Abstract
Recent phylogenomic analyses within the insect superfamily Coreoidea (Hemiptera: Heteroptera) have begun to challenge previous phylogenetic hypotheses of the Coreidae and Alydidae based on more traditional cladistic and non-cladistic studies. Phylogenomic studies have found the coreid subfamilies Hydarinae and Pseudophloeinae to be more closely related to a potentially paraphyletic Alydidae (an “AHP” clade) in contrast with traditional cladistic studies. However, taxon sampling within these higher-level groups has remained sparse in current phylogenetic analyses, and the taxonomic positions and monophyly of some of these taxa continue to be unclear. Here, we expand upon previous phylogenomic studies using ultraconserved element loci by increasing taxon sampling within the AHP clade. Using concatenation and summary coalescent approaches, we specifically tested previous support for an AHP clade, the paraphyly of Alydidae, the phylogenetic position of Hydarinae, and the monophyly of the two tribes of Pseudophloeinae. Our results robustly support an AHP clade and resolved the position of Hydarinae as the sister group to a clade consisting of a paraphyletic Alydidae and Pseudophloeinae, regardless of analytical method and locus/gene tree filtering strategies we employed. We also found support for the monophyly of the pseudophloeine tribes Clavigrallini and Pseudophloeini, but generic relationships within each of these tribes varied across analyses. We discuss past non-cladistic morphological studies that have suggested the potential for an AHP clade in light of our results, and we highlight further systematic work needed to discern the AHP clade as a morphologically diagnosable group for future re-classification of the Alydidae and Coreidae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Coreidae (Hemiptera: Heteroptera: Coreoidea) include ~ 2600 extant species of phytophagous insects (CoreoideaSF Team, 2021), some of which are agriculturally important (Mitchell, 2000) or are used as models for studies of microevolution and behavior (e.g., Procter et al., 2012; Woodman et al., 2021). The family is divided into four extant subfamilies (Coreinae, Hydarinae, Meropachyinae, and Pseudophloeinae) and numerous tribes (CoreoideaSF Team, 2021). Currently, there is no consensus on the phylogenetic relationships within the Coreidae, with just a few hypotheses proposed based on modern cladistic approaches that have analyzed a small sample of taxa with limited morphological (Li, 1996, 1997) or traditional Sanger data (Pan et al., 2008).
Recent phylogenomic analyses of Coreidae using sequence capture data comprised of hundreds of ultraconserved elements (UCEs) have begun to test previously proposed hypotheses on relationships within the Coreidae. Such analyses include family and subfamily level relationships within the superfamily Coreoidea (Forthman et al., 2019) and subfamily to generic level relationships within the Coreinae + Meropachyinae clade (Forthman et al., 2020). These studies, which utilized greater taxon sampling and/or more molecular data than past cladistic studies, have corroborated some previously proposed phylogenetic hypotheses, recovered novel sets of relationships challenging hypotheses based on other cladistic (Li, 1996, 1997) and non-cladistic (e.g., Ahmad, 1970, 1979; Kumar, 1965; Schaefer, 1965) studies, and found evidence of para- and polyphyletic subfamilies and tribes of Coreidae.
The UCE phylogenomic analyses by Forthman et al. (2019, 2020) additionally found, with robust support, that Alydidae and Coreidae are not monophyletic families. Although taxon sampling was limited, the coreid subfamilies Hydarinae and Pseudophloeinae were consistently recovered in a clade comprised of a paraphyletic Alydidae. While this relationship has not been supported in past cladistic analyses (Li, 1996, 1997) (summarized in Fig. 1), with the exception of a mitochondrial DNA analysis of Pentatomomorpha (Zhao et al., 2018), some non-cladistic morphological studies have suggested some similarities among these taxa (Ahmad, 1970; Ahmad & Shadab, 1975; Cobben, 1968; Kumar, 1965; Pluot-Sigwalt & Moulet, 2020; Schaefer, 1964, 1965, 1980; Shadab, 1972). However, given that the taxon sampling by Forthman et al. (2019) was very limited for members of this clade, and additional taxa within these higher-level groups were not included in their subsequent study that expanded sampling for the Coreinae (Forthman et al., 2020), additional analyses with greater sampling are needed to further test the hypotheses of an Alydidae + Hydarinae + Pseudophloeinae (AHP) clade (Forthman et al., 2019), as well as the relationships within it. For example, Li (1997) found the Pseudophloeini to be paraphyletic with respect to the Clavigrallini (one representative of the latter tribe was included in the analysis), but this has remained the only cladistic study to test relationships within the Pseudophloeinae. Furthermore, the phylogenetic position of the Hydarinae remains uncertain within this clade; Forthman et al. (2019) recovered it as the sister group to Micrelytrinae or Alydidae + Pseudophloeinae but with weak support. Thus, increased taxon sampling of Hydarinae and Pseudophloeinae, as well as Alydidae, may further clarify phylogenetic relationships.
Here, we expand upon the taxon sampling by Forthman et al. (2019, 2020) to investigate relationships among and within the Alydidae, Hydarinae, and Pseudophloeinae using UCEs. Our taxon sampling strategy aimed to increase generic diversity, include representatives of several major biogeographic regions, and sample the same species or genera within these higher-level taxa as in Li (1997), when suitable tissues were available. With our improved taxon sampling, our goals were to (1) test whether we supported an AHP clade; (2) if the AHP clade was found, determine the phylogenetic position of Hydarinae within the clade; (3) test whether Alydidae was paraphyletic; and (4) test monophyly of the two tribes of Pseudophloeinae.
Material and methods
Taxon sampling
We sampled 212 taxa for this study. Previously published sequence data for 160 of these taxa were retrieved from Kieran et al. (2019), Forthman et al. (2019, 2020), and Emberts et al. (2020). Our sampling expanded on the taxa sampled in these past phylogenomic studies by including more members of the Hydarinae (5 of 9 genera sampled), Pseudophloeinae (17 of 28 genera; 3 of 4 sampled for Clavigrallini; 14 of 24 sampled for Pseudophloeini), and Alydidae (~ 30% of described genera sampled), as well as additional outgroup taxa in Rhopalidae. Our new sampling primarily targeted freshly preserved material (i.e., recently collected material preserved frozen or in ethanol or isopropyl), but three samples were taken from pinned, degraded museum material (see Table S1, Online Resource 1 for information about newly sampled taxa).
DNA extraction, sequence capture, and UCE identification and alignment
For the 52 new taxa sampled in this analysis, genomic DNA was extracted using a Gentra Puregene Tissue Kit, Qiagen DNeasy Blood and Tissue kit, or Qiagen DNeasy Blood and Tissue kit coupled with a Qiagen QIAquick PCR purification kit (see Knyshov et al. (2019); and Forthman et al. (2019, 2020) for details regarding DNA extraction and isolation protocols) (Table S1, Online Resource 1). We extracted DNA from the head, thorax, legs, and/or abdomen or the whole body depending on specimen size and quality to sample similar amounts of tissue across our samples. DNA quality was visually assessed by gel electrophoresis, and concentration was quantified using a Qubit 2.0 fluorometer. When possible, samples were normalized (10–20 ng/µL), and those with high molecular weight were fragmented into 200–1000 bp using a Covaris M220 Focused-ultrasonicator (20–60 s) or Bioruptor UCD-300 sonicator (4–10 cycles, 30 s on/30 s off). Degraded DNA from pinned museum samples were repaired using a PreCR Repair Mix kit, with a 3 × SPRI clean-up. Libraries were constructed, amplified (14–16 cycles), and pooled following Forthman et al. (2019). We used Forthman et al.'s (2019) custom myBaits kit, which includes the pentatomomorphan UCE probes designed by Faircloth (2017). We followed Forthman et al.'s (2019) enrichment protocol for seven samples, and we followed Forthman et al.'s (2020) touchdown enrichment protocol for nine (Table S1). For the remaining 36 samples, we made the following modifications to Forthman et al.'s (2020) touchdown enrichment protocol: (1) a hybridization mixture with 1/4 or 1/2 volume of baits was used (for dried or fresh material, respectively); (2) baits hybridized with library pools at 65 °C for 12 h followed by 12 h at 62 °C and then by 12 h at 60 °C; and (3) bait-target hybrids were bound to Dynabeads M-280 Streptavidin beads and washed four times at 60 °C (Table S1). Enriched library pools were quantified with Qubit, pooled in equimolar amounts, and sequenced on a single Illumina HiSeq3000 lane (2 × 100) at the University of Florida’s Interdisciplinary Center for Biotechnology Research. New and previously published sequence read data were processed following Forthman et al. (2019) and assembled into contigs using SPAdes v3.13.0 (single-cell and auto coverage cutoff options) (Prjibelski et al., 2020).
The procedure in Forthman et al. (2019) was followed to identify and align UCEs from contigs. We generated locus alignments with at least 50% and 70% of taxa (referred to as “50p” and “70p” datasets, respectively) and also subsampled each dataset for the 25% most parsimony-informative UCEs since this filtering strategy can improve topological support for UCEs, as these loci often have limited variation (Hosner et al., 2016; Meiklejohn et al., 2016). Thus, four datasets were generated for analyses.
Phylogenetic inference
A recent phylogenomic study of the subfamily Coreinae found concatenation and multispecies coalescent approaches to be largely congruent, but there were still differences in topology and support for some nodes (Forthman et al., 2020). Thus, we analyzed our data using both concatenation and coalescent approaches to make robust phylogenetic inferences about the AHP clade. Unless otherwise stated, default settings were used in analyses.
For concatenation, locus alignments were concatenated in PHYLUCE v1.5.0 (Faircloth, 2016) for each dataset. We used IQ-Tree v2.0.3 (Minh et al., 2020) to estimate the best partitioning scheme and models of evolution for each partition (settings: -m MF + MERGE, -rcluster 10) (Kalyaanamoorthy et al., 2017). However, where the proportion of invariant sites and gamma-distributed rate parameters were both included in a partition’s model, we excluded the former as it is not independent from the latter (Sullivan et al., 1999; Yang, 2006). For each partitioned dataset, three separate maximum likelihood analyses were performed in IQ-Tree (-p option; Chernomor et al., 2016), with the Shimodaira-Hasegawa-like approximate likelihood ratio test (SH-aLRT) on 1000 replicates (-alrt; Guindon et al., 2010) and 1000 ultrafast bootstrap replicates (-B; Hoang et al., 2018) further optimized by nearest neighbor interchange based on bootstrap alignment (-bnni). For each partitioned dataset, the analysis that resulted in a topology with the best log-likelihood was selected. We also performed the above analysis after removing partitions violating stationarity and/or homogeneity assumptions (–symtest-remove-bad; Naser-Khdour et al., 2019).
We also inferred species trees under the multispecies coalescent using individual gene trees for each dataset (Degnan & Rosenberg, 2006, 2009). IQ-Tree was used to select the best-fit model of sequence evolution for each locus alignment (-m TESTONLY). We used GARLI v2.01 (Zwickl, 2006) to perform 10 maximum likelihood optimal gene tree searches using the models selected by IQ-Tree (excluding the invariant site parameter when the gamma parameter was included in the model), with zero-length gene tree branches collapsed into polytomies (Zhang et al., 2017). We inferred species trees from optimal gene trees in ASTRAL-III v5.7.7 (Mirarab et al., 2014; Sayyari & Mirarab, 2016; Zhang et al., 2018), with local posterior probabilities to assess clade support (Sayyari & Mirarab, 2016). We additionally excluded gene trees based on loci that violated stationarity and/or homogeneity assumptions (using IQ-Tree’s symtest), and then re-estimated the species tree as described above.
Lastly, two taxa in the subfamily Pseudophloeinae (Bathysolen nubilus (Fallén, 1807) and Coriomeris nigricornis (Stål, 1870)) were recovered in a few different positions in our resulting phylogenies (see “Results”). We believed these results could be due to poor UCE locus recovery in these taxa compared to others included in this study, which is likely attributable to the old and degraded quality of these pinned museum samples (Table S1). Thus, we performed the above analyses after excluding both taxa to determine if phylogenetic relationships would become more stable across analyses.
Results
For the 52 newly sequenced taxa in our study, 47.43% of the targeted UCE loci were recovered on average per taxon (range: 20–1,598 loci per taxon; median = 1,400 [52.38%] loci per taxon; Table S2, Online Resource 2). Two dried museum samples yielded drastically fewer UCE loci compared to all other taxa sampled (the latter group having ≥ 723 loci for a given taxon): B. nubilus (20 UCE loci) and C. nigricornis (23 UCE loci). The number of parsimony-informative sites and number of UCE loci in each concatenated and summary coalescent dataset are given in Tables S3 and S4, respectively (Online Resources 3 and 4, respectively). Given that our analyses excluding B. nubilus and C. nigricornis had results largely congruent with our full taxon sampling, we report the results from the latter and only reference results from analyses excluding taxa when relevant; for phylogenetic trees from analyses with the reduced taxon sampling, see the “Data availability” section.
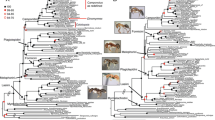
Across all analytical methods and data filtering strategies, we recovered Alydidae, Hydarinae, and Pseudophloeinae as a clade (i.e., the AHP clade) (Figs. 2, S1–S15), consistent with Forthman et al. (2019). Support for this relationship was consistently high (100%). This AHP clade was sister to the Coreinae + Meropachyinae with moderate to high support (see “Data availability” section for supplementary tree files). Within the AHP clade, Hydarinae were consistently recovered as the sister group to the remaining members of the AHP clade with robust support.
Maximum likelihood best tree generated from the 50p concatenated ultraconserved element alignment containing all partitions (outgroups and representatives of the subfamilies Coreinae and Meropachyinae pruned for visualization; see “Data availability” for tree with all taxa). Values at nodes represent Shimodaira-Hasegawa-like approximate likelihood ratio test/ultrafast bootstraps support where at least one of these measures is less than 100%. Dorsal habitus images of select representatives of higher-level taxa (not to scale): 1, Hydarella chiangdaoensis Brailovsky, 1994; 2, Maduranoides chemsaki Brailovsky, 1988; 3, Micrelytra fossularum (Rossi, 1790); 4, Dulichius trispinosus Stål, 1866; 5, Leptocorisa bipunctata Costa, 1863; 6, Stenocoris furcifera (Westwood, 1842); 7, Hyalymenus pulcher (Stål, 1854); 8, Apidaurus conspersus Stål, 1870; 9, Burtinus luteomarginatus Maldonado, 1953; 10, Alydus pilosulus Herrich-Schӓffer, 1847; 11, Nariscus cinctiventris (Germar, 1838); 12, Melanacanthus sp.; 13, Clavigralloides acantharis (Fabricius, 1803); 14, Clavigralla pusilla Dolling, 1979; 15, Clavigralla elongata Signoret, 1861; 16, Vilga westwoodi (Kolenati, 1845); 17, Myla sp.; 18, Mevanidea hystrix (Gerstaecker, 1873); 19, Psilolomia brunneofusca Dolling, 1986; 20, Pseudomyla cornuta (Hsiao, 1965); 21, Ceraleptus obtusus (Brullé, 1839); 22, Nemocoris fallenii Sahlberg, 1848; 23, Arenocoris waltlii (Herrich-Schӓffer, 1835); 24, Bathysolen nubilus (Fallén, 1807); 25, Strobilotoma typhaecornis (Fabricius, 1803); 26, Coriomeris denticulatus (Scopoli, 1763)
In all analyses, the family Alydidae was paraphyletic. The monophyletic subfamily Alydinae was more closely related to a monophyletic coreid subfamily, Pseudophloeinae, than to the other alydid subfamily, Micrelytrinae, with high support. This Alydinae + Pseudophloeinae clade was recovered as the sister group to the Micrelytrinae with low to high support (though support was often high). Within the Pseudophloeinae, we also found consistent, high support for the monophyly of both tribes (Clavigrallini and Pseudophloeini).
Relationships within the Hydarinae (except in one ASTRAL analysis (Fig. S13)) and Micrelytrinae were congruent across analyses. Relationships within the Alydinae were also largely congruent among most of our analyses with low to high support (often high), but six analyses (five based on our reduced taxon dataset) recovered three alternative topologies with similar support (Fig. S13; also see “Data availability”). These alternative relationships (mostly from summary coalescent analyses) largely involved a different phylogenetic placement of Apidaurus, as well as the placement of the Tenosius + Heegeria + Nariscus + Melanacanthus clade and the relationships within it.
We also observed several incongruences within the Pseudophloeinae. Within Clavigrallini, many analyses resulted in a moderately to highly supported Clavigralla (Figs. 2, S1–S9, S11–S13), but five of our analyses (mostly from summary coalescent analyses) found relatively low support for the paraphyly of the genus with respect to Clavigralloides + Gralliclava (Figs. S10, S14, S15; also see “Data availability”). With respect to the Pseudophloeini, three alternative relationships were recovered within the Mevanieda + Myla + Paramyla clade, all with low to moderate support and without one topology being predominately recovered. One large clade, in particular, had several different phylogenetic hypotheses recovered, i.e., the Anoplocerus + Arenocoris + Bathysolen + Ceraleptus + Coriomeris + Nemocoris + Strobilotoma clade. In our full taxon dataset, the incongruences we observed in this clade involved the phylogenetic placements of Bathysolen and C. nigricornis. Excluding these two poor quality samples resulted in relationships within this clade that were congruent to that found in Fig. 2, except for one analysis that recovered a slightly different position for the Anoplocerus + Arenocoris clade.
Discussion
The limited body of cladistic analyses involving the Coreoidea has not offered clarity on the phylogenetic positions of Hydarinae and Pseudophloeinae, which have been hindered by a relatively small taxon sampling, either for members of these subfamilies and/or across the Coreidae and Alydidae more generally. Here, using the largest taxon sampling of Coreidae and Alydidae to date, we robustly reconstructed relationships among the Hydarinae, Pseudophloeinae, and Alydidae, confirming results of recent phylogenomic studies (Forthman et al., 2019, 2020). Our study clarified the phylogenetic position of Hydarinae as the sister group to a paraphyletic Alydidae + a monophyletic Pseudophloeinae regardless of analytical approach and data filtering strategies. We further found robust support for the monophyly of the pseudophloeine tribes Clavigrallini and Pseudophloeini. It is evident that the taxonomic status of the Hydarinae and Pseudophloeinae should be evaluated further — with an emphasis on morphological studies building off past contributions — to formally revise the Alydidae and Coreidae classification. While the subfamily and tribal classification of the Coreidae and Alydidae appears to be based on highly variable external characters (e.g., see Forthman et al., 2019), internal morphological studies could provide valuable characters to inform future classification changes for these groups (e.g., see Pluot-Sigwalt & Moulet, 2020). The rest of our discussion focuses on clades of interest.
AHP clade
An AHP clade has been recovered only from recent phylogenomic analyses (Emberts et al., 2020; Forthman et al., 2019, 2020) in contrast to previous cladistic studies (e.g., Li, 1997), with the exception of a mitochondrial DNA analysis of Pentatomomorpha (Zhao et al., 2018). In evaluating a limited number of morphological characters in the phylogenomic analysis of Coreoidea, Forthman et al. (2019) found only the presence of non-pseudoperculate eggs to support the AHP clade (which was coded at higher taxonomic levels based on findings from Cobben’s (1968) study on heteropteran egg structures).
Several non-cladistic morphological studies, often based on very limited taxon sampling among higher taxonomic ranks (as well as within some of these ranks), have found similarities among the Alydidae, Hydarinae, and Pseudophloeinae that would suggest a potentially close relationship among them; however, many of these studies have also suggested that similarities observed among these taxa are probably plesiomorphic traits and that the Hydarinae and Pseudophloeinae are likely early diverging lineages within the Coreidae (Schaefer, 1965; Ahmad, 1970; Ahmad & Shadab, 1975; but see Kumar (1965), and Pluot-Sigwalt & Moulet, 2020). Kumar (1965) studied the genitalia of several coreoid taxa, in which he concluded that the female genital plates and several aspects of the male internal genitalia (e.g., the vesica and ejaculatory reservoir) in Pseudophloeinae are similar to the Alydidae (or at least the Leptocorisini [Micrelytrinae]) (also see Schaefer (1964) for a brief, general discussion on the structure of the ejaculatory reservoir in Coreoidea). Shadab (1972) concluded that the pseudophloeine paramere is more similar to Alydidae than to other coreids, while Schaefer (1980, 1982) noted some inconsistent similarities in the male pygophore of the Alydidae and Hydarinae. Ahmad and Shadab (1975) examined external cephalic morphology and concluded that the Pseudophloeinae and Alydinae have similar sutures that are distinctly different from other coreid taxa examined.
Perhaps one of the more investigated internal structures within the Coreoidea is the female spermatheca. Ahmad (1970) stated that the development of the spermathecal coils and the absence of a proximal flange in Hydarinae and Pseudophloeinae were conditions different from most other coreids. Shadab (1972) also briefly highlighted these similarities between Hydarinae, Pseudophloeinae, and Alydidae. Recently, Pluot-Sigwalt and Moulet (2020) documented the spermatheca in one of the most broadly and densely sampled morphological studies within the Coreoidea. They confirmed observations from previous studies (e.g.., Ahmad, 1970; Kumar, 1965; Schaefer, 1965) while emphasizing the presence of a bipartite spermatheca in the Alydidae, Hydarinae, and Pseudophloeinae, a morphological feature yet to be documented in other coreid subfamilies and coroeid families (these being tripartite). Thus, the spermatheca may become a useful character system for exploring putative synapomorphies of the AHP clade.
Paraphyly of Alydidae and the Alydinae + Pseudophloeinae clade
The higher taxonomic groups within the Alydidae (i.e., currently recognized subfamilies and tribes) have been variously treated as subfamilies and tribes (e.g., Leptocorisini treated as Leptocorisinae), but these groups have always been considered distinct from one another. However, relationships among the Alydidae have rarely been tested with cladistic approaches and, thus, remained unclear. The main support for the monophyly of the Alydidae comes from Li’s (1996) morphological phylogenetic analysis of the Coreoidea, but he did not list any apomorphic characters uniting the taxa within this clade. A monophyletic Alydidae has also been supported by molecular phylogenetic studies that included few alydid representatives as part of a larger sampling testing relationships among other taxonomic groups (Li et al., 2006, 2016; Wang et al., 2016); however, none of these included species of Hydarinae and Pseudophloeinae, some lacked sampling of the Micrelytrinae, and some also recovered conflicting hypotheses under different analytical conditions. While Li and Zheng (1993) also claimed that their morphological phylogenetic analysis supported the monophyly of the family, there is no indication of outgroup taxa being included that would permit a test of alydid monophyly.
Recently, phylogenetic analyses based on mitogenomes (Zhao et al., 2018) and UCE data (Forthman et al., 2019, 2020) have recovered the Alydidae as poly- or paraphyletic (often with high support). Some non-cladistic studies have found several differences in the head (Ahmad & Shadab, 1975) and male genital morphology (Kumar, 1965; Schaefer, 1980) among the alydid subfamilies Micrelytrinae and Alydinae, leading some authors to state that the Alydidae are likely not monophyletic (e.g., Dolling, 1978; Schaefer, 1980). Our results support the paraphyly of Alydidae, which are congruent with Forthman et al.'s (2019, 2020) results that found the subfamily Alydinae to be more closely related to Pseudophloeinae. In contrast, Li (1996) recovered Pseudophloeinae as the sister to all other coreoids sampled (i.e., Rhopalidae + Alydidae + Hydarinae + Coreinae + Meropachyinae), but Li (1997), who included only Coreidae, recovered Pseudophloeinae as sister to all coreids. While the possibility of a close relationship between Pseudophloeinae and the family Alydidae has been suggested in several non-cladistic studies (Ahmad, 1970; Cobben, 1968; Kumar, 1965; Pluot-Sigwalt & Moulet, 2020; Schaefer, 1965), only one has specifically suggested a close relationship between the Pseudophloeinae and the subfamily Alydinae based on male genital morphology (Kumar, 1965).
The composition of Micrelytrinae has differed among workers (e.g., Ahmad, 1965; CoreoideaSF Team, 2021; Schaefer, 1999), and the phylogenetic position of the tribes Micrelytrini or Leptocorisini within the Alydidae has not always been clear (e.g., see Schaefer, 1972, 1980). Our results support a monophyletic Micrelytrinae, inclusive of the Leptocorisini and Micrelytrini, which is congruent with previous morphological cladistic analyses (Li, 1996; Li & Zheng, 1993). Furthermore, we find support for the monophyly of each tribe, congruent with Li (1996). Li and Zheng (1993) reported several characters uniting the Micrelytrinae, most of which were supported by Schaefer (1999) and have been incorporated into diagnoses of the subfamily (e.g., Schuh & Weirauch, 2020): presence of a deep mid-cephalic sulcus, elongated body, slender legs, and M vein basally coriaceous in the membrane, as well as several traits of the male aedeagus.
Lastly, morphological and molecular cladistic analyses by Li and Zheng (1993), Li (1996), and Forthman et al. (2019, 2020) have also supported the monophyly of the Alydinae. Only one molecular analysis based on cytochrome b and a limited sample of Alydinae did not recover this subfamily as a clade (Pan et al., 2008). Li and Zheng (1993) listed a few hind leg and fore wing characters as supporting the Alydinae, while Schaefer (1999) identified several more apomorphies for Alydinae based on his re-evaluation of Li and Zheng’s (1993) analysis. Forthman et al.'s (2019) re-evaluation of Li’s (1996) characters also found several apomorphies, including the constricted abdomen, trilateral head shape, and closer proximity of the ocelli to each other than to the eyes.
Phylogenetic position of the monophyletic Hydarinae
The phylogenetic position of the small subfamily Hydarinae has been controversial, and few morphological studies have included representatives of the subfamily. Hydarinae was proposed as an early diverging lineage among coreoids and coreids in cladistic and non-cladistic morphological studies (e.g., Ahmad, 1970; Ahmad & Shadab, 1975; Li, 1996, 1997; Schaefer, 1965, 1982). From prior morphological phylogenetic studies, this subfamily has been recovered as the sister group of Rhopalidae + Alydidae + the remaining Coreidae (excluding Pseudophloeinae) (Li, 1996) or Coreinae + Meropachyinae (Li, 1997), with few, if any, characters listed as apomorphies for these relationships. Phylogenetic analyses with mitochondrial DNA have recovered Hydarinae in several other positions, including being sister to the Coreinae with weak support (Valero et al., 2017), Alydinae + Pseudophloeinae (Zhao et al., 2018; Bayesian analysis with moderate to high support) and the Alydinae (Zhao et al., 2018; ML analysis with poor support). Forthman et al. (2019, 2020) phylogenomic analyses suggested the Hydarinae to be a member of a clade including Pseudophloeinae and Alydidae, with weakly supported, conflicting hypotheses about the position of the subfamily within it (sister to Alydidae + Pseudophloeinae or sister to Micrelytrinae, the latter relationship not having any apomorphies in their analysis). Here, we find robust, consistent support for the Hydarinae as sister to the Alydidae + Pseudophloeinae, congruent with some of Zhao et al. (2018) and Forthman et al.'s (2019, 2020) findings.
While the position of Hydarinae within the Coreoidea has been relatively contentious, its status as a monophyletic group has not. Despite the uncontroversial recognition of Hydarinae as a natural group, there has been disagreement on which taxonomic rank should be accorded to it. While some modern works have treated Hydarinae as a subfamily since Ahmad (1970), there are still others (e.g., Brailovsky, 1998, 2011; Fernandes et al., 2015; Packauskas, 2010) that recognize it as a tribe within the subfamily Coreinae. Our phylogenetic findings, as well as others (e.g., Li, 1996, 1997; Zhao et al., 2018), provide evidence that the Hydarinae cannot be considered a tribe within the Coreinae. Until further systematic work is done on the AHP clade — hopefully leading to a revision of the Alydidae and Coreidae — the Hydarinae should be treated as a subfamily of Coreidae.
Monophyly of Pseudophloeinae, Clavigrallini, and Pseudophloeini
The monophyly of Pseudophloeinae has not been questioned in past studies. Li (1997) identified several apomorphies for the subfamily (some of which were not coded in his matrix), including abdominal and male genital morphology. In Forthman et al.'s (2019) analysis, they identified other apomorphies involving the head and abdomen. Other non-cladistic studies have also suggested a suite of characters to separate the Pseudophloeinae from the other members of Coreidae, including morphology of the hind wing (Stål, 1867; Štys, 1977), metathoracic scent glands (Shadab, 1972; Stål, 1867), legs (Schaefer, 1965; Stål, 1867), and male and female genitalia (Ahmad, 1965; Kumar, 1965; Pluot-Sigwalt & Moulet, 2020; Schaefer, 1965), among others (see Dolling (1978, 1986) for most recent diagnosis of the subfamily). However, some of these traits are not consistently observed across all species of Pseudophloeinae (e.g., the antevannal vein is absent in five genera (Dolling, 1986)), or restricted to the subfamily (e.g., the non-sulcate tibiae (Ahmad, 1970)). Furthermore, many of these studies often suffer from poor taxon sampling of Pseudophloeinae, with most studies rarely sampling species from both subfamilies. As such, many of these traits remain to be tested in cladistic analyses with a much larger taxon sampling to determine whether they are apomorphies of the subfamily.
To our knowledge, no studies have explicitly tested the monophyly of each of the tribes within the Pseudophloeinae (Li (1997) only included one representative of Clavigrallini but did recover a paraphyletic Pseudophloeini; Fig. 1), making our study the first cladistic analysis which robustly supports the monophyly of the Clavigrallini and Pseudophloeini. Dolling (1978, 1986) provided the most comprehensive treatments of the tribes to date. In those treatments, Dolling reviewed the traits Stål (1873) used to characterize the Clavigrallini and Pseudophloeini, identified a few characters that together would distinguish Clavigrallini from taxa within the Pseudophloeini, and concluded that the Pseudophloeini are more difficult to diagnosis due to a lack of universal traits among the species and the presence of some characters found in the Clavigrallini, which he attributed as plesiomorphies. Aside from Dolling’s studies, the only comparative morphological study to have a relatively broad sample of both pseudophloeine tribes (compared to other prior morphological studies) is Pluot-Sigwalt and Moulet’s (2020) investigation of the female spermatheca. While both tribes possess a “Type II” spermatheca, Pluot-Sigwalt and Moulet’s (2020) descriptions of the seminal receptacle and spermathecal duct suggested some trait differences among the Clavigrallini and Pseudophloeini that may be worth examining further with more generic representation and testing in a phylogenetic analysis. Given our results and past studies, it is clear that this poorly studied subfamily and its tribes should be a focus of future morphological studies, which need to include a large taxon sampling to better determine and evaluate putative synapomorphies of these clades.
Conclusion
Our study, the first phylogenetic analysis to have a broader sample of Hydarinae and Pseudophloeinae, found robust support for a clade comprised of the Alydidae, Hydarinae, and Pseudophloeinae. While we found that many of the family-level groupings are monophyletic, it appears that the Alydidae are paraphyletic. While previous comparative morphological studies have provided some insights on characters that may be apomorphies of the relationships we recovered, many of these studies suffered from sparse sampling of our ingroup taxa. Such morphological traits, among others, should be examined in more species where suitable material is available to determine apomorphies that can diagnose the AHP clade and clades within it. The results of this study, as well as recent phylogenomic analyses highlighting the non-monophyly of Coreinae and Meropachyinae, indicates that the classification of both the Alydidae and Coreidae needs further systematic attention and revision.
Data availability
Sequence read files of newly generated data are available on NCBI’s Sequence Read Archive under BioProject PRJNA774038. Alignments, gene trees, and concatenation and species trees are available from FigShare under the project titled “Phylogenomic analysis with improved taxon sampling corroborates an Alydidae + Hydarinae + Pseudophloeinae clade (Heteroptera: Coreoidea: Alydidae, Coreidae).”
Code availability
Not applicable.
References
Ahmad, I. (1965). The Leptocorisinae (Heteroptera: Alydidae) of the world. Bulletin of the British Museum (Natural History) Entomology Supplement, 5, 1–156.
Ahmad, I. (1970). Some aspects of the female genitalia of Hygia Uhler 1861 (Coreidae: Colpurinae) and their bearing on classification. Pakistan Journal of Zoology, 2, 235–243.
Ahmad, I. (1979). A revision of the check list of Coreidae and Pentatomidae of the superfamilies Coreoidea and Pentatomoidea (Heteroptera: Pentatomomorpha) from Pakistan, Azad Kashmir and Bangladesh. Part 1: Additions and corrections of coreid and pentatomid fauna with phylogenetic considerations. Supplement of the Entomological Society of Karachi, 4, 1–113.
Ahmad, I., & Shadab, M. U. (1975). Comparative external cephalic morphology of some coreoids (Heteroptera: Coreoidea) with reference to their phylogeny. Pakistan Journal of Scientific and Industrial Research, 18, 133–142.
Brailovsky, H. (1988). La tribu Hydarini Stål, en el continente americano con descripción de dos nuevos géneros, una nueva especie y una nueva subespecie (Hemiptera-Heteroptera-Coreidae). Anales del Instituto de Biología Universidad Nacional Autónoma de México. Serie Zoología, 58, 623–650.
Brailovsky, H. (1994). Revision of the tribe Hydarini in the Oriental region (Hemiptera, Heteroptera: Coreidae). Insect Systematics & Evolution, 24, 383–390.
Brailovsky, H. (1998). Systematics of the southern African Hydarini (Insecta: Heteroptera: Coreidae: Coreinae). Reichenbachia, 32, 165–174.
Brailovsky, H. (2011). Faune de Madagascar 94. Insecta Hemiptera Heteroptera Coreidae. Institut de recherche pour le développement, Éditions Quæ, Publications scientifiques du Muséum, Paris et Marseille.
Brullé, M. (1839). Insectes. In: P. B. Webb & S. Berthelot (Eds.) Histoire Maturelle des Îles Canaries. Tome Deuxième. (pp. 55–119). Paris : Béthune
Chernomor, O., von Haeseler, A., & Minh, B. Q. (2016). Terrace aware data structure for phylogenomic inference from supermatrices. Systematic Biology, 65, 997–1008. https://doi.org/10.1093/sysbio/syw037
Cobben, R. H. (1968). Evolutionary trends in Heteroptera. Part I. Eggs, architecture of the shell, gross embryology, and eclosion. Wageningen, Netherlands: Centre for Agricultural Publishing and Documentation.
CoreoideaSF Team. (2021). Coreoidea Species File Online. Version 5.0/5.0. Retrieved February 22, 2022, from http://coreoidea.speciesfile.org
Costa, A. (1863). Illustrazione di alcuni Emitteri stranieri all'Europa. Rendiconto della Adunaze e de' Lavori della Reale accademia delle Scienze di Napoli, 2, 250–261.
Degnan, J. H., & Rosenberg, N. A. (2006). Discordance of species trees with their most likely gene trees. PLoS Genetics, 2, e68. https://doi.org/10.1371/journal.pgen.0020068
Degnan, J. H., & Rosenberg, N. A. (2009). Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends in Ecology and Evolution, 24, 332–340. https://doi.org/10.1016/j.tree.2009.01.009
Dolling, W. (1979). A revision of the African pod bugs of the tribe Clavigrallini (Hemiptera: Coreidae) with a checklist of the world species. Bulletin of the British Museum (Natural History) Entomology, 39, 1–84.
Dolling, W. R. (1978). A revision of the Oriental pod bugs of the tribe Clavigrallini (Hemiptera: Coreidae). Bulletin of the British Museum (Natural History). Entomology, 36, 281–321.
Dolling, W. R. (1986). The tribe Pseudophloeini (Hemiptera: Coreidae) in the Old World tropics with a discussion on the distribution of the Pseudophloeinae. Bulletin of the British Museum (Natural History). Entomology, 53, 151–212.
Emberts, Z., Mary, C. M., Howard, C. C., Forthman, M., Bateman, P. W., Somjee, U., Hwang, W. S., Li, D., Kimball, R. T., & Miller, C. W. (2020). The evolution of autotomy in leaf-footed bugs. Evolution, 74, 897–910. https://doi.org/10.1111/evo.13948
Fabricius, J. C. (1803). Systema Rhyngotorum: Secundum Ordines, Genera, Species: Adiectis Synonymis, Locis, Observationibus, Descriptionibus. Brunsvigae: C. Reichard.
Faircloth, B. C. (2016). PHYLUCE is a software package for the analysis of conserved genomic loci. Bioinformatics, 32, 786–788. https://doi.org/10.1093/bioinformatics/btv646
Faircloth, B. C. (2017). Identifying conserved genomic elements and designing universal bait sets to enrich them. Methods in Ecology and Evolution, 8, 1103–1112. https://doi.org/10.1111/2041-210X.12754
Fallén, C. F. (1807). Monographia Cimicum Sveciae. Hafniæ, apud C. G. Proft, literis directoris, J. F. Schulzii.
Fernandes, J. A. M., Mitchell, P. L., Livermore, L., & Nikunlassi, M. (2015). Leaf-footed bugs (Coreidae). In A. R. Panizzi & J. Grazia (Eds.), True bugs (Heteroptera) of the Neotropics (pp. 549–605). Entomology in focus.
Forthman, M., Miller, C. W., & Kimball, R. T. (2019). Phylogenomic analysis suggests Coreidae and Alydidae (Hemiptera: Heteroptera) are not monophyletic. Zoologica Scripta, 48, 520–534. https://doi.org/10.1111/zsc.12353
Forthman, M., Miller, C. W., & Kimball, R. T. (2020). Phylogenomics of the leaf-footed bug subfamily Coreinae (Hemiptera: Coreidae). Insect Systematics and Diversity, 4, 2. https://doi.org/10.1093/isd/ixaa009
Germar, E. F. (1838). Hemiptera Heteroptera promontorii bonae spei, nondum descripta, quae collegit C.F. Drège. Revue Entomologique, 5, 121–192.
Gerstaecker, A. (1873). Baron Carl Claus von der Decken's Reisen in Ost-Africa. Gliederthiere (Insecten, Arachniden, Myriapoden und Isopoden). Leipzig und Heidelberg : C.F. Winter'ische Verlagshandlung
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., & Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Systematic Biology, 59, 307–321. https://doi.org/10.1093/sysbio/syq010
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., & Vinh, L. S. (2018). UFBoot2: Improving the ultrafast bootstrap approximation. Molecular Biology and Evolution, 35, 518–522. https://doi.org/10.1093/molbev/msx281
Herrich-Schäffer. (1835). Favnae insectorvm Germanicae initia, oder, Deutschlands Insecten. Heft 127. Nürnberg: In den Felseckerschen Buchhandlung.
Herrich-Schäffer. (1847). Die wanzenartigen Insecten. Getreu nach der Natur abgebildet und beschrieben. Achter Band. Nürnberg: J. L. Lotzbeck.
Hosner, P. A., Faircloth, B. C., Glenn, T. C., Braun, E. L., & Kimball, R. T. (2016). Avoiding missing data biases in phylogenomic inference: An empirical study in the landfowl (Aves: Galliformes). Molecular Biology and Evolution, 33, 1110–1125. https://doi.org/10.1093/molbev/msv347
Hsaio, T.-Y. (1965). New coreids from China (Hemiptera, Heteroptera) IV. Acta Zoologica Sinica, 17, 421–434.
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., & Jermiin, L. S. (2017). ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods, 14, 587–589. https://doi.org/10.1038/nmeth.4285
Kieran, T. J., Gordon, E. R., Forthman, M., Hoey-Chamberlain, R., Kimball, R. T., Faircloth, B. C., Weirauch, C., & Glenn, T. C. (2019). Insight from an ultraconserved element bait set designed for hemipteran phylogenetics integrated with genomic resources. Molecular Phylogenetics and Evolution, 130, 297–303. https://doi.org/10.1016/j.ympev.2018.10.026
Knyshov, A., Gordon, E. R. L., & Weirauch, C. (2019). Cost-efficient high throughput capture of museum arthropod specimen DNA using PCR-generated baits. Methods in Ecology and Evolution, 10, 841–852. https://doi.org/10.1111/2041-210X.13169
Kolenati, F. A. (1845). Hemiptera Caucasi Tesseratomidae: Monographice Dispositae. Petropoli: Imperialis Academiae Scientiarum.
Kumar, R. (1965). Aspects of the morphology of Coreoidea and their value in its higher classification. Proceedings of the Royal Society of Queensland, 76, 27–91.
Li, X. (1996). Cladistic analysis and higher classification of Coreoidea. Entomologia Sinica, 3, 283–292.
Li, X. Z. (1997). Cladistic analysis of the phylogenetic relationships among the tribal rank taxa of Coreidae (Hemiptera-Heteroptera: Coreoidea). Acta Zootaxonomica Sinica, 22, 60–68.
Li, H. M., Deng, R. Q., & Wang, X. Z. (2006). Phylogenetic relationships of the Pentatomomorpha (Hemiptera: Heteroptera) inferred from nuclear 18S rDNA sequences. Zoological Research, 27, 307–316.
Li, M., Wang, Y. H., Xie, Q., Tian, X. X., Li, T., Zhang, H. F., & Bu, W. J. (2016). Reanalysis of the phylogenetic relationships of the Pentatomomorpha (Hemiptera: Heteroptera) based on ribosomal, Hox and mitochondrial genes. Entomotaxonomia, 38, 81–91.
Li, X., & Zheng, L. (1993). Preliminary study on the phylogeny of Alydidae (Hemiptera: Coreoidea). Acta Zootaxonomica Sinica, 18, 330–343.
Maldonado, J. (1953). Redescription of the genus Burtinus Stal and description of a new species from Puerto Rico. Proceedings of the Entomological Society of Washington, 58, 40–44.
Meiklejohn, K. A., Faircloth, B. C., Glenn, T. C., Kimball, R. T., & Braun, E. L. (2016). Analysis of a rapid evolutionary radiation using ultraconserved elements: Evidence for a bias in some multispecies coalescent methods. Systematic Biology, 65, 612–627. https://doi.org/10.1093/sysbio/syw014
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., & Lanfear, R. (2020). IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution, 37, 1530–1534. https://doi.org/10.1093/molbev/msaa015
Mirarab, S., Reaz, R., Bayzid, M. S., Zimmermann, T., Swenson, M. S., & Warnow, T. (2014). ASTRAL: Genome-scale coalescent-based species tree estimation. Bioinformatics, 30, i541–i548. https://doi.org/10.1093/bioinformatics/btu462
Mitchell, P. L. (2000). Leaf-footed bugs (Coreidae). In C. W. Schaefer & A. R. Panizzi (Eds.), Heteroptera of economic importance (pp. 337–403). CRC Press LLC.
Naser-Khdour, S., Minh, B. Q., Zhang, W., Stone, E. A., & Lanfear, R. (2019). The prevalence and impact of model violations in phylogenetic analysis. Genome Biology and Evolution, 11, 3341–3352. https://doi.org/10.1093/gbe/evz193
Packauskas, R. (2010). Catalog of the Coreidae, or leaf-footed bugs, of the New World. Fort Hays Studies, Fourth Series, 5, 1–270.
Pan, X., Su, F., & Song, Y. (2008). Phylogeny of partial species of Coreidae based on mitochondrial cytochrome b gene sequence. Sichuan Journal of Zoology, 27, 21–25.
Pluot-Sigwalt, D., & Moulet, P. (2020). Morphological types of spermatheca in Coreidae: bearing on intra-familial classification and tribal-groupings (Hemiptera: Heteroptera). Zootaxa, 4834, 451–501. https://doi.org/10.11646/zootaxa.4834.4.1
Prjibelski, A., Antipov, D., Meleshko, D., Lapidus, A., & Korobeynikov, A. (2020). Using SPAdes de novo assembler. Current Protocols in Bioinformatics, 70, e102. https://doi.org/10.1002/cpbi.102
Procter, D. S., Moore, A. J., & Miller, C. W. (2012). The form of sexual selection arising from male–male competition depends on the presence of females in the social environment. Journal of Evolutionary Biology, 25, 803–812. https://doi.org/10.1111/j.1420-9101.2012.02485.x
Rossi, P. (1790). Fauna Etrusca Sistens Insecta Quae In Provinciis Florentina Et Pisana Praesertim, Tomus Secundus. Typis Thomae Masi et Sociorum.
Sahlberg, R. F. (1848). Monographia Geocorisarum Fenniae. Ex officina typographica Frenckelliana.
Sayyari, E., & Mirarab, S. (2016). Fast coalescent-based computation of local branch support from quartet frequencies. Molecular Biology and Evolution, 33, 1654–1668. https://doi.org/10.1093/molbev/msw079
Schaefer, C. W. (1964). The morphology and higher classification of the Coreoidea (Hemiptera-Heteroptera): Parts I and II. Annals of the Entomological Society of America, 57, 670–684. https://doi.org/10.1093/aesa/57.6.670
Schaefer, C. W. (1965). The morphology and higher classification of the Coreoidea (Hemiptera-Heteroptera). Part III. The Families Rhopalidae, Alydidae, and Coreidae. Miscellaneous Publications of the Entomological Society of America, 5, 1–76.
Schaefer, C. W. (1972). Degree of metathoracic scent-gland development in the trichophorous Heteroptera (Hemiptera). Annals of the Entomological Society of America, 65, 810-821.
Schaefer, C. W. (1980). The host plants of the Alydinae, with a note on heterotypic feeding aggregations (Hemiptera: Coreoidea: Alydidae). Journal of the Kansas Entomological Society, 53, 115–122.
Schaefer, C. W. (1982). The genital capsule of the Hydarinae (Hemiptera: Coreidae). Uttar Pradesh Journal of Zoology, 2, 1–6.
Schaefer, C. W. (1999). The higher classification of the Alydidae (Hemiptera: Heteroptera). Proceedings of the Entomological Society of Washington, 101, 94–98.
Schuh, R. T., & Weirauch, C. (2020). True bugs of the world (Hemiptera: Heteroptera): Classification and natural history (2nd ed.). Siri Scientific Press.
Scopoli, G. A. (1763). Entomologia Carniolica : Exhibens Insecta Carnioliae Indigena et Distributa in Ordines, Genera, Species, Varietates : Methodo Linnaeana. Vindobonae : Typis Ioannis Thomae Trattner.
Shadab, M. U. (1972). A new genus of pseudophloeine bugs from the Democratic Republic of the Congo (Heteroptera, Coreoidea). American Museum Novitates, 2493, 1–11.
Signoret, V. (1861). Faune des Hémiptères de Madagascar. (Suite et fin.). Hétéroptères. Annales de la Société Entomologique de France, 8, 917–972.
Sullivan, J., Swofford, D. L., & Naylor, G. J. P. (1999). The effect of taxon sampling on estimating rate heterogeneity parameters of maximum-likelihood models. Molecular Biology and Evolution, 16, 1347–1356.
Stål, C. (1854). Bidrag till Hemipterernas systematik. Öefversigt Af Kongliga Vetenskaps-Akademiens Förhandlingar, 11, 231–255.
Stål, C. (1866). Hemiptera Africana. Tomus Secundus. Holmiae: Ex officina Norstedtiana.
Stål, C. (1867). Bidrag till Hemipterernas systematik. Öefversigt Af Kongliga Vetenskaps-Akademiens Förhandlingar, 24, 491–560.
Stål, C. (1870). Enumeratio Hemipterorum: bidrag till en förtechkning öfver alla hittills kӓnda Hemiptera, jemte systematiska meddelanden. Kongliga Svenska Vetenskaps-Akademiens Handlingar, 9, 1–232.
Stål, C. (1873). Enumeratio Hemipterorum. Bidrag till en förteckning öfver alla hittills kända Hemiptera, jemte systematiska meddelanden. 3. Kongliga Svenska Vetenskaps-Akademiens Handlingar, 11(2), 1–163.
Štys, P. (1977). Hind wing venation of Coreidae (Heteroptera): A history of misinterpretation. Acta Entomologica Musei Nationalis Pragae, 39, 263–269.
Valero, M. C., Ojo, J. A., Sun, W., Tamò, M., Coates, B. S., & Pittendrigh, B. R. (2017). The complete mitochondrial genome of Anoplocnemis curvipes F. (Coreinea, Coreidae, Heteroptera), a pest of fresh cowpea pods. Mitochondrial DNA Part B, 2, 421–423. https://doi.org/10.1080/23802359.2017.1347829
Wang, Y. H., Cui, Y., Rédei, D., Baňař, P., Xie, Q., Štys, P., Damgaard, J., Chen, P., Yi, W., Wang, Y., Dang, K., Li, C., & Bu, W. (2016). Phylogenetic divergences of the true bugs (Insecta: Hemiptera: Heteroptera), with emphasis on the aquatic lineages: The last piece of the aquatic insect jigsaw originated in the Late Permian/Early Triassic. Cladistics, 32, 390–405. https://doi.org/10.1111/cla.12137
Westwood, J. O. (1842). A Catalogue of Hemiptera, in the Collection of the Rev. F. W. Hope, with Short Latin Descriptions of the New Species. Part II. London: J. C. Bridgewater.
Woodman, T. E., Chen, S., Emberts, Z., Wilner, D., Federle, W., & Miller, C. W. (2021). Developmental nutrition affects the structural integrity of a sexually selected weapon. Integrative and Comparative Biology, 61, 723–735. https://doi.org/10.1093/icb/icab130
Yang, Z. (2006). Computational Molecular Evolution. Oxford University Press.
Zhang, C., Rabiee, M., Sayyari, E., & Mirarab, S. (2018). ASTRAL-III: Polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics, 19, 153.
Zhang, C., Sayyari, E., & Mirarab, S. (2017). ASTRAL-III: Increased scalability and impacts of contracting low support branches. In J. Meidanis & L. Nakhleh (Eds.), Comparative Genomics: 15th International Workshop, RECOMB CG 2017, Barcelona, Spain, October 4–6, 2017, Proceedings (pp. 53–75). Springer International Publishing.
Zhao, Q., Wang, J., Wang, M., Cai, B., Zhang, H., & Wei, J. (2018). Complete mitochondrial genome of Dinorhynchus dybowskyi (Hemiptera: Pentatomidae: Asopinae) and phylogenetic analysis of Pentatomomorpha species. Journal of Insect Science, 18, 44. https://doi.org/10.1093/jisesa/iey031
Zwickl, D. J. (2006). Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Ph.D. dissertation, The University of Texas at Austin.
Acknowledgements
Christiane Weirauch, Paul Masonick, Joe Eger, Petr Kment, Marcos Roca-Cusachs, Vasily Grebennikov, Field Museum of Natural History, Florida State Collection of Arthropods, and California Academy of Sciences contributed specimens. Bob McCleery, Cebisile N. Magagula, and the Savannah Research Center facilitated specimen collection in eSwatini. South African specimens were acquired under Ezemvelo KZN Wildlife permit #OP 172/2018 and the iSimangaliso Wetland Park Authority (World Heritage Site). Additional samples were collected with funding by University of Florida Research Abroad for Doctoral Students, Lee Kong Chian Natural History Museum fellowship, and National Science Foundation (NSF) OISE‐1614015 to Zach Emberts (Australia permit #01‐000204‐1; Singapore permit #NP/RP17‐012); the Society of the Study of Evolution Rosemary Grant; Smithsonian Tropical Research Institute Short Term Fellowship; and Systematics, Evolution, and Biodiversity Endowment Award (Entomological Society of America) to Ummat Somjee; and NSF DEB-0542864 to Michael Sharkey and Brian Brown. Caroline Miller prepared habitus images. Nathan Friedman, Caroline Miller, and Min Zhao assisted with Molecular Benchwork.
Funding
This study was funded by the National Science Foundation IOS‐1553100 (awarded to C.W. Miller).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
13127_2022_548_MOESM1_ESM.pdf
Supplementary file1 Supplementary figures of maximum likelihood best trees (Figs. S1–S7) and ASTRAL-III species trees (Figs. S8–S15) estimated from various datasets used in this study. (PDF 3904 KB)
13127_2022_548_MOESM4_ESM.xlsx
Supplementary file4 Table S3 Summary of parsimony-informative sites and number of UCE loci in each concatenated dataset. (XLSX 12 KB)
13127_2022_548_MOESM5_ESM.xlsx
Supplementary file5 Table S4 Summary of parsimony-informative sites and number of UCE loci in each dataset used for summary coalescent analyses. (XLSX 13 KB)
Rights and permissions
About this article
Cite this article
Forthman, M., Miller, C.W. & Kimball, R.T. Phylogenomic analysis with improved taxon sampling corroborates an Alydidae + Hydarinae + Pseudophloeinae clade (Heteroptera: Coreoidea: Alydidae, Coreidae). Org Divers Evol 22, 669–679 (2022). https://doi.org/10.1007/s13127-022-00548-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13127-022-00548-w