Abstract
Streptomyces sp. isolated from marine sediment collected from Palk Strait, Bay of Bengal was investigated for its antagonistic potential. The isolate exhibited antimicrobial activity against selected bacterial strains of clinical importance such as Staphylococcus aureus MTCC 3160, Bacillus pumilus NCIM 2327, S. aureus (methicillin resistant), Escherichia coli MTCC 1698, E. coli (ESBL), Shigella flexneri MTCC 1457, Proteus vulgaris and Enterobacter cloacae. Phenotypic and molecular characterization ascertained the isolate BDK01 as Streptomyces chumphonensis. Media optimization with one variable-at-a-time strategy was attempted to identify the ideal concentrations of starch (5–15 g/l), casein (0.01–0.05 g/l), NaCl 1.0–3.0 g/l, pH (4.0–9.0 g/l), temperature (25–45 °C) and inoculum level (0.5–5 ml) towards achieving maximum antimicrobial compound production. Statistical optimization of production media was carried by establishing an 11 variables 17 run experiment through PB model which evinced starch, calcium carbonate, pH and inoculum concentration that highly influenced bioactive compound production. Spectral data of active ethyl acetate extract revealed the presence of various bioactive compounds such as Salicyl alcohol, N-phenyl benzamide. 6-Octadecenoic acid, (Z), 1,3,5-Cycloheptatriene. Antiproliferation activity of active fraction against MCF-7 Cell line exhibited IC50 value of 9.5 µg/ml. Overall, it is observed that the marine actinomycete S. chumphonensis BDK01 could be employable as promising strain for novel antimicrobial and cytotoxic metabolites.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Marine habitats are potential and promising region unravelling novel biomolecules due to its diverse physical, chemical, and biological entities compared to terrestrial environment (Romano et al. 2017). Often, shallow and deep-water sediments are proven source for antimicrobial and antitumor compounds-producing microorganisms (Zheng et al. 2000; Gu et al. 2004; Kwon et al. 2006). Generally, Actinobacteria accounts for more than 7000 bioactive compounds as reported in the Dictionary of Natural Products. Among them, Streptomyces genus alone contribute for more than 80% of the actinobacterial compounds (Jensen et al. 2005; Bull and Stach 2007). Dalisay et al. (2013) found that 25% of Streptomyces isolated from marine environment showed antimicrobial activity, which increased when tested with seawater. On the other hand, multiple drug-resistant bacteria and other clinical pathogens are evolving as big threat to human community leading to high morbidity and mortality especially in developing countries (Campfield et al. 2014). Hence, the present investigation was focused on discovery of novel bioactive compounds for the treatment of such burgeoning diseases.
Bay of Bengal is well known potential source for marine-derived bacteria rich in bioactive compounds (Peela et al. 2005; Arumugam et al. 2010; Saurav et al. 2013). Marine sediments of these regions predominantly contain the genus Streptomyces, which is reportedly active against a range of clinically important bacteria and fungi (Peela et al. 2005). Arumugam et al. (2010) reported an unusual metabolite 2-allyloxyphenol, with substantial antimicrobial activity from Streptomyces M1/7 isolated from the Sundarbans of Bay of Bengal. Similarly, a larvicidal compound, 5-(2,4-dimethylbenzyl) pyrrolidin-2-one, was found to be produced by Streptomyces VITSVK strain from Marakkanam coast of Bay of Bengal (Saurav et al. 2013). All such reports exposes enormous metabolites from this region that could serve as valuable source for exploring products of pharmaceutically importance.
In the present study, antimicrobial compound producing actinobacteria Streptomyces chumphonensis strain BDK01 was isolated from marine sediment sample collected from Palk Strait Region of Bay of Bengal. Taxonomic affiliation of the isolate was carried by incorporating various morphological, chemotaxonomic and molecular methods. Media used for antimicrobial compound production was optimized statistically through one variable-at-a-time strategy and Plackett–Burman model. Liquid–liquid extraction was employed for compound separation and subsequent distilled product was subjected to spectral studies to identify the biologically active compounds from fermented broth. Antiproliferative property of active ethyl acetate fraction was also investigated against MCF7 cell line through MTT assay.
Materials and methods
Sample collection and isolation of antagonistic Actinobacteria
Totally, 60 marine sediment samples were collected from 27 different locations of coastal Tamil Nadu and Kerala during pre-monsoon periods of 2012–2013. Few marine sediment samples were also collected from deep sea using the facilities extended by Centre for Advanced Studies in Marine Biology, Annamalai University. All the collected samples were dried in hot air oven at 55 °C for 30 min, serially diluted, and plated on starch casein agar and Actinomycete isolation agar (Himedia). The plates were incubated for 7–10 days under room temperature, and the matured colonies were sub-cultured on YEME agar. For genus level identification, the isolates were subjected to cultural, biochemical, physiological, and microscopic examinations according to Shirling and Gottlieb (1966) and Bergey’s Manual of Systematic Bacteriology (Goodfellow et al. 2012). To analyze the antagonistic ability of the isolates, each strain was inoculated into starch casein broth and incubated in rotary shaker at 28 °C for 7 days. After fermentation, the supernatant was collected by centrifugation and the test bacterial strains (Staphylococcus aureus MTCC 3160, Bacillus pumilus NCIM 2327, S. aureus (methicillin resistant), Escherichia coli MTCC 1698, E. coli (ESBL), Shigella flexneri MTCC 1457 and P. vulgaris) were challenged against it (SM 1). The obtained results were recorded and those mean values were subjected to post hoc (Tukey’s test) analysis. Based on comparative analysis amongst the isolates BDK01 from Dhanushkodi (Lat—09°09′07.29″; Lon—79°26′35.82″) was chosen for further studies.
Taxonomic characterization of BDK01 by polyphasic approach
Cultural characteristics of the potential isolate BDK01 was analyzed as suggested by Shirling and Gottlieb (1966). All the microscopic, metabolic, and physiological examinations, which include biochemical, carbohydrate utilization, and optimization of growth conditions, were carried out as per Bergey’s Manual of Systematic Bacteriology (Goodfellow et al. 2012). For micromorphological profiling, the isolate was grown on starch casein agar by coverslip culture technique (Pridham et al. 1958) and the matured colonies were observed under scanning electron microscope JEOL JSM-5610 (Mitra et al. 2008). For whole cell sugar and amino acid investigation, the sample was prepared by the methods of Suput et al. (1967) and Becker et al. (1964), respectively. Furthermore, the cell hydrolysate was analyzed by thin layer chromatography (Staneck and Roberts 1974) against suitable standard sugars and amino acids. For cell wall Fatty acid methyl ester (FAME) investigation, sample was prepared using the standard protocol of MIDI (Sherlock Microbial Identification System, version 4.0) and analyzed through TRACE Ultra Ver: 5.0, MS DSQ gas chromatographic mass system (Thermo Fisher Scientific, Waltham, MA, USA) (Lu et al. 2013).
The extracted genomic DNA of isolate BDK01 was amplified for its 16S rRNA gene using the 27F 5′-GAG TTT GAT CCT GGC TCA G-3′—forward and 1530R 5′-GTT ACC TTG TTA CGA CTT-3′—reverse primers (Yukphan et al. 2004). The reaction volume of 25 µl consists of 1X Taq buffer contains 25 mM MgCl2, 4 mM dNTPs, 0.5 IU Taq polymerase (Fermentas, Thermo Scientific, Vilnius, Lithuania), 10 pmol of forward and reverse primers and 50–100 ng template DNA. The PCR conditions included initial denaturation at 94 °C for 10 min followed by 30 cycles of denaturation at 94 °C for 15 s, annealing at 55 °C for 15 s, extension at 72 °C for 1 min, and a final extension of 72 °C for 10 min using MyCycler™ Thermal Cycler (Bio-Rad). The purified PCR product was sequenced and the taxonomical affiliation of the isolate was assigned through BLAST search against GenBank database (http://www.ncbi.nlm.nih.gov). The phylogenetic tree was constructed for the isolate BDK01 with the closest identified type strains following pairwise and multiple sequence analysis by neighbour joining method. The phylogeny inference, maximum parsimony, and maximum likelihood methods were applied to assess dendrogram reliability and stability using MEGA 5.0 (Tamura et al. 2011).
Statistical optimization of medium for antimicrobial compound production
To select suitable media for antimicrobial compound production, three different media named as PM1 (Remya and Vijayakumar 2008), PM2 (Ilić et al. 2005), and PM3 (Badji et al. 2006) were prepared in 50% marine water (125 ml/500 ml flask). PM1 contained the following ingredients: soluble starch 10.0 g/l, casein 0.03 g/l, KNO3 2.0 g/l, NaCl 2.0 g/l, K2HPO4 2.0 g/l, MgSO4 7H2O 0.05 g/l, CaCO3 0.02 g/l, FeSO4 7H2O 0.01 g/l, and pH 7.2. Another media PM2 contained glucose 2.0 g/l, yeast extract 3.0 g/l, NaCl 0.8 g/l, NH4Cl 1.0 g/l, KCl 0.1 g/l, KH2PO4 0.1 g/l, MgSO4 7H2O 0.2 g/l, CaCl2 0.04 g/l, marine water 50%, and pH 7.3. PM3 contained yeast extract 4.0 g/l, malt extract 10.0 g/l, dextrose 4.0 g/l, marine water 50%, and pH 7.2. All the three media were inoculated with 1% of 0.6 O.D (600 Å) inoculum of BDK01 and the flasks were kept in a rotary shaker at 28 °C for 7 days at 180 rpm. After incubation, the fermented broth was collected, centrifuged at 6000 rpm for 10 min, and supernatant was tested for antimicrobial activity against the selected test organisms by well diffusion method (Schillinger and Lücke 1989).
On the basis of zone formation, PM1 was selected as suitable media for production and subjected for statistical optimization, which includes one variable-at-a-time strategy and Plackett–Burman (PB) design through Design expert 8.0. One variable-at-a-time strategy was used to identify the effective concentrations and conditions of starch, casein, NaCl, pH, temperature, and inoculum concentrations on antimicrobial compound production. Those best productive conditions were subjected to PB design to find the significant factors that influence bioactive compound production. In addition, concentrations of other variables such as KNO3, K2HPO4, MgSO4 7H2O, CaCO3, and FeSO4 7H2O were also included. With the selected low (−) and high (+) levels of variable concentrations (Table 1), a 11 factor 17 run experiment was generated through Design Expert 8.0 (Stat-Ease, Minneapolis, MN, USA). All experiments were conducted as triplicate and the collected data were analyzed at 5% significant level (P < 0.05), were considered to have substantial effect on the antimicrobial production. The predicted regression equation based on the results of Plackett–Burman model was as follows:
where Y is the predicted response, β0 is constant (intercept), β1, β2, to βn are the coefficient of the variables and X1, X2, to Xn are the coded values of independent variables.
By applying the best obtained results of PB design, mass production was carried for 10 l in a series of 500 ml (1/4 working volume) conical flasks added with 6-mm diameter glass beads. After incubation, the culture was centrifuged (C-24 BL, REMI) at 6000 rpm for 10 min and supernatant was subjected for partial purification by liquid–liquid extraction.
Partial purification and identification of active compound
The supernatant was subjected to solvent extraction employing various solvents such as petroleum ether, butanol, chloroform, ethyl acetate, and methanol in a 250 ml flasks. Fifty mL of each solvent were mixed with equal volume of cell-free broth and left on a shaker for 60 min. Then, the solvent layer was separated, concentrated by distillation apparatus at 60 °C, and investigated against the test bacteria by well diffusion method. An extra well loaded with each pure solvent served as control against all test organisms. Obtained results were subjected to one sample t test to identify the significance of solvents in extraction process. Accordingly, ethyl acetate was identified as ideal solvent at the ratio of 1:4. Later, solvent phase was collected and distilled in a distillation flask at 60 °C (Parthasarathi et al. 2013). For partial purification, the gummy crude compound was subjected to isocratic silica gel (60–120 mesh) column chromatography using ethyl acetate as mobile phase (Praveen et al. 2008). The active ethyl acetate fraction was subjected for spectral and mass analysis. The maximum absorbance (λmax) spectra of the partially purified compound were analyzed between 800 and 200 nm in PerkinElmer Lambda 25 UV/Vis Spectrophotometer. Similarly, the functional group analysis of ethyl acetate fraction was carried out using Shimadzu IR Affinity FT-IR spectrophotometer (3500–600 cm−1). Metabolic profiling of active fraction was done in gas chromatography mass spectrophotometer (PerkinElmer Clarus 500, Software: Turbomass ver 5.2.0) with the Capillary Column Elite-5MS (5% Phenyl 95% dimethylpolysiloxane). The oven program was set at 50 °C at 7 °C/min to 200 °C (3 min) @ 7 °C/min to 280 °C (15 min), and 1.0 µl (25 µg/ml) of crude sample was injected using helium (carrier gas) at the rate of 1 ml/min. The mass range of 40–600 amu was analyzed at 70 eV at the temperature of 200 °C.
Antiproliferative activity of the active ethyl acetate fraction against MCF 7 cell line
The human breast cancer cell line (MCF-7) was obtained from National Centre for Cell Science (NCCS), Pune, India, and grown in Eagles Minimum Essential Medium (EMEM) containing 10% foetal bovine serum (FBS). The monolayer cells were detached with trypsin—EDTA to make single cell suspensions, and the viable cells were diluted with medium containing 5% FBS to give final density of 1 × 105 cells/ml. Cell suspension (100 µl) was seeded into 96-well plate (103 cells/well) and incubated to facilitate cell attachment. After 24 h, the cells were treated with serial concentrations of the test samples (0, 3.12, 6.25, 12.5, 25, and 50 µg/ml), which were dissolved in dimethyl sulfoxide (DMSO). Furthermore, with appropriate control, the plates in triplicate were maintained for all concentrations. After 48 h of incubation, 15 µl of MTT (5 mg/ml) in phosphate-buffered saline (PBS) was added to each well and kept at 37 °C for 4 h. The medium with MTT was then flicked off and formed formazan crystals were solubilized in 100 µl of DMSO which was measured at absorbance of 570 nm using microplate reader. Mean and IC50 values were calculated by nonlinear regression analysis, and a graph was plotted between percentage of cell inhibition and Log10 concentration using GraphPad Prism software.
Results
Totally, 85 morphologically distinct actinobacterial strains were isolated from marine sediment samples on different media, which were subjected to phenotypic characterization to identify the genus. Among them, 34 (40%) isolates were Streptomyces, 15 (17.6%) were Micromonospora, 12 (14.1%) were Saccharopolyspora, 6 (7%) were Actinopolyspora, 7 (8.24%) were Nocardia, 2 (2.35%) Nocardioides, 5 (5.8%) were Kitasatospora, and 4 (4.71%) remained as ‘unidentified’. The well diffusion assay of fermented broth had exposed 34 (40%) isolates for its antimicrobial activity against test and clinical bacterial strains. Among them, isolate BDK01, which is identified as Streptomyces sp., was splendid in antagonistic ability with enormous zone of inhibition and active against all employed test organisms.
Taxonomic characterization of the isolate BDK01
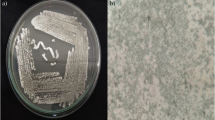
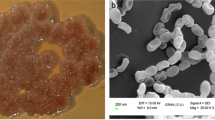
Colonies were circular and umbonate with ability to produce melanin on starch casein agar. The isolate showed excellent growth in Actinomycete isolation agar, Bennet’s agar, nutrient agar, and ISP media except on ISP 5 and ISP 7. Pale orange colour aerial and dark brown colour substrate mycelia were the dominant growth features on all media. Isolate was identified as positive for hydrogen sulphide production, catalase, and oxidase, and negative for indole, methyl red, Voges–Proskauer, citrate, and urease. The strain had the ability to degrade starch, casein, and gelatin, and utilizing sugars like glucose and mannitol. Physiological examinations revealed that the strain can grow well between the pH 6.0 and 9.0 at 25–37 °C and optimal NaCl concentration was 1–5%. Scanning electron microscopic (SEM) studies revealed that aerial and substrate mycelia of the strain BDK01 were well developed without fragmentation. Oval, smooth surfaced rectiflexibiles long spore chain measured more than 5 µm in the length contains more than 10 spores per chain measuring 840.0 × 508.2 nm (Fig. 1). No sugar was diagnosed in the cell wall, whereas amino acid analysis revealed the presence of glycine and LL 2,6 diaminopimelic acid. FAME analysis indicated the presence of various straight and branched chain of iso and ante-iso fatty acids, mainly C15:0 antesio (12-methyltetra decanoic acid), C16:0 (hexadecanoic acid, methyl ester), and C18:2 w6c (9,12-Octadecadienoic acid). BLAST analysis of BDK01 16S rRNA gene sequence revealed more than 23 Streptomyces strains were sharing 98–99% sequence homology (Fig. 2). In the phylogenetic analysis, BDK01 clustered with strain S. chumphonensis KK1-2T (NCBI accession no: AB738401) carry 99% sequence homology. The 16S rDNA sequence of BDK01 was deposited in NCBI under the name of S. chumphonensis strain BDK01 (NCBI accession no. JX486779).
Statistical optimization and production of antimicrobial compound
Among the three different media employed, PM1 (Table 2) was observed as an ideal media for antimicrobial compound production. The one variable-at-a-time strategy reveals the effective concentrations of selected variables on antimicrobial compound production (Fig. 3). The variables such as starch, casein, sodium chloride, temperature, pH, and inoculum concentrations expressed antimicrobial compound in all ranges served with difference in production volume. Based on the results of One variable-at-a-time strategy Plackett–Burman experimental model (Table 3) was attempted, where the eighth run produced maximum amount of antimicrobial compound followed by eleventh run. ANOVA test revealed F value of 21.39 and a low probability value of 0.0047, thereby implying that the model is significant (Tables 4, 5). The p values of A, G, K, and L were significant as they are less than 0.05, whereas other values were greater and not significant (Li et al. 2008). The predicted R2 value 0.9832 variability in response, is in reasonable agreement with the adjusted R2 value 0.9408. The lack of fit value 0.4545 for the design is insignificant, suggested that the obtained data were good in fit. The Pareto chart indicates that the pH (K), calcium carbonate (G), starch (A), and inoculum concentration (L) were highly influencing the antimicrobial compound production (Fig. 4). Particularly, calcium carbonate and pH were influencing the production positively, whereas starch and inoculum concentration were influencing negatively. The antimicrobial compound production could be predicted with the regression equations derived through Plackett–Burman model as follows:
where Y was the response, whereas A, B, C, D, E, F, G, H, J, K, and L were the coded values of starch, casein, KNO3, NaCl, K2HPO4, MgSO4 7H2O, CaCO3, FeSO4 7H2O, pH, temperature, and inoculum concentration, respectively.
Pareto chart showing effects of factors above and below the ‘Bonferroni Limit’ (absolute significance) and ‘t-value Limit’ (line of significance) on antimicrobial compound production. A (Starch), B (Casein), C (KNO3), D (NaCl), E (KHPO4), F (MgSO4 7H2O) (F), G (CaCO3), H (FeSO ∙4 7H2O), K (pH), J (temperature) and L (inoculum concentration). Blue and orange bars indicates positive and negative effects, respectively
Extraction and spectral identification of potential bioactive compound
Among the five different solvents studied for solvent extraction, ethyl acetate (P ≤ 0.001) was found to be the best for extraction of antimicrobial compounds (SM 2). Ethyl acetate fraction (25 µg/ml) was potentially active against 16 reference bacterial and fungal strains there was no activity against Aspergillus niger (SM 3). Antiproliferative activity against MCF7 cell line by MTT assay resulted in significant IC50 value of 9.5 µg/ml (Fig. 5). Maximum peak value was observed at 324.30 nm and 273.12 nm in UV–Vis spectrum analysis. Absorption peak 273.12 indicated possible presence of polyene compounds (Ilić et al. 2005). Multiple sharp and broad peaks were observed between 3500 and 600 cm−1 in FT-IR indicated the presence of functional groups such as C=C alkenes, C=O group of acids, C≡C alkynes, –C–H stretch (alkane H), and O–H (hydrogen bond, intermolecular, polymeric association). GC–MS results of active ethyl acetate extract indicated the presence of more than 44 different possible compounds, of which 18 compounds were already reported for several biological activities. Salicyl alcohol was in maximum proportion (16.32%) followed by N-phenylbenzamide (10.73%). Other important biologically potential compounds such as 6-octadecenoic acid, (Z) (8.12%), 1,3,5-cycloheptatriene (3.29%), 2-pyrrolidinone (0.083%), and nomifensine acetate (0.48%) were also observed. Other bioactive compounds with complete volatile metabolic profiling are presented in Table 6.
Discussion
Marine Actinobacteria are well known for its bioactivities, such as antimicrobial and cytotoxic activities as evinced by many previous investigations (Sujatha et al. 2005; Remya and Vijayakumar 2008; Suthindhiran and Kannabiran 2010; Parthasarathi et al. 2013). In the present study, 85 morphologically distinct actinobacteria, dominantly Streptomyces (40%), were isolated from sediment samples collected from coastal Tamil Nadu and Kerala. Post hoc (Tukey’s test) analysis of results obtained from well diffusion assay showed that the isolate BDK01 had excellent activity against the test pathogens based on average mean performance value.
The isolate BDK01 was subjected to polyphasic taxonomic characterization for species level identification. Morphological, microscopical, biochemical, carbohydrate utilization, and physiological examination suggested that the strain BDK01 could be a Streptomyces species (Shirling and Gottlieb 1966; Goodfellow et al. 2012). Lack of cell wall sugar and the presence of LL isomeric form of 2,6 diaminopimelic acid is the unique taxonomic feature of type I cell wall. The cell wall fatty acid methyl esters of S. chumphonensis KK1-2T, a strain which is phylogenetically related to the isolate BDK01, were investigated by Phongsopitanun et al. (2014). The strain contains C15:0 anteiso, C16:0 iso, and C15:0 iso in cell wall, whereas the isolate BDK01 of present investigation contains C15:0 antesio, C16:0, and C18:2 w6c, which indicates that the chemical composition of these two strains had subtle variation. Phylogenetic analysis revealed that the isolate BDK01 was closely related to strain S. chumphonensis KK1-2T (NCBI accession no: AB738401) (Fig. 1). A recently reported study, S. chumphonensis KK1-2T was isolated from marine sediments of Chumphon Province, Thailand (Phongsopitanun et al. 2014). Hence, it is strongly believed that the strain BDK01 is a marine habitant.
Among 3 media compared, PM1 showed the maximum appropriateness for antimicrobial compound production than PM2 and PM3 (Table 2). This agrees with the investigation of Vijayakumar et al. (2012), who reported the elevated antimicrobial activity of Actinobacteria when grown in medium containing starch. PM2 containing glucose failed to produce quantifiable amount of antimicrobial compound. Similar findings were observed by Gupte and Kulkarni (2002) assuming that the complex polysaccharides were the best constituent for production media than any other monosaccharides. While the readily available carbon source like glucose is utilized for microbial growth, complex carbohydrate like starch was assimilated slowly for growth and compound production (Remya and Vijayakumar 2008; Durairaj and Ramasamy 2013).
PB model evinced that the variables such as pH (K), calcium carbonate (G), starch (A), and inoculum concentration (L) were influencing the antimicrobial compound production (Fig. 4). Earlier, Sathiyanarayanan et al. (2014) proposed that pH is a significant variable in antimicrobial compound production by Streptomyces sp. MAPS15. Besides, other earlier findings also suggested that minerals such as KNO3, NaCl, K2HPO4, MgSO4, CaCO3, and FeSO4 had influenced antimicrobial compound production either positively or negatively (Guo et al. 2012; Sathiyanarayanan et al. 2014). In the present study, the R2 value of antimicrobial compound 0.9832 indicates that the model is significant to enhance the production (Tables 4, 5). Even, a regression model having an R2 value higher than 0.90 was considered as a high correlation factor (Chen et al. 2009). The standard error counter plot was very circular and flat, indicating that the design is very significant. Conclusively, PB design identified the significant factors that influence antimicrobial compound production. The obtained variables through central composite design may be utilized to identify the precise medium for enhanced production of antimicrobial compounds.
Ethyl acetate was concluded as best solvent for extraction of antimicrobial compounds (SM 2) (Parthasarathi et al. 2012; Vijayakumar et al. 2012). Nithyanand et al. (2011) reported, besides ethyl acetate chloroform also good for bioactive compound extraction. Ethyl acetate with polarity index 4.4 was immiscible in water phase which had no effect on test pathogens making it as the best solvent for extraction of antimicrobial compounds.
Results of UV and FT-IR were compared with the Bordoloi et al. (2001), Wu et al. (2007), and Dhanasekaran et al. (2008) to identify peak points and their functional group which indicated the possible presence of various biologically active functional groups. GC–MS analysis disclosed more than 18 different compounds with various biological activities, whereas rest of the compounds was not. Significantly, salicyl alcohol in vast quantity (16.32%), was repeatedly reported for antibacterial and antifungal activities (Harun et al. 2005).
Salicyl alcohol or Saligenin is a phenolic alcohol usually produced as a derivative by hydrolysis of salicin. Generally, this compound is a derivative of benzyl alcohol with substitution of an OH group at the ortho position. In FT-IR analysis of the current investigation, peak value of 3327 indicates the presence of O–H stretching frequencies. Saligenin is used as multipurpose drug that can act as analgesic and antipyretic agents. Earlier, the antimicrobial activity of salicyl alcohol against 20 bacterial strains were studied by Harun et al. (2005). Results showed promising antagonistic effects against Proteus mirabilis, Pseudomonas aeruginosa, Escherichia coli, Enterobacter sp, and S. aureus, which are in highly accordance with current investigation. Ghasemzadeh et al. (2012) and Vejselova and Kutlu (2015) reported the anticancer potential of salicylic acid against MCF7 cell line and A549 human lung adenocarcinoma, respectively. Another study carried by El-Shemy et al. (2007) suggested that salicyl alcohol, contains antitumor activity, which can eliminate immature white blood cells through apoptosis associated DNA damage. Moreover, salicyl alcohol can induce resistance to multiple antibiotics in bacteria, notably the quinolone group of antimicrobial agents (Cohen et al. 1993).
The second major constituent N-phenylbenzamide (10.73%), was already reported for strong antiviral activity against Enterovirus (Ji et al. 2013), Hepatitis C Virus and Enterovirus 71 (strain SZ-98) (Jiang et al. 2015). Another study carried by Pasha et al. (2008) suggested that the compound is highly active against a range of Gram-positive and Gram-negative bacteria. Besides, antimalarial and antifungal properties of N-phenylbenzamide was endorsed by various investigators (Niewiadomy et al. 2001; Ertan et al. 2009; Desai et al. 2011).
Another biologically active compound 6-Octadecenoic acid, (Z) was detected in the GC–MS spectrum. Earlier, Raghad and Jalill (2014) found that the compound 6-Octadecenoic acid extracted from Calendula officinalis leaves widely known for its ability to suppress Human epidermoid larynx carcinoma (Hep-2). The compound has several bioactivities such as antimicrobial (Gheda et al. 2013), anti-inflammatory, antiandrogenic, anticancer, dermatitigenic, hypocholesterolemic, 5-alpha reductase inhibitor, anemiagenic, insectifuge, and flavouring agents for food/pharmaceutical industries (Basu et al. 2013). 9-Hexacosene known for antinociceptive and anti-inflammatory activities (Githinji et al. 2012) was also reported in considerable quantity. Another major constituent 1,3,5-Cycloheptatriene (3.29%) commercially called Triprolidine, is an antibacterial compound, which is currently in usage (Trust and Bartlett 1975).
Overall, many bioactive compounds were detected in GC–MS profile of ethyl acetate extract recovered from S. chumphonensis BDK01 fermented broth. Although many compounds are not reported for any biological activities, certain structurally related compounds have significant activities. Hence, we strongly believe that the S. chumphonensis strain BDK01 could be a novel source for several new antimicrobial compounds.
American National Cancer Institute recommends that any crude compound IC50 value less than 30 μg/ml could be a promising anticancer drug (de Oliveira et al. 2016). The MTT assay of the current study revealed that the compounds present in the crude could be a potential drug against cancer cells as it expressed the IC50 value of 9.5 μg/ml (Fig. 5). The GC–MS analysis revealed the presence of many compounds in various concentrations with different antagonistic effects against microbial and cancer cell lines. Especially the phenolic compounds (Dr. Duke’s Phytochemical and Ethnobotanical Databases), 2-pyrrolidinone (Thangam et al. 2013), 2,4-bis(1,1-dimethylethyl) phenol (Varsha et al. 2015), 6-octadecenoic acid (Z) (Basu et al. 2013), and longifolene-(V4) (Murugesan et al. 2013) were also present in considerable quantity. Consequently, the present investigation confirms that the crude extract contains many anticancer compounds at various concentrations, which could be a promising agent to control the cancer proliferation. Moreover, anticancer analysis against other transformed cell lines could reveal potential information about the compounds.
Conclusion
Our study on isolation, taxonomic characterization, and identification of bioactive compounds from marine Actinobacteria from coastal Tamil Nadu and Kerala resulted in the discovery of S. chumphonensis BDK01. The strain BDK01 was phylogenetically closer to identified as S. chumphonensis KK1-2T, a novel species recently reported by Phongsopitanum et al. (2014) and yet to be studied for any bioprospecting potential. Hence, to our knowledge, the present study serve as a first report for antimicrobial activity in this species. The isolate S. chumphonensis strain BDK01 exhibited significant antagonistic activity against test bacterial and fungal strains. Moreover, the crude compound displayed remarkable IC50 value of 9.5 µg/ml against MCF-7 (Breast Cancer) cell line, which demonstrate its activity to combat cancer cell proliferation. Optimization of production media was done statistically towards the maximization of antimicrobial compound production. Metabolic profiling of the active compounds was reported through spectral studies. Further exploration of S. chumphonensis strain BDK01 will throw light on its pharmacological potential which could benefit drug industries.
References
Arockia J, Veerabahu R, Mohan (2014) GC-MS analysis of bioactive components on the stem extract of Bacolepsis nervosa decne. Ex. Moq. (Periplocaceae). World J Pharm Pharm Sci 3(4):1044–1059.
Arumugam M, Mitra A, Jaisankar P et al (2010) Isolation of an unusual metabolite 2-allyloxyphenol from a marine actinobacterium, its biological activities and applications. Appl Microbiol Biotechnol 86:109–117. https://doi.org/10.1007/s00253-009-2311-2
Badji B, Zitouni A, Mathieu F et al (2006) Antimicrobial compounds produced by Actinomadura sp. AC104 isolated from an Algerian Saharan soil. Can J Microbiol 52:373–382. https://doi.org/10.1139/w05-132
Basu S, Choudhury UR, Das M, Datta G (2013) Identification of bioactive components in ethanolic and aqueous extracts of Amorphophallus campanulatus tuber by GC-MS analysis. Int J Phytomed 5:243–251
Becker B, Lechevalier MP, Gordon RE, Lechevalier HA (1964) Rapid Differentiation between Nocardia and Streptomyces by paper chromatography of whole-cell hydrolysates. Appl Microbiol 12:421–423
Bordoloi GN, Kumari B, Guha A et al (2001) Isolation and structure elucidation of a new antifungal and antibacterial antibiotic produced by Streptomyces sp. 201. Biosci Biotechnol Biochem 65:1856–1858. https://doi.org/10.1271/bbb.65.1856
Bull AT, Stach JEM (2007) Marine actinobacteria: new opportunities for natural product search and discovery. Trends Microbiol 15:491–499
Campfield B, Chen K, Kolls JK (2014) Vaccine approaches for multidrug resistant Gram negative infections. Curr Opin Immunol 28:84–89
Chen X-C, Bai J-X, Cao J-M et al (2009) Medium optimization for the production of cyclic adenosine 3′,5′-monophosphate by Microbacterium sp. no. 205 using response surface methodology. Bioresour Technol 100:919–924. https://doi.org/10.1016/j.biortech.2008.07.062
Cohen SP, Levy SB, Foulds J, Rosner JL (1993) Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J Bacteriol 175:7856–7862. https://doi.org/10.1128/jb.175.24.7856-7862.1993
Corre J, Lucchini JJ, Mercier GM, Cremieux A (1990) Antibacterial activity of phenethyl alcohol and resulting membrane alterations. Res Microbiol 141:483–497. https://doi.org/10.1016/0923-2508(90)90074-Z
Dalisay DS, Williams DE, Wang XL et al (2013) Marine sediment-derived Streptomyces bacteria from british columbia, canada are a promising microbiota resource for the discovery of antimicrobial natural products. PLoS One. https://doi.org/10.1371/journal.pone.0077078
de Oliveira PF, Damasceno JL, Bertanha CS et al (2016) Study of the cytotoxic activity of Styrax camporum extract and its chemical markers, egonol and homoegonol. Cytotechnology 68:1597–1602. https://doi.org/10.1007/s10616-015-9864-y
Desai KR, Shaikh MS, Coutinho EC (2011) Molecular modeling studies, synthesis and biological evaluation of derivatives of N-phenylbenzamide as Plasmodium falciparum dihydroorotate dehydrogenase (PfDHODH) inhibitors. Med Chem Res 20:321–332. https://doi.org/10.1007/s00044-010-9323-4
Dhanasekaran D, Thajuddin N, Panneerselvam A (2008) An Antifungal compound: 4′ phenyl-1-naphthyl -phenyl acetamide from Streptomyces sp. DPTB16. Facta Univ Med Biol 15:7–12
Durairaj T, Ramasamy V (2013) A potent fish pathogenic bacterial killer Streptomyces sp. isolated from the soils of east coast region, South India. J Coast Life Med 1:175–180. https://doi.org/10.12980/JCLM.1.2013C1086
El-Shemy HA, Aboul-Enein AM, Aboul-Enein KM, Fujita K (2007) Willow leaves’ extracts contain anti-tumor agents effective against three cell types. PLoS One 2:e178. https://doi.org/10.1371/journal.pone.0000178
Ertan T, Yildiz I, Tekiner-Gulbas B et al (2009) Synthesis, biological evaluation and 2D-QSAR analysis of benzoxazoles as antimicrobial agents. Eur J Med Chem 44:501–510. https://doi.org/10.1016/j.ejmech.2008.04.001
Friedman M, Henika PR, Mandrell RE (2002) Bactericidal activities of plant essential oils and some of their isolated constituents against Campylobacter jejuni, Escherichia coli, Listeria monocytogenes, and Salmonella enterica. J Food Prot 65:1545–1560. https://doi.org/10.4315/0362-028X-65.10.1545
Ghasemzadeh A, Jaafar HZE, Karimi E (2012) Involvement of salicylic acid on antioxidant and anticancer properties, anthocyanin production and chalcone synthase activity in ginger (zingiber officinale roscoe) varieties. Int J Mol Sci 13:14828–14844. https://doi.org/10.3390/ijms131114828
Gheda SF, Khalil MA, Gheida SF (2013) In vitro and in vivo preliminary results on Spirulina platensis for treatment of impetigo: topical cream application. Afr J Biotechnol 12:2498–2509. https://doi.org/10.5897/AJB12.1640
Githinji CG, Mbugua PM, Kanui TI, Kariuki DK (2012) Analgesic and anti-inflammatory activities of 9-hexacosene and stigmasterol isolated from Mondia whytei. Phytopharmacology 2:212–223
Goodfellow M, Kämpfer P, Busse HJ, Trujillo ME, Suzuki K, Ludwig W, Whitman WB (eds) (2012) Bergey’s manual of systematic bacteriology, vol 5. The Actinobacteria, part A and B. Springer, New York, NY
Gu Q, Lu J, Cui C et al (2004) Recent researches of bioactive metabolites in marine organisms-associated microorganisms. J Ocean Univ China 3:150–156. https://doi.org/10.1007/s11802-004-0026-7
Guo Z, Shen L, Ji Z, Wu W (2012) Enhanced production of a novel cyclic hexapeptide antibiotic (NW-G01) by Streptomyces alboflavus 313 using response surface methodology. Int J Mol Sci 13:5230–5241. https://doi.org/10.3390/ijms13045230
Gupte MD, Kulkarni PR (2002) A study of antifungal antibiotic production by Streptomyces cattanoogensis MTCC 3423 using full factorial design. Lett Appl Microbiol 35:22–26. https://doi.org/10.1046/j.1472-765X.2002.01119.x
Hema R, Kumaravel S, Alagusundaram K (2011) GC/MS determination of bioactive components of Murraya koenigii. J Am Sci 7:2009–2012
Hsouna AB, Trigui M, Mansour RB et al (2011) Chemical composition, cytotoxicity effect and antimicrobial activity of Ceratonia siliqua essential oil with preservative effects against Listeria inoculated in minced beef meat. Int J Food Microbiol 148:66–72. https://doi.org/10.1016/j.ijfoodmicro.2011.04.028.
Huang CB, Alimova Y, Myers TM, Ebersole JL (2011) Short- and medium-chain fatty acids exhibit antimicrobial activity for oral microorganisms. Arch Oral Biol 56:650–654. https://doi.org/10.1016/j.archoralbio.2011.01.011
Ilić SB, Konstantinović SS, Todorović ZB (2005) UV/VIS analysis and antimicrobial activity of Streptomyces isolates. Med Biol 12:44–46. https://doi.org/10.1142/S0217595914500213
Jensen PR, Mincer TJ, Williams PG, Fenical W (2005) Marine actinomycete diversity and natural product discovery. Antonie van Leeuwenhoek Int J Gen Mol Microbiol 87:43–48. https://doi.org/10.1007/s10482-004-6540-1
Ji XY, Wang HQ, Hao LH et al (2013) Synthesis and antiviral activity of N-phenylbenzamide derivatives, a novel class of Enterovirus 71 inhibitors. Molecules 18:3630–3640. https://doi.org/10.3390/molecules18033630
Jiang Z, Wang H, Li Y et al (2015) Synthesis and antiviral activity of a series of novel N-phenylbenzamide and N-phenylacetophenone compounds as anti-HCV and anti-EV71 agents. Acta Pharm Sin B 5:201–209. https://doi.org/10.1016/j.apsb.2015.03.013
Kwon HC, Kauffman CA, Jensen PR, Fenical W (2006) Marinomycins A-D, antitumor-antibiotics of a new structure class from a marine actinomycete of the recently discovered genus “Marinispora”. J Am Chem Soc 128:1622–1632. https://doi.org/10.1021/ja0558948
Li Y, Jiang H, Xu Y, Zhang X (2008) Optimization of nutrient components for enhanced phenazine-1-carboxylic acid production by gacA-inactivated Pseudomonas sp. M18G using response surface method. Appl Microbiol Biotechnol 77:1207–1217. https://doi.org/10.1007/s00253-007-1213-4
López A, Dong SM, Towers GHN (2002) Antifungal activity of benzoic acid derivatives from Piper lanceaefolium. J Nat Prod 65:62–64. https://doi.org/10.1021/np010410g
Lu Y, Wang J, Deng Z et al (2013) Isolation and characterization of fatty acid methyl ester (FAME)-producing Streptomyces sp. S161 from sheep (Ovis aries) faeces. Lett Appl Microbiol 57:200–205. https://doi.org/10.1111/lam.12096
Mitra A, Santra SC, Mukherjee J (2008) Distribution of actinomycetes, their antagonistic behaviour and the physico-chemical characteristics of the world’s largest tidal mangrove forest. Appl Microbiol Biotechnol 80:685–695. https://doi.org/10.1007/s00253-008-1626-8
Murugesan S, Senthilkumar N, Rajeshkannan C, Vijayalakshmi KB (2013) Phytochemical characterization of Melia dubia for their biological properties. Der Chem Sin 4:36–40
Niewiadomy A, Matysiak J, Mącik-Niewiadomy G (2001) In vitro evaluation of 2,4-dihydroxythiobenzanilides against various moulds. Eur J Pharm Sci 13:243–248. https://doi.org/10.1016/S0928-0987(01)00126-9
Nithyanand P, Manju S, Karutha Pandian S (2011) Phylogenetic characterization of culturable actinomycetes associated with the mucus of the coral Acropora digitifera from Gulf of Mannar. FEMS Microbiol Lett 314:112–118
Parthasarathi S, Sathya S, Bupesh G et al (2012) Isolation and characterization of antimicrobial compound from marine Streptomyces hygroscopicus BDUS 49. World J Fish Mar Sci 4:5–7. https://doi.org/10.5829/idosi.wjfms.2012.04.03.5658
Parthasarathi S, Sathya S, Bupesh G et al (2013) Isolation, characterization and extraction of antimicrobial compound from marine actinomycete Streptomyces hygroscopicus BDUS 49. Res J Biotechnol 8:40–48
Pasha FA, Muddassar M, Lee C, Cho SJ (2008) Mechanism based QSAR studies of N-phenylbenzamides as antimicrobial agents. Environ Toxicol Pharmacol 26:128–135. https://doi.org/10.1016/j.etap.2008.01.005
Peela S, Kurada VVSNB, Terli R (2005) Studies on antagonistic marine actinomycetes from the Bay of Bengal. World J Microbiol Biotechnol 21:583–585. https://doi.org/10.1007/s11274-004-3493-5
Phongsopitanun W, Thawai C, Suwanborirux K et al (2014) Streptomyces chumphonensis sp. nov., isolated from marine sediments. Int J Syst Evol Microbiol 64:2605–2610. https://doi.org/10.1099/ijs.0.062992-0
Pietro Z, Maurizio S, Maurizio B, Antonella, M, Sergio R, Carmen F, Felice S (2010) Essential oil composition of stems and fruits of Caralluma europaea N.E.Br. (Apocynaceae). Molecules 15:627–638
Praveen V, Tripathi CKM, Bihari V, Srivastava SC (2008) Production of actinomycin-D by the mutant of a new isolate of Streptomyces sindenensis. Braz J Microbiol 39:689–692. https://doi.org/10.1590/S1517-83822008000400017
Prescott LM, Klein DA, Harley JP (2002) Microbiology, 5th edn. McGraw-Hill, New York
Pridham TG, Hesseltine CW, Benedict RG (1958) A guide for the classification of Streptomycetes according to selected groups. J Biol Chem Can J Microbiol 189:109–114
Raghad DH, Jalill A (2014) GC-MS analysis of Calendula officinalis and cytotoxic effects of its flower crude extract on human epidermoid larynx carcinoma (HEP-2). World J Pharm Pharm Sci 3:237–275
Rowe RC, Sheskey PJ, Quinn ME (2009) Handbook of pharmaceutical excipients, 6th edn. Pharmaceutical Press and American Pharmacists Association. ISBN 978 0 85369 792 3
Remya M, Vijayakumar R (2008) Isolation and characterization of marine antagonistic actinomycetes from west coast of India. Med Biol 15:13–19. https://doi.org/10.1117/1.1801431
Romano G, Costantini M, Sansone C et al (2017) Marine microorganisms as a promising and sustainable source of bioactive molecules. Mar Environ Res 128:58–69. https://doi.org/10.1016/j.marenvres.2016.05.002
Rowe RC, Sheskey PJ, Quinn ME (2009) Handbook of pharmaceutical excipients, 6th edn. Pharmaceutical Press and American Pharmacists Association. ISBN 978 0 85369 792 3
Sathiyanarayanan G, Gandhimathi R, Sabarathnam B et al (2014) Optimization and production of pyrrolidone antimicrobial agent from marine sponge-associated Streptomyces sp. MAPS15. Bioprocess Biosyst Eng 37:561–573. https://doi.org/10.1007/s00449-013-1023-2
Saurav K, Rajakumar G, Kannabiran K et al (2013) Larvicidal activity of isolated compound 5-(2,4-dimethylbenzyl) pyrrolidin-2-one from marine Streptomyces VITSVK5 sp. against Rhipicephalus (Boophilus) microplus, Anopheles stephensi, and Culex tritaeniorhynchus. Parasitol Res 112:215–226. https://doi.org/10.1007/s00436-011-2682-z
Schillinger U, Lücke FK (1989) Antibacterial activity of Lactobacillus sake isolated from meat. Appl Environ Microbiol 55:1901–1906. https://doi.org/10.1016/S0168-1605(03)00051-5
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340. https://doi.org/10.1099/00207713-16-3-313
Staneck JL, Roberts GD (1974) Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol 28:226–231
Sujatha P, Bapi Raju KVVSN, Ramana T (2005) Studies on a new marine streptomycete BT-408 producing polyketide antibiotic SBR-22 effective against methicillin resistant Staphylococcus aureus. Microbiol Res 160:119–126. https://doi.org/10.1016/j.micres.2004.10.006
Suput J, Lechevalier MP, Lechevalier HA (1967) Chemical composition of variants of aerobic actinomycetes. Appl Microbiol 15:1356–1361
Suthindhiran K, Kannabiran K (2010) Diversity and exploration of bioactive marine actinomycetes in the Bay of Bengal of the Puducherry coast of India. Indian J Microbiol 50:76–82. https://doi.org/10.1007/s12088-010-0048-3
Tamura K, Peterson D, Peterson N et al (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. https://doi.org/10.1093/molbev/msr121
Tayung K, Barik B, Jha D, Deka DC (2011) Identification and characterization of antimicrobial metabolite from an endophytic fungus, Fusarium solani isolated from bark of Himalayan yew. Mycosphere 2:203–213
Thangam R, Suresh V, Rajkumar M et al (2013) Antioxidant and in vitro anticancer effect of 2-pyrrolidinone rich fraction of Brassica oleracea var. capitata through induction of apoptosis in human cancer cells. Phytother Res 27:1664–1670. https://doi.org/10.1002/ptr.4908
Trust TJ, Bartlett KH (1975) Antibacterial activity of tropilidine and tropone. Antimicrob Agents Chemother 8:381–383
Üçüncü H, Aktafi AE, Yazgi H et al (2005) Assessment of antibacterial activity of some topical otological solutions. Turk J Ear Nose Thorat 14:97–100
Ulrich S, Wolfgang H (1974) Effect of nomifensine (Hoe 984), a new antidepressant, on uptake of noradrenaline and serotonin and on release of noradrenaline in rat brain synaptosomes. Pharmacology 23(24):3413–3422
Varsha KK, Devendra L, Shilpa G et al (2015) 2,4-Di-tert-butyl phenol as the antifungal, antioxidant bioactive purified from a newly isolated Lactococcus sp. Int J Food Microbiol 211:44–50. https://doi.org/10.1016/j.ijfoodmicro.2015.06.025
Vejselova D, Kutlu HM (2015) Inhibitory effects of salicylic acid on A549 human lung adenocarcinoma cell viability. Turk J Biol 39:1–5. https://doi.org/10.3906/biy-1401-7
Vijayakumar R, Panneerselvam K, Muthukumar C et al (2012) Optimization of antimicrobial production by a marine actinomycete Streptomyces afghaniensis VPTS3-1 isolated from palk strait, East Coast of India. Indian J Microbiol 52:230–239. https://doi.org/10.1007/s12088-011-0138-x
Wu X-C, Chen W-F, Qian C-D et al (2007) Isolation and identification of newly isolated antagonistic Streptomyces sp. strain AP19-2 producing chromomycins. J Microbiol 45:499–504
Yukphan P, Potacharoen W, Tanasupawat S et al (2004) Asaia krungthepensis sp. nov., an acetic acid bacterium in the α-proteobacteria. Int J Syst Evol Microbiol 54:313–316. https://doi.org/10.1099/ijs.0.02734-0
Zheng Z, Zeng W, Huang Y et al (2000) Detection of antitumor and antimicrobial activities in marine organism associated actinomycetes isolated from the Taiwan Strait, China. FEMS Microbiol Lett 188:87–91. https://doi.org/10.1016/S0378-1097(00)00215-9
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Manikandan, M., Gowdaman, V., Duraimurugan, K. et al. Taxonomic characterization and antimicrobial compound production from Streptomyces chumphonensis BDK01 isolated from marine sediment. 3 Biotech 9, 167 (2019). https://doi.org/10.1007/s13205-019-1687-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-019-1687-7