Abstract
The diversity of rhizobia that establish symbiosis with Lotus corniculatus has scarcely been studied. Several species of Mesorhizobium are endosymbionts of this legume, including Mesorhizobium loti, the type species of this genus. We analysed the genetic diversity of strains nodulating Lotus corniculatus in Northwest Spain and ten different RAPD patterns were identified among 22 isolates. The phylogenetic analysis of the 16S rRNA gene showed that the isolated strains belong to four divergent phylogenetic groups within the genus Mesorhizobium. These phylogenetic groups are widely distributed worldwide and the strains nodulate L. corniculatus in several countries of Europe, America and Asia. Three of the groups include the currently described Mesorhizobium species M. loti, M. erdmanii and M. jarvisii which are L. corniculatus endosymbionts. An analysis of the recA and atpD genes showed that our strains belong to several clusters, one of them very closely related to M. jarvisii and the remanining ones phylogenetically divergent from all currently described Mesorhizobium species. Some of these clusters include L. corniculatus nodulating strains isolated in Europe, America and Asia, although the recA and atpD genes have been sequenced in only a few L. corniculatus endosymbionts. The results of this study revealed great phylogenetic diversity of strains nodulating L. corniculatus, allowing us to predict that even more diversity will be discovered as further ecosystems are investigated.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The genus Lotus, which belongs to the tribe Loteae, is distributed worldwide being able to adapt to different environmental stresses. It has been introduced into non-native areas by human activities (Escaray et al. 2012). Lotus corniculatus is a perennial legume used for pasture and silage production in many temperate countries (Grant and Small 1996). It has a natural distribution in Western Europe and North Africa (http://www.fao.org/Ag/agp/agpc/doc/Gbase/DATA/pf000344.htm) where it establishes a nitrogen-fixing symbiosis with rhizobia that have been scarcely studied despite of the value of L. corniculatus as fodder for animals (Ramírez-Restrepo and Barry 2005). The endosymbiont of L. corniculatus was initially named Rhizobium loti (Jarvis et al. 1982) but was later reclassified as Mesorhizobium loti (Jarvis et al. 1997). Recently it has been shown that the type strains of this species held in different culture collections represent three species, Mesorhizobium loti, Mesorhizobium erdmanii and Mesorhizobium jarvisii having phylogenetically divergent 16S rRNA and housekeeping genes (Martínez-Hidalgo et al. 2015).
Although there are only a few studies of the L. corniculatus endosymbionts, they show high phylogenetic diversity among strains isolated in Europe (Ampomah and Huss-Danell 2011; Lorite et al. 2010; De Meyer et al. 2011; Gossmann et al. 2012) and South America (Binde et al. 2009; Sotelo et al. 2011). They form different clusters within the genus Mesorhizobium. Several endosymbionts of L. corniculatus have also been isolated from a province of South Spain and found to comprise two different phylogenetic lineages clustering with Mesorhizobium albiziae and with Mesorhizobium alhagi, on the basis of the analyses of the core genes 16S rRNA, atpD and recA (Lorite et al. 2010). In Spain, L. corniculatus is widely distributed in many regions as part of the natural plant cover. Nevertheless, there are no studies about the endosymbionts of this legume in most Spanish soils.
Therefore, the aims of the present study were to examine the genetic diversity of strains isolated from L. corniculatus in Salamanca (Northwest Spain), using RAPD fingerprinting and phylogenetic analysis of their 16S rRNA, recA and atpD genes, and then to compare the results with those from L. corniculatus endosymbiont strains isolated in other geographic locations.
2 Materials and methods
2.1 Bacterial strains and nodulation experiments
The strains were isolated from 35 effective nodules (pink colour) of three L. corniculatus plants in flowering stage collected in an alfisol soil from Carbajosa de la Sagrada (a Mediterranean region from Salamanca, NW Spain, latitude 40° 55´59″N and 5° 39´05´´O, altitude 789 mamsl) using the standard method of Vincent (1970) on YMA plates at 28 °C. Re-infection experiments in L. corniculatus were performed using the previously published conditions (Robledo et al. 2008). Seeds were scarified with sulfuric acid for 12 min and then rinsed several times with sterile distilled water. After that, seeds were surface-disinfected with 2.5 % sodium hypochlorite for 10 min and then, rinsed five times with sterile distilled water. Seeds were germinated in 1 % (w/v) agar plates for 24 h in dark. One day-old seedlings were transferred to tubes and inoculated with 1 ml of a suspension containing approximately 1x108 UFC/ml. Tubes were placed in a growth chamber at 24 °C, 20 °C day-night cycle, 16 h photoperiod, and 60 % relative humidity. N-free and N-fed uninoculated treatments were performed in the same conditions. Roots were examined and the number of nodules was counted 30 days after inoculation.
2.2 DNA extraction and RAPD fingerprinting
Total genomic DNA from the isolates was extracted according to Rivas et al. (2001). Briefly, strains were grown for 48 h in TY medium (0.4 % tryptone, 0.3 % yeast extract and 0.09 % Ca2Cl) and cells were collected by centrifugation at room temperature in a microspin centrifuge at 5000 xg and then washed with 100 μl of an aqueous solution of 0.1 % (w/v) sarkosyl. The DNA was extracted with 100 μl of 0.05 M NaOH (DNA-free) heating at 100°C for 4 min. Samples were then placed in an ice bath and 900 μl of water was added to each microtube and mixed thoroughly. After an additional centrifugation at 5000 xg, 700 μl of the supernatants were harvested and frozen at −20 °C. The RAPD profiles were obtained as was previously described (Rivas et al. 2006) using the M13 primer (5′-CAGGGTGGCGGTTCT-3′) and an AmpliTaq reagent kit (Perkin-Elmer Biosystems, California, USA). PCR conditions were as follows: preheating at 95 °C for 9 min; 35 cycles of denaturing at 95 °C for 1 min; annealing at 45 °C for 1 min and extension at 75 °C for 2 min, and a final extension at 72 °C for 7 min. A total of 17 μl of each PCR amplification product were electrophoresed in 1.5 % agarose gel in TBE buffer (100 mM Tris, 83 mM boric acid, 1 mM EDTA, pH 8.5) during 2 h at 6 V/cm and stained with ethidium bromide (0.5 μg ml−1) and visualized using the Molecular Imager Chemidoc XRS System (Biorad, U.S.A). The RAPD patterns were obtained in the same electrophoretic conditions and using the same molecular weigth standard as size marker (Standard VI; Hofmann-La Roche, Switzerland).
2.3 Phylogenetic analyses
The amplification and sequencing of 16S rRNA gene were carried out according to Rivas et al. (2007) and atpD and recA gene sequences were obtained according to Gaunt et al. (2001). PCR amplifications were performed with a REDExtract-N-Amp™ PCR Kit (Sigma Co., USA) following the manufacturer’s instructions. Bands corresponding to the different genes were purified directly from the gel by room temperature centrifugation using a DNA gel extraction device (Millipore Co., USA) for 10 min at 5.000 xg, according to the manufacturer’s instructions. Sequencing reactions were performed on an ABI PRISM® 3100 sequencer using a BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems Inc., USA), as supplied by the manufacturer. The obtained sequences were compared with those from EzTaxon-e server (Kim et al. 2012). Sequences were aligned using the Clustal X software (Thompson et al. 1997). The distances were calculated according to Kimura’s two-parameter model (Kimura 1980). A phylogenetic tree was inferred using the neighbor-joining analysis (Saitou and Nei 1987). MEGA5 software (Tamura et al. 2011) was used for all analyses.
3 Results and discussion
3.1 Genetic biodiversity of strains
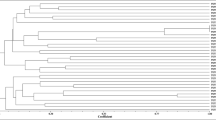
The 22 strains isolated from L. corniculatus nodules in Salamanca were able to induce nodules in this host (Table 1). Their genetic diversity was analysed by RAPD fingerprinting that allowed differentiation among strains from Mesorhizobium (Rivas et al. 2006; Armas-Capote et al. 2014). The strains isolated in this study displayed 10 different RAPD patterns (Fig. 1, Table 1) that showed genetic diversity allowing us to select a representative strain from each RAPD type for the analysis of the 16S rRNA gene.
RAPD profiles of strains isolated in this study CSLC01N (lane 1), CSLC03N (lane 2), CSLC06N (lane 3), CSLC07N (lane 4), CSLC09N (lane 5), CSLC14N (lane 6), CSLC15N (lane 7), CSLC17N (lane 8), CSLC18N (lane 9), CSLC19N (lane 10), CSLC22N (lane 11), CSLC24N (lane 12), CSLC28N (lane 13), CSLC30N (lane 14), CSLC31N (lane 15), CSLC35N (lane 16), CSLC36N (lane 17), CSLC37N (lane 18), CSLC38N (lane 19), CSLC42 N (lane 20), CSLC115N (lane 21), CSLC116N (lane 22). MW: molecular size markers with 2176, 1766, 1230, 1033, 653, 517, 453, 394, 298, 234 and 154 bp
3.2 Phylogenetic analysis of the 16S rRNA gene
The complete 16S rRNA gene sequences were obtained for the 10 strains representing the different RAPD types and compared with those from the EzTaxon-e database, which contains the type strains from all described species. The results obtained showed that the strains isolated in this study were closely related to different species of the genus Mesorhizobium with similarity values higher than 99 % in all cases (Table 1).
Considering the closeness of several Mesorhizobium species, in the phylogenetic analysis of the 16S rRNA gene we only included the strains isolated from L. corniculatus in other studies when the available sequences have more than 1200 nt. The strains isolated by De Meyer et al. (2011) from Belgium were not included because their sequences were too short. Some species that do not have identical 16S rRNA gene sequences, appear to be identical in the phylogenetic tree due to the lack of the initial nucleotides in the available 16S rRNA gene sequences. This is the case, for example in M. gobiense, M. metallidurans and M. tarimense, where the differences are located in the first 70 bp that are absent in the available 16S rRNA gene sequences. For this reason, differences observed after the analysis of the EzTaxon-e database, which performed pairwise analyses, are not in some cases observed in the phylogenetic trees.
The phylogenetic analysis of the 16S rRNA gene showed that the strains isolated in Northwest Spain clustered into four groups (fig. 2). The strains CSLC01N, CSLC28N, CSLC14N, CSLC36N and CSLC42N representing RAPD types I, II, IV, VIII and IX, respectively, belong to a wide cluster (cluster I) which includes the type strains of M. huakuii IAM 14158T, M. jarvisii ATCC 33669T, M. amorphae ACCT19665T, M. septentrionale SDW014T and M. waimense ICMP 19557T. The strain ATCC 33669T (before named M. loti) was also isolated from L. corniculatus nodules and it has been recently reclassified into a new species called M. jarvisii (Martínez-Hidalgo et al. 2015). Several strains isolated from L. corniculatus in European countries, for example Sweden (Ampomah and Huss-Danell 2011), Norway (Gossmann et al. 2012) and Spain (Lorite et al. 2010), and in American countries, such as Uruguay (Binde et al. 2009; Sotelo et al. 2011) and Brazil (Binde et al. 2009), belong to the same cluster. It should be noted that the strains SEMIA 848 and R6 isolated in Uruguay are incorrectly named as M. amorphae and M. loti, respectively. In the case of strain R6, this may be due to the fact that the strain ATCC 33669T was named M. loti for decades before its reclassification as M. jarvisii (Martínez-Hidalgo et al. 2015).
Neighbour-joining phylogenetic tree based on 16S rRNA gene sequences showing the position of strains isolated from L. corniculatus nodules in Northwest Spain and other geographical locations with respect to the type strains of the currently described species from genus Mesorhizobium. Bootstrap values calculated for 1000 replications are indicated. Bar, 1 nt substitution per 100 nt. The accession numbers for this gene in GenBank are: CSLC01N (KT899875), CSLC14N (KT899877), CSLC28N (KT899880), CSLC36N (KT899882), CSLC42N (KT899884), CSLC22N (KT899879), CSLC19N (KT899878), CSLC37N (KT899883), CSLC115N (KT899885) and CSLC30N (KT899881)
The strain CSLC22N representing RAPD pattern type V clustered with the type strain of M. erdmanii USDA 3471T (cluster III). This cluster also included strains isolated in Sweden (Ampomah and Huss-Danell 2011), Norway (Gossmann et al. 2012). Uruguay (Sotelo et al. 2011) and two strains nodulating L. corniculatus, R7A and MAFF 303099, isolated in New Zealand and whose genome has been sequenced (Kelly et al. 2014; Kaneko et al. 2000). The strains isolated in Sweden are also misnamed as M. loti because the strain USDA 3471T was considered to be the type strain of this species before its reclassification as M. erdmanii (Martínez-Hidalgo et al. 2015).
The strains CSLC19N, CSLC37N and CSLC115N, representing RAPD types III, VI and X, respectively, clustered with the type strains of M. caraganae CCBAU 11299T, M. gobiense CCBAU 83330T, M. metallidurans STM 2683T, M. tarimense CCBAU83306T and M. tianshanense A1BST (cluster IV). This cluster also contained strains nodulating L. corniculatus isolated in Sweden (Ampomah and Huss-Danell 2011), Northern Mexico (Qian and Parker 2002) and Uruguay (Sotelo et al. 2011).
Finally, the strain CSLC30N representing RAPD group VII belongs to the 16S rRNA cluster VII, which included the type strain of M. loti NZP 2213T, M. ciceri UPMCa7T, M. sangaii SCAU27T, M. qinshensii CCBAU 33460T, M. australicum WSM2073T, M. shangrilense CCBAU65327T and M. cantuariense ICMP 19515T. This cluster contains several strains isolated in Sweden, most of them incorrectly named M. loti (Ampomah and Huss-Danell 2011). This also includes two strains isolated in Norway (Gossmann et al. 2012) and two strains isolated in New Zealand, R88b and CJ3sym, whose genomes have been sequenced (Reeve et al. 2014).
Only cluster I contains strains isolated from both Northwest and South Spain (Lorite et al. 2010). The other strains isolated in South Spain belong to a cluster including the type strains of M. alhagi CCNWXJ12-2T and M. camelthorni CCNWXJ40-4T (cluster VI) and to an independent cluster formed by two strains only isolated in South Spain (cluster V).
The strain N3 isolated in Uruguay (Sotelo et al. 2011) formed an independent lineage related to the cluster II, which also includes several type strains of Mesorhizobium species nodulating legumes other than Lotus. The cluster III contains the strain CSLC22N isolated in this study and several strains isolated from L. corniculatus nodules in Sweden (Ampomah and Huss-Danell 2011), the strain N33 in Uruguay (Sotelo et al. 2011) and the strains MAFF303099 and R7A isolated in New Zealand (Kaneko et al. 2000; Kelly et al. 2014). Moreover, this cluster includes the type strain M. erdmanii USDA 3471T isolated from L. corniculatus nodules.
In summary, cluster I is the most widely distributed group due to its presence in Europe (South and Northwest Spain, Sweden and Norway), America (Brazil and Uruguay) and Asia (New Zealand), followed by the cluster III present in Europe (Northwest Spain and Sweden), America (Uruguay) and Asia (New Zealand). After this comes cluster IV, present in Europe (Northwest Spain and Sweden) and America (Northern Mexico and Uruguay) and cluster VII, present in Europe (Northwest Spain, Sweden and Norway) and Asia (New Zealand).
It is remarkable that some L. corniculatus strains are phylogenetically related with the type strains of the three Mesorhizobium species which are endosymbionts of this legume. These are now classified as M. loti, M. erdmanii and M. jarvisii, but were initially considered to be the same species. This highlights the need of a correct classification and naming of the type strains held in different culture collections in order to ensure that the type strains of a species is the same in all of them. This will avoid erroneous conclusions based on comparisons with type strains that are different but have been assigned to the same species.
3.3 Phylogenetic analysis of the recA and atpD genes
The 16S rRNA genes of several species from genus Mesorhizobium are very closely related as was showed above. However, they can be differentiated by their housekeeping genes, such as recA and atpD that are available for most of Mesorhizobium species, which allow the identification of new isolates. The recA gene has been analysed in the strains isolated from L. corniculatus in Belgium (De Meyer et al. 2011) and South Spain (Lorite et al. 2010) and the atpD gene was analysed in strains isolated in Spain and in Uruguay (Sotelo et al. 2011). The analysis of these two genes in our strains allowed us to know their identities and to analyse their phylogentic relationships with other L. corniculatus endosymbionts.
The analysis of recA gene showed that our strains belong to three different clusters within the genus Mesorhizobium (fig. 3). The strains from the 16S rRNA gene cluster I do not have the same recA gene sequences, with the exception of strains CSLC01N, CSLC28N and CSLC36N, but all of them formed a cluster including the type strains of the two species closely related in the 16S rRNA gene analysis, M. jarvisii ATCC 33669T, isolated from L. corniculatus nodules, and M. huakuii IAM 14158T. The strains CSLC01N, CSLC28N and CSLC36N with recA genes identical to those of M. jarvisii ATCC 33669T and the strain CSLC42N with 99.2 % similarity in this gene, probably belong to M. jarvisii. However, the strain CSLC14N probably do not belong to this species since it has 96.5 % similarity with respect to M. jarvisii ATCC 33669T (Table 1). This recA cluster also contains the strain CSLC22N belonging to the 16S rRNA gene cluster III, showing 98.2 % similarity with respect to M. jarvisii ATCC 33669T. Since higher similarity values are presented by other species of the genus Mesorhizobium, such as M. huakuii and M. qingsenghii, we cannot assign the strain CSLC22 to the species M. jarvisii. This cluster also contains four strains that are phylogenetically divergent to our strains and to M. jarvisii ATCC 33669T. These four strains, MAFF303099, R7A, R88b and CJ3sym, nodulate L. corniculatus and were isolated in New Zealand (Kaneko et al. 2000; Kelly et al. 2014; Reeve et al. 2014). One L. corniculatus strain isolated in Belgium (De Meyer et al. 2011) is also related to this cluster but it was phylogenetically divergent to our strains. Since this strain has less than 96 % similarity with the type strains of M. jarvisii ATCC 33669T and M. huakuii IAM 14158T, it probably does not belong to these species nor to other species in this cluster, such as M. australicum and M. qingshensii.
Neighbour-joining phylogenetic tree based on partial recA gene sequences showing the position of strains isolated from L. corniculatus nodules in Northwest Spain and other geographical locations with respect to the type strains of the currently described species from genus Mesorhizobium. Bootstrap values calculated for 1000 replications are indicated. Bar, 1 nt substitution per 100 nt
The strains from the 16S rRNA gene cluster IV, CSLC19N, CSLC37N and CSLC115N, have identical recA gene sequences and occupy a phylogenetically divergent branch within the cluster that contains the type strains of M. caraganae and M. metallidurans. They were closely related in the 16S rRNA gene analysis, but this recA cluster also contains M. amorphae, M. septentrionale and M. waimense. The closest related species to the three strains from cluster IV is M. metallidurans with 96.4 % similarity, suggesting that they do not belong to this species (Table 1). This cluster includes several strains isolated from L. corniculatus in Belgium (De Meyer et al. 2011), but they are phylogenetically divergent from eachother and from our strains.
The strain CSLC30N from 16S rRNA gene cluster VII formed a recA gene branch belonging to a cluster containing its closest species in 16S rRNA gene analysis, M. loti, isolated from L. corniculatus nodules, M. ciceri and M. cantuariense, being the type strain of M. ciceri USDA 3383T the most closely related with 98.3 % similarity (Table 1). This cluster does not contain other isolates of L. corniculatus. We do not know if the strains from this host isolated in Norway (Gossmann et al. 2012) and Sweden (Ampomah and Huss-Danell 2011), which belonged to the same cluster as the strain CSLC30N in the 16S rRNA gene analysis, belong to this recA gene cluster since this gene is not available in databases for these strains.
The analysis of the atpD gene showed that our strains are divided into four clusters with some differences in their distribution with respect to that found after recA gene analysis (fig. 4). The strains from 16S rRNA gene cluster I, have different atpD gene sequences being particularly divergent in the atpD gene of the strain CSLC14N. This strain formed an independent lineage related to the type strain of M. caraganae CCBAU 11299T with 92.5 % similarity. The remaining strains were phylogenetically related to the type strains of two species nodulating L. corniculatus, M. jarvisii ATCC 33669T and M. erdmanii USDA 3471T. The phylogenetic lineage formed by the strain CSLC14N belongs to a cluster that also included two strains, S1302 and S789 isolated from L. corniculatus nodules in Uruguay (Sotelo et al. 2011). The strains CSLC01N, CSLC36N and CSLC28N were equidistant between M. jarvisii ATCC 33669T and M. erdmanii USDA 3471T (similarity higher than 98.5 % in all cases) (Table 1). This makes difficult the identification of these strains as M. jarvisii, as suggested the recA gene analysis. The strain CSLC42N was also related to the type strain of M. jarvisii ATCC 33669T with 98.3 % similarity, but in this case M. erdmanii USDA 3471T was less closely related, with 96.7 % similarity (Table 1). This atpD gene cluster, also contains the strain CSLC22N from 16S rRNA gene cluster III, which present 97.8 % similarity with respect to its closest relative M. jarvisii ATCC 33669T (Table 1). The L. corniculatus nodulating strains R7A and MAFF303099 isolated in New Zealand (Kaneko et al. 2000; Kelly et al. 2014) and the strain N105 isolated in Uruguay (Sotelo et al. 2011) also belong to this cluster. However, the strains R88b and CJ3sym, isolated from L. corniculatus nodules in New Zealand, formed two different lineages that are phylogenetically divergent.
Neighbour-joining phylogenetic tree based on partial atpD gene sequences showing the position of strains isolated from L. corniculatus nodules in Northwest Spain and other geographical locations with respect to the type strains of the currently described species from genus Mesorhizobium. Bootstrap values calculated for 1000 replications are indicated. Bar, 2 nt substitution per 100 nt
The strains from the 16S rRNA cluster IV, CSLC19N, CSLC37N and CSLC115N, have identical atpD gene sequences. They occupied a phylogenetically divergent branch within a cluster that also contains the type strain of the species closest to these strains in the 16S rRNA gene analysis, i.e. M. metallidurans STM2683T. This divergent branch does not include M. caraganae CCBAU11299T or those of the species related to our strains in the analysis of the recA gene. The similarity with respect to the closest type strain of M. metallidurans STM 2683T was 97.4 % (Table 1), which is a similarity value found for other Mesorhizobium species such as M. ciceri and M. loti or M. shangrilense and M. qingshengii. Therefore, in agreement with the results of the recA gene, the strains from the cluster IV probably belong to a new species of the genus Mesorhizobium. Related to this cluster is the strain N362 isolated in Uruguay (Sotelo et al. 2011) which belongs to a cluster containing the type strains of M. septentrionale and M. amorphae.
The strain CSLC30N from 16S rRNA gene cluster VII represents an atpD gene phylogenetic lineage clustering with the same strains than in 16S rRNA and recA clusters which were M. sangaii, M. ciceri, M. loti, M.shangrilense and M. qingshensii. From them, the closest related species to the strain CSLC30N is M. loti with 95.4 % similarity whereas in the recA gene analysis, the most closely related species was M. ciceri. As occurred in the case of recA gene, the atpD gene is not available for the strains isolated in Norway and Sweden, which belong to the same cluster as the strain CSLC30N in the 16S rRNA gene analysis, and so we are unsure if these strains belong to the same atpD gene cluster.
In conclusion, the results of the analysis of the recA and atpD genes revealed that some strains isolated in Belgium, Uruguay and New Zealand belong to clusters that also contain strains isolated in Northwest Spain. Nevertheless, the strains isolated in South Spain from L. corniculatus nodules, in agreement with the results found after 16S rRNA gene analysis, were found to belong to two different clusters that are phylogenetic divergent. The results of the 16S rRNA, recA and atpD gene analyses showed great phylogenetic diversity in strains nodulating L. corniculatus in different continents and countries. However, the endosymbionts of L. corniculatus have been analysed from rather few geographical locations and further studies of rhizobial strains nodulating this legume are needed to increase knowledge of this symbiosis.
References
Ampomah OY, Huss-Danell K (2011) Genetic diversity of root nodule bacteria nodulating Lotus corniculatus and Anthyllis vulneraria in Sweden. Syst Appl Microbiol 34:267–275
Armas-Capote N, Pérez-Yépez J, Martínez-Hidalgo P, Garzón-Machado V, del Arco-Aguilar M, Velázquez E, León-Barrios M (2014) Core and symbiotic genes reveal nine Mesorhizobium genospecies and three symbiotic lineages among the rhizobia nodulating Cicer canariense in its natural habitat (La palma, Canary Islands). Syst Appl Microbiol 37:140–148
Binde DR, Menna P, Bangel EV, Barcellos FG, Hungria M (2009) Rep-PCR fingerprinting and taxonomy based on the sequencing of the 16S rRNA gene of 54 elite commercial rhizobial strains. Appl Microbiol Biotechnol 83:897–908
De Meyer SE, Van Hoordea K, Vekeman B, Braeckmana T, Willems A (2011) Genetic diversity of rhizobia associated with indigenous legumes in different regions of Flanders (Belgium). Soil Biol Biochem 43:2384–2396
Escaray FJ, Menéndez AB, Garriz A, Pieckenstain FL, Estrella MJ, Castagno LN, Carrasco P, Sanjuán J, Ruiz OA (2012) Ecological and agronomic importance of the plant genus Lotus. Its application in grassland sustainability and the amelioration of constrained and contaminated soils. Plant Sci 182:121–133
Gaunt MW, Turner SL, Rigottier-Gois L, Lloyd-Macgilp SA, Young JWP (2001) Phylogenies of atpD and recA support the small subunit rRNA-based classification of rhizobia. Int J Syst Evol Microbiol 51:2037–2048
Gossmann JA, Markmann K, Brachmann A, Rose LR, Parniske M (2012) Polymorphic infection and organogenesis patterns induced by a Rhizobium leguminosarum isolate from Lotus root nodules are determined by the host genotype. New Phytol 196:561–573
Grant WF, Small E (1996) The origin of the Lotus corniculatus (fabaceae) complex: a synthesis of diverse evidence. Can J Bot 74:975–989
Jarvis BDW, Pankhurst CE, Patel JJ (1982) Rhizobium loti, a new species of legume root nodule bacteria. Int J Syst Bacteriol 32:378–380
Jarvis BDW, van Berkum P, Chen WX, Nour SM, Fernandez MP, Cleyet-Marel JC, Gillis M (1997) Transfer of Rhizobium loti, Rhizobium huakuii, Rhizobium ciceri, Rhizobium mediterraneum, and Rhizobium tianshanense to Mesorhizobium gen. nov. Int J Syst Bacteriol 47:895–898
Kaneko T, Nakamura Y, Sato S, Asamizu E, Kato T, Sasamoto S, Watanabe A, Idesawa K, Ishikawa A, Kawashima K, Kimura T, Kishida Y, Kiyokawa C, Kohara M, Matsumoto M, Matsuno A, Mochizuki Y, Nakayama S, Nakazaki N, Shimpo S, Sugimoto M, Takeuchi C, Yamada M, Tabata S (2000) Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res 7:331–338
Kelly S, Sullivan J, Ronson C, Tian R, Bräu L, Munk C, Goodwin L, Han C, Woyke T, Reddy T, Huntemann M, Pati A, Mavromatis K, Markowitz V, Ivanova N, Kyrpides N, Reeve W (2014) Genome sequence of the Lotus spp. microsymbiont Mesorhizobium loti strain R7A. Stand Genomic Sci 9:6
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Lorite MJ, Muñoz S, Olivares J, Soto MJ, Sanjuán J (2010) Characterization of strains unlike Mesorhizobium loti that nodulate Lotus spp. in saline soils of Granada, Spain. Appl Environ Microbiol 76:4019–4026
Martínez-Hidalgo P, Ramírez-Bahena MH, Flores-Félix JD, Rivas R, Igual JM, Mateos PF, Martínez-Molina E, León-Barrios M, Peix A, Velázquez E (2015) Revision of the taxonomic status of type strains of Mesorhizobium loti and reclassification of strain USDA 3471T as Mesorhizobium erdmanii sp. nov. and ATCC 33669T Mesorhizobium jarvisii sp. nov. Int J Syst Evol Microbiol 65:1703–1708
Qian J, Parker MA (2002) Contrasting nifD and ribosomal gene relationships among Mesorhizobium from Lotus oroboides in northern Mexico. Syst Appl Microbiol 25:68–73
Ramírez-Restrepo CA, Barry TN (2005) Alternative temperate forages containing secondary compounds for improving sustainable productivity in grazing ruminants. Anim Feed Sci Technol 120:179–201
Reeve W, Sullivan J, Ronson C, Tian R, Bräu L, Davenport K, Goodwin L, Chain P, Woyke T, Lobos E, Huntemann M, Pati A, Mavromatis K, Markowitz V, Ivanova N, Kyrpides N (2014) Genome sequence of the Lotus corniculatus microsymbiont Mesorhizobium loti strain R88B. Stand Genomic Sci 9:3
Rivas R, Velázquez E, Valverde A, Mateos PF, Martínez‐Molina E (2001) A two primers random amplified polymorphic DNA procedure to obtain polymerase chain reaction fingerprints of bacterial species. Electrophoresis 22:1086–1089
Rivas R, Peix A, Mateos PF, Trujillo ME, Martínez-Molina E, Velázquez E (2006) Biodiversity of populations of phosphate solubilizing rhizobia that nodulates chickpea in different Spanish soils. Plant Soil 287:23–33
Rivas R, García-Fraile P, Mateos PF, Martínez-Molina E, Velázquez E (2007) Characterization of xylanolytic bacteria present in the bract phyllosphere of the date palm Phoenix dactylifera. Lett Appl Microbiol 44:181–187
Robledo M, Jiménez-Zurdo JI, Velázquez E, Trujillo ME, Zurdo-Piñeiro JL, Ramírez-Bahena MH, Ramos B, Díaz-Mínguez JM, Dazzo F, Martínez-Molina E, Mateos PF (2008) Rhizobium cellulase CelC2 is essential for primary symbiotic infection of legume host roots. PNAS, USA 105:7064–7069
Saitou N, Nei M (1987) A neighbour-joining method: a new method for reconstructing phylogenetics trees. Mol Biol Evol 4:406–425
Sotelo M, Irisarri P, Lorite MJ, Casaretto E, Rebuffo M, Sanjuán J, Monza J (2011) Diversity of rhizobia nodulating Lotus corniculatus grown in northern and southern regions of Uruguay. Appl Soil Ecol 49:197–207
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The clustal_X windows interface: flexible strategies for multiple sequence alignement aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Vincent JM (1970) The cultivation, isolation and maintenance of rhizobia. In: Vincent JM (ed) A manual for the practical study of root-nodule. Blackwell Scientific, Oxford, pp. 1–13
Acknowledgments
This work was funded by the “Junta de Castilla y León” (Regional Government, Grant SA169U14. MM thanks for PhD fellowship from the “Miguel Casado San José” foundation (Spain) and XCG acknowledges a research grant from the “Kinesis” foundation (Puerto Rico, US).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Marcos-García, M., Menéndez, E., Cruz-González, X. et al. The high diversity of Lotus corniculatus endosymbionts in soils of northwest Spain. Symbiosis 67, 11–20 (2015). https://doi.org/10.1007/s13199-015-0368-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13199-015-0368-5