary anthocyanin-rich plants: biochemical basis and recent progress in health benefits studies. Mol Nutr Food Res 56:159–170
Upadhyay AK, Chacko AR, Gandhimathi A, Ghosh P, Harini K, Joseph AP, Joshi AG, Karpe SD, Kaushik S, Kuravadi N, Lingu CS, Mahita J, Malarini R, Malhotra S, Malini M, Mathew OK, Mutt E, Naika M, Nitish S, Pashant SN, Singh HR, Sukhwal A, Margaret SS, Manojkumar S, Ramaswamy S, Gowda M, Ramanathan S (2015) Genome sequencing of herb Tulsi (Ocimum tenuiflorum) unravels key genes behind its strong medicinal properties. BMC Plant Biol 15:212
Wu H, Yang K, Chiang P (2018) Roselle anthocyanins: antioxidant properties and stability to heat and pH. Molecules 23:1357
Yamazaki M, Makita Y, Springob K, Saito K (2003) Regulatory mechanisms for anthocyanin biosynthesis in chemotypes of Perillafrutescens var. crispa. Biochem Eng J 14(3):191–197
Yoshida CM, Maciel VBV, Mendonc MED, Franco TT (2014) Chitosan biobased and intelligent films: monitoring pH variations. LWT Food Sci Technol 55(1):83–89
Zhai X, Shi J, Zou X, Wang S, Jiang C, Zhang J, Huang X, Zhang W, Holmes M (2017) Novel colorimetric films based on starch/polyvinyl alcohol incorporated with roselle anthocyanins for fish freshness monitoring. Food Hydrocoll 69:308–317
Zhang H, He H, Wang X, Wang X, Yang X, Li L, Deng XW (2011) Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J 65:346–358
Zhu F (2018) Anthocyanins in cereals: composition and health effects. Food Res Int 109:32–249
Acknowledgement
We are thankful to ICAR-DMAPR, India for the financial support.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
Conceptualization [Manish Kumar Suthar], Methodology [Manish Kumar Suthar and ParmeshawarLal Saran], Writing—original draft [Manish Kumar Suthar] and Writing—review & editing [Manish Kumar Suthar and Parmeshawar Lal Saran].
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Rights and permissions
About this article
Cite this article
Suthar, M.K., Saran, P.L. Anthocyanins from Ocimum sanctum L., a promising biomolecule for development of cost-effective and widely applicable pH indicator. 3 Biotech 10, 388 (2020). https://doi.org/10.1007/s13205-020-02380-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-020-02380-5