Abstract
The driver of plant community assembly in riparian marshes is vital but still unclear. In this study, we report the results of sixty plots located along the downstream Yellow River to identify the driver of plant community assemblages. Seven functional traits were measured and compiled to calculate functional beta diversity, and phylogenetic tree was constructed to assess phylogenetic dissimilarity. The analyses show that all of traits had weak phylogenetic signals using the Blomberg’s K statistic. The phylogenetic and functional beta diversity present clustering patterns, with the exception of leaf nitrogen concentration and leaf phosphorus concentration; and the standardized effect sizes based on multi-traits, maximum height, leaf dry matter content are well correlated with the environmental gradient. The findings imply that environment filtering was responsible for the variation of beta diversity, and drove community assembly in the riparian marsh. Of all environmental factors, soil salinity is the most crucial indicator to dominate the pattern of beta diversity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Community assembly studies are imperative in explaining the formation and maintenance of biodiversity, which have been continually pursued in the field of ecology (Rosindell et al. 2011). Two major theories have been proposed to elucidate the mechanism of species assemblages, the niche theory based on deterministic processes (Diamond 1975) and the neutral theory based on stochastic processes (Hubbell 2001). As the relative importance of ecological processes varies largely with different plant communities, however, the quantification and assessment of individual assembly processes are of vital importance.
The original explication of assembly mechanisms is often based on taxonomic identity. But it has proven to be poorly responsive to the process of species assemblage though it was a reasonable starting point (Siefert et al. 2013; Jiang et al. 2018). An alternative approach, using phylogenetic and functional diversity to test which deterministic or stochastic processes drive the assembly of species, is widely applied and offers another justification of testing ecological processes in different communities (Cornwell and Ackerly 2009; Purschke et al. 2013). For instance, the lower functional beta diversity occurred in a scale of less than 600 km and the higher functional beta diversity was shown in a scale of more than 1800 km in the eastern North American forest community (Siefert et al. 2013). In the grassland regions of China, community assembly processes are remarkably driven by environmental filtering using phylogenetic relatedness among species (Chi et al. 2014). Such observations highlight that beta diversity, reflecting the dissimilarity of phylogenetic and functional components between communities, is an effective tool for ecologists to analyze and interpret the mechanisms of community formation and maintenance (Anderson et al. 2011).
Nonetheless, the mechanisms of community assembly are still a hotly debatable topic. Under niche-based processes, environmental filtering and the species interactions are considered as the most fundamental drivers of community assembly (Cornwell and Ackerly 2009; López-Angulo et al. 2018). Environmental filtering processes are shown to sort the potential species from a regional species pool to a suitable habitat (Diamond 1975). A common view is shared that there is a potential relationship between phylogenetic or functional diversity and environmental gradients (Spasojevic and Suding 2012; Satdichanh et al. 2015; Cadotte and Tucker 2017). In particular, environmental filtering is frequently determined as the driving force of community assemblage processes along an extensive environmental gradient (Swenson et al. 2012a; Cadotte and Tucker 2017; López-Angulo et al. 2018). Conversely, the definition and inappropriate use of environmental filtering have been questioned in recent researches (Cavender-Bares et al. 2009; Maire et al. 2012; Kraft et al. 2015). Phylogeny and trait convergence are no longer merely the result of environmental filtering processes, as biotic interactions and dispersal processes may lead to the same outcomes (Mayfield and Levine 2010; Kraft et al. 2015).
It is well recognized that environmental filtering as a deterministic pattern of community assemblages, inevitably leads to a decreasing range of traits value and increasing niche overlapping (Weiher and Keddy 1995; Cornwell and Ackerly 2009). From the perspective of phylogeny and traits, environmental filtering leads to a tighter correlation of coexisting species or more similarity of specific traits, i.e., phylogenetic clustering or trait convergence. Determining a clustered pattern of community dispersion, therefore, is the first step in detecting environmental filtering processes. But such determination alone is not comprehensive, since certain species with higher intrinsic growth rate having a fitness advantage over those with lower intrinsic growth rate shall bring about a competitive exclusion which is similar with the consequence drove by environmental filtering. Moreover, the modern coexistence theory argues that the differences between population growth rate and niche overlap are vital indicators inferring species coexistence and persistence pattern (Chesson 2000; Cadotte and Tucker 2017). It is understood that species may have different growth rates in the optimal and suboptimal habitats as there is variation of fitness along environmental gradient. So it is observable a pattern that the population growth rate is continuously varying along the environmental gradient (Cadotte and Tucker 2017). It is thus crucial to ensure that the observed relationships between the growth rates and phylogenetic and functional similarity are revealed by the environment and is not the result of other processes, such as plastic traits. In consequence, confirming the existence of such non-independence in traits requires to quantify the phylogenetic signals to acquire a powerful piece of evidence in the trait data to combine phylogeny, traits, and environment.
In this work, we conduct studies concerning the high environmental pressures in the riparian marsh along the downstream of Yellow River (high salinity and low nutrients in soil). We postulate that environmental filtering is the driver of community assemblages in the study area, and adopt the guidelines proposed by Cadotte and Tucker (2017) to confirm the leading role of environmental filtering with community phylogenetic and functional beta diversity. Based on the hypothesis, a variation partitioning approach is applied to distinguish edaphic, climatic, and spatial variables to explicate the variation of beta diversity. From previous studies, the variation partitioning approach of trait-based response variables proves to advance our understanding of deterministic ecological processes (Legendre et al. 2009; Siefert et al. 2013; Jiang et al. 2018). Furthermore, we endeavor to find out key environmental factors that drive the variation of phylogenetic and functional beta diversity separately in the riparian marsh of Yellow River. We focus on three issues: (1) Are there any significant phylogenetic signals in functional trait data? (2) Does environmental filtering drive the community assembly processes along the environmental gradient? (3) What factor is the most crucial in phylogenetic and functional beta diversity in the riparian marsh of Yellow River?
Materials and Methods
Study Site
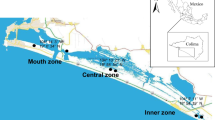
The study area is located in the central and eastern China (34°56'–37°45'N; 113°29'–119°09'E). The mean annual temperature is 16.3 °C throughout the study area, with January and July being the coldest (−1.6 °C) and warmest (28.2 °C) months, respectively. The mean annual precipitation is about 650 mm, with a peak during the summer and a dip between December and February. A large amount of sediment is deposited and formed sandbars resulting from the decrease of river’s velocity, providing habitats for wetland species within the channel downstream Yellow River.
Field Sampling
60 plots (10 m × 10 m) were established along 786 km of the Yellow River channel (Fig. 1), from the Peach Valley to the estuary, from August to September 2016. Within each plot, we set up five 1 m2 quadrats to investigate plant communities using random sampling methods. We recorded the species name and coverage of the species occurring in each quadrat. The plant communities were mainly composed of herbs and immature shrubs. We sorted out the species covered at least 80% of the plant community per plot and included a sufficient range of traits (Pakeman and Quested 2007; Raevel et al. 2012). Finally, 48 species were selected for measurement of functional traits representing a regional species pool. Seven functional traits, quantitatively describing certain independent axes of a species niche, were measured and compiled: maximum height (Hmax, cm), leaf thickness (LT, mm), leaf dry matter content (LDMC, g/g), specific leaf area (SLA, cm2/g), seed mass (SM, g), leaf nitrogen concentration (LNC, %), and leaf phosphorus concentration (LPC, %) (Table 1). We measured plant traits on 10–20 individuals of each species per plot following a standard protocol (Cornelissen et al. 2003) with the exception of LT for which another method (Seelig et al. 2012) was followed (See supplementary material S1). Subsequently, we created a species × traits matrix with mean trait value and generated trait dendrograms by the R software performing a hierarchical clustering to implement the same structure as phylogeny (Swenson et al. 2012b; Yang et al. 2014) with multi-traits and univariate trait, respectively. The trait data were log1p-transformed prior to the construction of the trait dendrograms.
Environmental Data
To characterize the spatial distribution of the plots in the study area, we obtained the geo-location information in the form of latitude and longitude by GPS. It was not found that there is significant spatial autocorrelation in species distributional data analyzed by the SAM software (Dormann et al. 2007; Qin et al. 2017). One soil sample taken at a depth of 0–20 cm was collected in each quadrat using a 5 cm diameter soil auger, stored and sealed in a plastic bag, and taken back to the laboratory to determine its physical and chemical properties. Five soil samples in total from a plot were composited to measure nine different soil properties: soil moisture content (SMC, g/g), pH, soil salinity (SS, mg/L), total nitrogen (TN, %), total phosphorus (TP, %), total carbon (TC, %), available nitrogen (AN, %), extractable phosphorus (EP, %), and effective cation exchange capacity (CEC, cmol/kg) with three replicates. The parameters were obtained in the following way, SMC as the oven-dry mass of a soil sample divided by its fresh mass, pH measured using a pH meter (FE28, Mettler-Toledo Company, Switzerland), and SS calculated by the conductivity in soil measured by a conductivity meter (FE38, Mettler-Toledo Company, Switzerland). While the Kiadhal digestion method and the ammonium acetate method were used to measure TN and CEC in soil via automatic azotometer (9860, Hanon Company, China), respectively, TP was assessed by the Mo-Sb colorimetric method using a spectrophotometer (UV-9100, Unico Company, China) and TC by a carbon sulfur analyzer (CS2800, NCS Testing Technology Company, China). AN and EP were determined, separately, by a diffuser and ‘Avio 200 ICP-OES’ (PerkinElmer Avio Company, USA). In addition, the climate data of study area with 1 km spatial resolution was obtained from the WorldClim2 database (Fick and Hijmans 2017). We extracted climatic variables to characterize the climate conditions for each plot from the WorldClim2 database: annual mean temperature (AMT, °C), mean temperature of coldest quarter (MTC, °C), mean temperature of warmest quarter (MTW, °C), annual precipitation (AP, mm), precipitation of warmest quarter (PW, mm), precipitation of coldest quarter (PC, mm), temperature seasonality (TS), and precipitation seasonality (PS). Besides, we calculated annual biotemperature (AB, °C), warmth index (WI, °C), and coldness index (CI, °C) using monthly mean temperature data at the same spatial resolution (Fang et al. 2011) (refer to supplementary material S2 for detailed information).
Phylogenetic Tree
In order to infer the pattern of community assemblages in the riparian marsh of the Yellow River, we explicated the causes for species coexistence through species evolution. The classification information of all 48 species was retrieved and input into the Phylomatic software (http://www.phylodiversity.net/phylomatic), generating the megatrees according to Zanne et al. (2014) to identify the distribution of species on an existing phylogenetic framework. Followed was the construction of a species-level phylogenetic tree based on the APG III system.
Data Analysis
Following the guidelines of Cadotte and Tucker (2017), an appropriate approach is to quantify the phylogenetic and functional structures of the community via the standardized effect size. We first calculated the beta diversity (Dnn′) and standardized effect size (S.E.S. Dnn′) on the basis of phylogeny, multi-traits and all univariate traits. Then, we implemented an appropriate null model and randomized shuffled taxa names across the tips of the phylogenetic tree and trait dendrograms 999 times, while maintaining species occurrence frequency and abundance. This approach has been proven to have the best ability to discover filtering based on multivariate functional traits (Spasojevic and Suding 2012). Next, we used the Student’s t test to check whether the phylogenetic clustering/overdispersion or trait convergence/divergence is significant with the standardized effect size. The calculation was based on the equation below,
where Dnn′(obs) is the observed value of the weighted nearest neighbor distance between communities, \( Dnn{\prime}_{\left(\overline{\mathrm{null}}\right)} \) is the mean of the weighted nearest neighbor distance by the null model, and sd(null) is the standard deviation of the weighted nearest neighbor distance by the null model.
As the influence of the environment on species coexistence is both relevant in extraction of a species from the species pool to the habitat and in the competition process by covariation with the population growth rate and environment, we focused on the variation of population growth rate along environmental gradients, which is associated to community structure (Cadotte and Tucker 2017). Simultaneously, the principal component analysis was utilized to reduce redundant variables of edaphic and climatic variables. We defined the scores of the first principal component (37.4% of variance) as the environmental gradient of the study area (supplementary material S3), and observed the correlation of S.E.S. Dnn′ and the environmental gradient by linear regressions. Since it is necessary to explore whether the evolution of traits conform to the principle of phylogenetic conservatism, we searched the phylogenetic signals in every univariate trait using the K statistic (Blomberg et al. 2003). The three cases of the K value is distinguished, (1) K = 0 suggests that there are no phylogenetic signals in trait evolution; (2) K < 1 indicates that weaker similarities exist in closely related species than expected based on the Brownian motion model; and (3) K > 1 indicates more phylogenetic signals than expected (Blomberg et al. 2003). All of the calculations and tests were carried out with R package vegan, phytools, and picante (R Core Team 2017). Should the values of S.E.S. Dnn′ be negative and correlated significantly with the environmental gradient simultaneously, significant phylogenetic signals appeared in the univariate trait data. If it is the case, we may conclude that environmental filtering drives the community assembly processes.
In order to explore the relationship of beta diversity with environmental and spatial variables in depth, we used the method of variation partitioning over edaphic, climatic, and spatial variables as explanatory variables to dissect the beta diversity as the response variables (Siefert et al. 2013; Spasojevic et al. 2016; Jiang et al. 2018). For the spatial variables, we applied the principal coordinates of neighbor matrices eigenvectors, which are recurrently implemented as a tool for quantifying stochastic processes (Borcard and Legendre 2002) in terms of centralized geographic information data. In the present study, we extracted 20 positive eigenvalue variables as a proxy for total spatial variables. For the environmental variables, we used 9 edaphic variables and 11 climatic variables to represent the environmental conditions of the study area. Prior to variation partitioning, the redundancy analysis (Borcard et al. 1992) and the forward selection (Blanchet et al. 2008) were executed to reduce redundant components and retain variables which were significantly associated with beta diversity in edaphic, climatic, and spatial variables, respectively. All the calculations and analyses were accomplished by the software R (R Core Team 2017).
Results
Dispersions of Phylogenetic and Functional Beta Diversity
The standardized effect sizes of phylogeny and multi-traits were significantly lower than zero (P < 0.05) (Table 2). For univariate traits, all traits passed a significant test except for LPC, which had no significant difference from zero. Of interest, we found that LNC had a particular pattern in contrast to other traits, which was significantly greater than zero.
Standardized Effect Sizes of Beta Diversity (S.E.S. Dnn′) and Environmental Gradient
The functional beta diversity of multi-traits (Fig. 2b) showed a significant negative relationship with a continuous environmental gradient (P < 0.001, R2 = 0.209). In comparison, the relationship between phylogeny and the environment gradient was nonlinear (Fig. 2b). For individual traits, only the maximum height (P < 0.05, R2 = 0.086) (Fig. 2c) and leaf dry matter content (P < 0.001, R2 = 0.337) (Fig. 2e) showed a significant correlation. Remarkably, the maximum height increased with the environmental gradient, whereas the opposite trend was apparent for the multi-traits and leaf dry matter content.
Phylogenetic Signals
The K values of seven functional traits were less than one (Table 3), ranging from 0.129 (LPC) to 0.295 (SM), in which a mean weaker phylogenetic signal than expected was obtained during the process of trait evolution. The P values of the permutation test for univariate traits were all less than 0.05, indicating that all of the selected traits had significant phylogenetic signals.
Variance Partitioning of Phylogenetic and Functional Beta Diversity
We applied the forward selection and redundancy analysis preceding variance partitioning for explanatory variables (climatic, spatial and edaphic variables), which effectively reduces the collinearity between variables. We found that all the facets of beta diversity were associated with SS in edaphic variables (Table 4). The correlations were ubiquitous in the forward selection results, and even the most critical, shed light on most of the variance in each facet of beta diversity. The variables sets of phylogenetic beta diversity were consistent with that of multi-trait based beta diversity (Table 4). For individual traits, SS was a widespread and robust factor for all traits in the selected sets, accounting for the overall variance of the edaphic variables. In the sets of LT (R2 = 0.040) and LDMC (R2 = 0.034) and LPC (R2 = 0.028), CEC could be responsible for a little fraction of variance (Table 4). AN (R2 = 0.014) explained tiny fraction of LT and TN (R2 = 0.032) had little contribution to LPC (Table 4). Regarding the climatic variables, we found that TS represented the climatic variables in SLA (R2 = 0.324) and LNC (R2 = 0.352), whereas WI is an important climatic factor in LDMC (R2 = 0.413) and LPC (R2 = 0.534), CI (R2 = 0.354) is highly associated with LT. Because the spatial variables did not pass the test, all of them were utilized in variance partitioning.
We employed the approach of variance partitioning using selected variable sets. The results of phylogenetic beta diversity were similar to that of multi-traits functional beta diversity (Fig. 3). The sole edaphic variable fraction could explain most of the variance in phylogenetic (0.575) and multi-traits functional (0.488) beta diversity. But it had no significant correlation with phylogenetic and multi-traits functional beta diversity. For single traits (Fig. 3), the sole edaphic variables indicated the best explanatory power for all facets of functional diversity. The climatic variables had minute contribution to the justification of the univariate functional diversity. Moreover, the sole spatial variables were not associated to each functional diversity.
Discussion
In the present study we observed three lines of evidences that could be in the support of a community assembly process driven by environmental filtering, and then expounded key factors affecting the phylogenetic and functional turnover in the riparian marsh of the Yellow River. First, we found that the general phylogenetic and functional clustering patterns, apart from LNC and LPC, pointed to the fact that deterministic assembly processes played a core role of structuring a community. Second, a pertinent mechanism between the growth rate and environmental gradient was set up when the deterministic assembly process was taken into account. Third, we observed the presence of significant phylogenetic signals in the trait data. In consequence, we could come to the conclusion that the environment filtering drove the community assembly processes in the study area. Of the environmental factors, we found that the soil salinity in the edaphic variables was the most crucial factor in environmental filtering, leading to form the pattern of phylogenetic and functional dissimilarity between communities.
Assembly Processes Drove by Environmental Filtering
As shown in the negative S.E.S. Dnn′ values of phylogeny (Table 2) and multi-traits, the general phylogenetic and functional clustering were observed in the riparian marsh of the Yellow River, signifying that phylogenetic and multivariate functional turnover were driven by deterministic assembly processes. Still, the pattern of phylogenetic turnover was in accordance, though not perfectly aligned, with that of multi-traits functional turnover. The findings could be reasonably interpreted as a result of evolutionary lability of functional traits due to the phylogenetic signals in the trait data, as similar results reported in other studies (Swenson 2011; Yang et al. 2014). Compared to whole-plant traits and leaf morphological traits, our results of leaf nutrient concentrations showed inconsistent patterns, while LNC and LPC had divergence and random patterns, respectively. The underlying reason could be the relative high intraspecific trait variation in leaf nutrient traits responding to available resources in habitat (Siefert et al. 2015).
Along with the guidelines, we identified the relationships between phylogenetic and functional S.E.S. Dnn′ and environmental gradient to provide additional evidence of environmental filtering (Fig. 2). The result of the S.E.S. Dnn′ based on multi-traits was correlated with the environmental gradient (Fig. 2b), yet it is interesting to notice that phylogeny did not show a correlation with the environmental gradient (Fig. 2a). Since significant phylogenetic signals had been discovered in the trait data, a consistent pattern should be expected in the phylogenetic turnover and functional turnover (Webb et al. 2002; Swenson 2011), but the actual results were in contrary, instead. As a matter of fact, it has been argued that although phylogeny is habitually used as a tool to deduce the key ecological processes, it is not as perfect as traits (Cadotte et al. 2017; Xu et al. 2017) and even biased to apply the phylogenetic pattern to infer the community assemblages (Gerhold et al. 2015). Moreover, sympatric species might reflect the accordance of phylogenetic similarity and functional similarity, which not only contains conserved traits but also plastic traits (Pavoine and Bonsall 2011; Xu et al. 2017). In previous works on different ecosystems, some cases in reality showed inconsistent ecological processes for phylogenetic and functional inference. In the Panama and Puerto Rico tropical rain forests at different succession stages, phylogeny displayed a stochastic pattern and traits a deterministic one (Swenson et al. 2012b). For the grassland in Algeria, similar life form to our study area, researchers described phylogeny as a poor surrogate for traits (Pavoine et al. 2013). Manifestly, the relationship between phylogeny and the assemblage mechanism is complex. Contrariwise, we recognized that the environmental filtering process was essentially the screening of species with appropriate niche from the species pool into habitats (Kraft et al. 2008; Cornwell and Ackerly 2009), and plant functional traits were the appropriate proxies for species niches (McGill et al. 2006; Violle et al. 2007). Accordingly, we were convinced that the functional turnover revealed the essence of the assembly mechanism more than the phylogenetic turnover in the riparian marsh of the Yellow River. For the univariate traits, only Hmax (Fig. 2c) and LDMC (Fig. 2e) had significant correlations with the environmental gradient. The results proved that these two traits were the key ones in determining the community dispersion and population growth rates along the environmental gradient. Hmax is associated with the capability of tolerating environmental stress, and LDMC is related to the potential relative growth rate (Cornelissen et al. 2003). But SLA (Fig. 2f), though showing strongly correlation with potential relative growth rate and proven in many other ecosystems (Westoby et al. 2002; Iida et al. 2014; Gibert et al. 2016), did not give the same result as LDMC. We attribute the discordance to that the difficulty in measuring the leaf area of certain species reduced the accuracy of SLA (Cornelissen et al. 2003). For example, Suaeda glauca and Suaeda salsa in Suaeda genus, Typha angustata and Typha laxmannii in Typha genus are all have high leaf thicknesses depending on the fleshy leaves, which may affect the measurement of SLA (Wilson et al. 1999). In such a case, LDMC may yield a more meaning result.
All of the phylogenetic and functional analyses were based on the evolutionary conservatism assumption as proposed. Actually, we did find a significant phylogenetic signal in the univariate trait data (Table 3). In contrast, the phylogenetic signals were weak (K < 1), which is usually interpreted as evolutionary lability (Blomberg et al. 2003; Yang et al. 2014), and might lead to an ambiguous relationship between phylogeny and functional traits (Swenson 2011).
So far we have exposed, following the guidelines for multivariate functional beta diversity and part univariate functional beta diversity, all three aspects of evidences, (1) a clustered pattern of community dispersion, (2) a covariant relationship between the population growth rate and the environmental gradient, and (3) significant phylogenetic signals in trait data. Thus, the environmental filtering was determined to be dominating in the community assembly processes in the riparian marsh of the Yellow River.
Variance Partitioning of Phylogenetic and Functional Beta Diversity
In light of the result of variance partitioning, we found that the environment, especially edaphic variables, affected phylogenetic and functional beta diversity in the riparian marsh of the Yellow River. The sole edaphic variables could always be accountable for the most variances (Fig. 3). Such observation was in consistence with reports that edaphic variables were generally crucial at a local scale (John et al. 2007; Chi et al. 2014; Yang et al. 2014; López-Angulo et al. 2018), in the support of our inference. From the results of forward selection (Table 4), we revealed that SS was the most critical indicator in edaphic variables. As a result, soil salinization steered a most direct and intense environmental pressure for coexisting species. Besides, we found that climatic variables had a weaker explanatory power on phylogenetic and functional beta diversity (Fig. 3). Although the species were generally influenced by edaphic conditions at the fine scale and by climate at the broad spatial scale (Díaz et al. 1998), a harsh edaphic environment, which had strong environmental pressure of high soil salinity and low nutrient content, might perform a more rigorous filtering of species than climate for wetland species. With respect to the spatial variables, the sole spatial variable in variance partitioning analyses might represent the dispersal limitation or the spatial structure of unmeasured environment variables (Legendre et al. 2009; Siefert et al. 2013; Spasojevic et al. 2016; Jiang et al. 2018). Different from previous findings, our study showed that the spatial variables could not explain the phylogenetic and functional turnover (Fig. 3), which might result in the neglect of dispersal limitation in the neutral theory. We still could not rule out the effect of dispersal, however, because most of species are spread seeds through the wind depending on small and light seeds.
As given in Fig. 3a, b, we observed a similar pattern to multivariate functional and phylogenetic beta diversity in variance partitioning, that is, approximately half of the variance was explainable by edaphic variables and around 30% of variance by edaphic, climatic, and spatial variables together. The results revealed that the environment, particularly edaphic conditions, controlled the phylogenetic and multivariate functional turnover between the communities. In spite of the report that herbaceous species showed to be more responsive to edaphic variables than climatic variables (Murphy et al. 2016), we realized that about 30% of the variance had no reasonable explanation still, which might be prompted by unmeasured environmental factors and spatial structure or ecological drift (Myers et al. 2013; Chi et al. 2014). Also, we ascertained the relative importance of edaphic variables in univariate functional beta diversity. Clearly, these functional traits disclosed distinct dimensions representing different ecological strategies for sympatric species (Spasojevic and Suding 2012; Gross et al. 2013; Wellstein et al. 2014). Henceforth, we deliberated that edaphic conditions played a decisive role in diverse ecological strategies of plants in the riparian marsh of the Yellow River, comprising resource acquisition, environmental tolerance, competitive exclusion and reproduction. It is amazing to find that, the whole univariate functional beta diversity was well explicated by SS in the edaphic variables, or the soil salinity was a pivotal environmental factor in influencing diverse ecological strategies.
In conclusion, our results and analyses suggest that environmental filtering plays an indispensable role in the community assembly processes in the riparian marsh of the Yellow River. The phylogenetic clustering and trait convergence alone are insufficient to infer the driver of ecological processes. As a result, we have tested the relationships between S.E.S. Dnn′ and the environmental gradient, and phylogenetic signals in the trait data to determine the dominance of environmental filtering in the assembly processes. We have observed the soil salinity in edaphic variables as a core filter influences the community composition and species coexistence. Concurrently, some inconsistent patterns or non-significant relationships between community dispersions and environmental gradient, likely to be affected by other ecological processes, have been identified, which request further work to gauge their effects.
References
Anderson MJ, Crist TO, Chase JM, Vellend M, Inouye BD, Freestone AL, Sanders NJ, Cornell HV, Comita LS, Davies KF, Harrison SP, Kraft NJ, Stegen JC, Swenson NG (2011) Navigating the multiple meanings of β diversity: a roadmap for the practicing ecologist. Ecology Letters 14:19–28
Blanchet FG, Legendre P, Borcard D (2008) Forward selection of explanatory variables. Ecology 89:2623–2632
Blomberg SP, Garland T, Ives AR (2003) Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57:717–745
Borcard D, Legendre P (2002) All-scale spatial analysis of ecological data by means of principal coordinates of neighbour matrices. Ecological Modelling 153:51–68
Borcard D, Legendre P, Drapeau P (1992) Partialling out the spatial component of ecological variation. Ecology 73:1045–1055
Cadotte MW, Tucker CM (2017) Should environmental filtering be abandoned? Trends in Ecology & Evolution 32:429–437
Cadotte MW, Davies TJ, Peres-Neto PR (2017) Why phylogenies do not always predict ecological differences. Ecological Monographs 87:535–551
Cavender-Bares J, Kozak KH, Fine PVA, Kembel SW (2009) The merging of community ecology and phylogenetic biology. Ecology Letters 12:693–715
Chesson P (2000) Mechanisms of maintenance of species diversity. Annual Review of Ecology and Systematics 31:343–366
Chi X, Tang Z, Fang J (2014) Patterns of phylogenetic beta diversity in China's grasslands in relation to geographical and environmental distance. Basic and Applied Ecology 15:416–425
Cornelissen JHC, Lavorel S, Garnier E, Díaz S, Buchmann N, Gurvich DE, Reich PB, Hter S, Morgan HD, van der Heijden MGA, Pausas JG, Poorter H (2003) A handbook of protocols for standardised and easy measurement of plant functional traits worldwide. Australian Journal of Botany 51:335–380
Cornwell WK, Ackerly DD (2009) Community assembly and shifts in plant trait distributions across an environmental gradient in coastal California. Ecological Monographs 79:109–126
Diamond JM (1975) Assembly of species communities. In: Cody ML, Diamond JM (eds) Ecology and evolution of communities. Belknap Press, Harvard University Press, Cambridge, pp 342–444
Díaz S, Cabido M, Casanoves F (1998) Plant functional traits and environmental filters at a regional scale. Journal of Vegetation Science 9:113–122
Dormann CF, McPherson JM, Araújo MB, Bivand R, Bolliger J, Carl G, Davies RG, Hirzel A, Jetz W, Kissling WD, Kühn I, Ohlemüller R, Peres-Neto PR, Reineking B, Schröder B, Schurr FM, Wilson R (2007) Methods to account for spatial autocorrelation in the analysis of species distributional data: a review. Ecography 30:609–628
Fang J, Wang Z, Tang Z (2011) Atlas of woody plants in China: distribution and climate. High Education Press, Beijing
Fick SE, Hijmans RJ (2017) WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. International Journal of Climatology 37:4302–4315
Gerhold P, Cahill JF, Winter M, Bartish IV, Prinzing A (2015) Phylogenetic patterns are not proxies of community assembly mechanisms (they are far better). Functional Ecology 29:600–614
Gibert A, Gray EF, Westoby M, Wright IJ, Falster DS, Wilson S (2016) On the link between functional traits and growth rate: meta-analysis shows effects change with plant size, as predicted. Journal of Ecology 104:1488–1503
Gross N, Börger L, Soriano-Morales SI, Le Bagousse-Pinguet Y, Quero JL, García-Gómez M, Valencia-Gómez E, Maestre FT (2013) Uncovering multiscale effects of aridity and biotic interactions on the functional structure of Mediterranean shrublands. Journal of Ecology 101:637–649
Hubbell SP (2001) The unified neutral theory of biodiversity and biogeography (MPB-32). Princeton University Press, Princeton
Iida Y, Kohyama TS, Swenson NG, Su SH, Chen CT, Chiang JM, Sun IF (2014) Linking functional traits and demographic rates in a subtropical tree community: the importance of size dependency. Journal of Ecology 102:641–650
Jiang F, Xun Y, Cai H, Jin G (2018) Functional traits can improve our understanding of niche- and dispersal-based processes. Oecologia 186:783–792
John R, Dalling JW, Harms KE, Yavitt JB, Stallard RF, Mirabello M, Hubbell SP, Valencia R, Navarrete H, Vallejo M, Foster RB (2007) Soil nutrients influence spatial distributions of tropical tree species. Proceedings of the National Academy of Sciences of the United States of America 104:864–869
Kraft NJB, Valencia R, Ackerly DD (2008) Functional traits and niche-based tree community assembly in an Amazonian forest. Science 322:580–582
Kraft NJB, Adler PB, Godoy O, James EC, Fuller S, Levine JM (2015) Community assembly, coexistence and the environmental filtering metaphor. Functional Ecology 29:592–599
Legendre P, Mi X, Ren H, Ma K, Yu M, Sun IF, He F (2009) Partitioning beta diversity in a subtropical broad-leaved forest of China. Ecology 90:663–674
López-Angulo J, Swenson NG, Cavieres LA, Escudero A (2018) Interactions between abiotic gradients determine functional and phylogenetic diversity patterns in Mediterranean-type climate mountains in the Andes. Journal of Vegetation Science 29:245–254
Maire V, Gross N, Börger L, Proulx R, Wirth C, Da Silveira Pontes L, Soussana J-F, Louault F (2012) Habitat filtering and niche differentiation jointly explain species relative abundance within grassland communities along fertility and disturbance gradients. New Phytologist 196:497–509
Mayfield MM, Levine JM (2010) Opposing effects of competitive exclusion on the phylogenetic structure of communities. Ecology Letters 13:1085–1093
McGill BJ, Enquist BJ, Weiher E, Westoby M (2006) Rebuilding community ecology from functional traits. Trends in Ecology & Evolution 21:178–185
Murphy SJ, Salpeter K, Comita LS (2016) Higher β-diversity observed for herbs over woody plants is driven by stronger habitat filtering in a tropical understory. Ecology 97:2074–2084
Myers JA, Chase JM, Jiménez I, Jørgensen PM, Araujo-Murakami A, Paniagua-Zambrana N, Seidel R (2013) Beta-diversity in temperate and tropical forests reflects dissimilar mechanisms of community assembly. Ecology Letters 16:151–157
Pakeman RJ, Quested HM (2007) Sampling plant functional traits: what proportion of the species need to be measured? Applied Vegetation Science 10:91–96
Pavoine S, Bonsall MB (2011) Measuring biodiversity to explain community assembly: a unified approach. Biological Reviews 86:792–812
Pavoine S, Gasc A, Bonsall MB, Mason NWH (2013) Correlations between phylogenetic and functional diversity: mathematical artefacts or true ecological and evolutionary processes? Journal of Vegetation Science 24:781–793
Purschke O, Schmid BC, Sykes MT, Poschlod P, Michalski SG, Durka W, Kühn I, Winter M, Prentice HC (2013) Contrasting changes in taxonomic, phylogenetic and functional diversity during a long-term succession: insights into assembly processes. Journal of Ecology 101:857–866
Qin H, Dong G, Zhang Y, Zhang F, Wang M (2017) Patterns of species and phylogenetic diversity of Pinus tabuliformis forests in the eastern Loess Plateau, China. Forest Ecology and Management 394:42–51
R Core Team. (2017). R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing
Raevel V, Violle C, Munoz F (2012) Mechanisms of ecological succession: insights from plant functional strategies. Oikos 121:1761–1770
Rosindell J, Hubbell SP, Etienne RS (2011) The unified neutral theory of biodiversity and biogeography at age ten. Trends in Ecology & Evolution 26:340–348
Satdichanh M, Millet J, Heinimann A, Nanthavong K, Harrison RD (2015) Using plant functional traits and phylogenies to understand patterns of plant community assembly in a seasonal tropical forest in Lao PDR. PLoS One 10:e0130151
Seelig H-D, Stoner RJ, Linden JC (2012) Irrigation control of cowpea plants using the measurement of leaf thickness under greenhouse conditions. Irrigation Science 30:247–257
Siefert A, Ravenscroft C, Weiser MD, Swenson NG (2013) Functional beta-diversity patterns reveal deterministic community assembly processes in eastern North American trees. Global Ecology and Biogeography 22:682–691
Siefert A, Violle C, Chalmandrier L, Albert CH, Taudiere A, Fajardo A, Aarssen LW, Baraloto C, Carlucci MB, Cianciaruso MV, de L. Dantas V, de Bello F, Duarte LDS, Fonseca CR, Freschet GT, Gaucherand S, Gross N, Hikosaka K, Jackson B, Jung V, Kamiyama C, Katabuchi M, Kembel SW, Kichenin E, Kraft NJB, Lagerström A, Le Bagousse-Pinguet Y, Li Y, Mason N, Messier J, Nakashizuka T, Overton JM, Peltzer DA, Pérez-Ramos IM, Pillar VD, Prentice HC, Richardson S, Sasaki T, Schamp BS, Schöb C, Shipley B, Sundqvist M, Sykes MT, Vandewalle M, Wardle DA (2015) A global meta-analysis of the relative extent of intraspecific trait variation in plant communities. Ecology Letters 18:1406–1419
Spasojevic MJ, Suding KN (2012) Inferring community assembly mechanisms from functional diversity patterns: the importance of multiple assembly processes. Journal of Ecology 100:652–661
Spasojevic MJ, Turner BL, Myers JA (2016) When does intraspecific trait variation contribute to functional beta-diversity? Journal of Ecology 104:487–496
Swenson NG (2011) Phylogenetic beta diversity metrics, trait evolution and inferring the functional beta diversity of communities. PLoS One 6:e21264
Swenson NG, Enquist BJ, Pither J, Kerkhoff AJ, Boyle B, Weiser MD, Elser JJ, Fagan WF, Forero-Montaña J, Fyllas N, Kraft NJB, Lake JK, Moles AT, Patiño S, Phillips OL, Price CA, Reich PB, Quesada CA, Stegen JC, Valencia R, Wright IJ, Wright SJ, Andelman S, Jørgensen PM, Lacher TE Jr, Monteagudo A, Núñez-Vargas MP, Vasquez-Martínez R, Nolting KM (2012a) The biogeography and filtering of woody plant functional diversity in north and South America. Global Ecology and Biogeography 21:798–808
Swenson NG, Erickson DL, Mi X, Bourg NA, Forero-Montaña J, Ge X, Howe R, Lake JK, Liu X, Ma K, Pei N, Thompson J, Uriarte M, Wolf A, Wright SJ, Ye W, Zhang J, Zimmerman JK, Kress WJ (2012b) Phylogenetic and functional alpha and beta diversity in temperate and tropical tree communities. Ecology 93:112–125
Violle C, Navas ML, Vile D, Kazakou E, Fortunel C, Hummel I, Garnier E (2007) Let the concept of trait be functional! Oikos 116:882–892
Webb CO, Ackerly DD, McPeek MA, Donoghue MJ (2002) Phylogenies and community ecology. Annual Review of Ecology and Systematics 33:475–505
Weiher E, Keddy PA (1995) Assembly rules, null models, and trait dispersion: new questions from old patterns. Oikos 74:159–164
Wellstein C, Campetella G, Spada F, Chelli S, Mucina L, Canullo R, Bartha S (2014) Context-dependent assembly rules and the role of dominating grasses in semi-natural abandoned sub-Mediterranean grasslands. Agriculture, Ecosystems & Environment 182:113–122
Westoby M, Falster DS, Moles AT, Vesk PA, Wright IJ (2002) Plant ecological strategies: some leading dimensions of variation between species. Annual Review of Ecology and Systematics 33:125–159
Wilson PJ, Thompson K, Hodgson JG (1999) Specific leaf area and leaf dry matter content as alternative predictors of plant strategies. New Phytologist 143:155–162
Xu J, Chen Y, Zhang L, Chai Y, Wang M, Guo Y, Li T, Yue M (2017) Using phylogeny and functional traits for assessing community assembly along environmental gradients: a deterministic process driven by elevation. Ecology and Evolution 7:5056–5069
Yang J, Zhang G, Ci X, Swenson NG, Cao M, Sha L, Li J, Baskin CC, Slik JWF, Lin L (2014) Functional and phylogenetic assembly in a Chinese tropical tree community across size classes, spatial scales and habitats. Functional Ecology 28:520–529
Zanne AE, Tank DC, Cornwell WK, Eastman JM, Smith SA, FitzJohn RG, McGlinn DJ, O’Meara BC, Moles AT, Reich PB, Royer DL, Soltis DE, Stevens PF, Westoby M, Wright IJ, Aarssen L, Bertin RI, Calaminus A, Govaerts R, Hemmings F, Leishman MR, Oleksyn J, Soltis PS, Swenson NG, Warman L, Beaulieu JM (2014) Three keys to the radiation of angiosperms into freezing environments. Nature 506:89–92
Acknowledgements
We thank Junhua Wu, Jian Liu, Ning Du and Xiaona Yu for helpful comments on earlier versions of the manuscript, and Huiying Qin and Tong Wu for assistance in the fieldwork. This study is supported in part by the National Key R&D Program of China (No. 2017YFC0505905), Ministry of Science and Technology of China (No. 2015FY110300), National Natural Science Foundation of China (No. 31770361; 31470402), and Natural Science Foundation of Shandong Province (No. ZR2017BC090).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Supplementary material S1
(DOCX 12 kb)
Supplementary material S2
(DOCX 12 kb)
Supplementary material S3
(DOCX 13 kb)
Supplementary material S4
(XLSX 22 kb)
Supplementary material S5
(DOCX 350 kb)
Rights and permissions
About this article
Cite this article
Yi, S., Wu, P., Bai, F. et al. Environmental Filtering Drives Plant Community Assembly Processes in the Riparian Marsh of Downstream Yellow River, China. Wetlands 40, 287–298 (2020). https://doi.org/10.1007/s13157-019-01182-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13157-019-01182-7