Abstract
Pteropodidae constitutes one of the most diverse bat families. These bats have evolved a phytophagous diet, likely lost laryngeal echolocation capability, and attained the largest body sizes among bats. Previous phylogenetic studies suggested that the family might have experienced an explosive diversification at its origin. Here, we readdress this hypothesis using a macroevolutionary approach based on Bayesian statistics (BAMM), a sampling of 139 pteropodid species, and divergence date estimates obtained in a comprehensive phylogenetic study of Chiroptera with multiple fossil calibration points. We evaluated the effect of missing data and of a reduced outgroup by repeating the analyses across simulated complete phylogenies and across a comprehensive Yinpterochiroptera phylogeny, respectively. Additionally, we performed an alternative analysis to detect diversification-rate shifts through time, the birth-death-shift method. In contrast with a previous study, we found strong statistical signals of rapid diversification at the origin of Pteropodidae. BAMM also detected diversification-rate shifts (increases) at the origin of Pteropus, as well as at crown Hipposideridae and Rhinolophidae. The birth-death-shift method detected a shift at approximately 25 million years ago, the estimated crown ages of both Pteropodidae and Hipposideridae. Our results point to a complex dynamics in the evolution of bat families, likely related to key innovations, demographic factors, and environmental opportunity enhanced by global-scale climatic and geographic changes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
One fundamental question in biology is how and why diversity varies between groups of organisms (Cornwell and Nakagawa 2017). Differences in diversity are observed in a variety of groups and at different taxonomic levels such as among vertebrate classes, mammalian orders, bat families, and butterfly genera (Alfaro et al. 2009; Jones et al. 2005; Mullen et al. 2011; Stadler 2011a). The number of species is the most obvious aspect of diversity in which groups differ between each other, but disparities are also observed in morphology, diet types, mating systems, and virtually every other organismic feature. Some taxa have high diversification rates as a result of high speciation rates, low extinction rates, or a combination of both, which are products of complex processes involving both extrinsic and intrinsic factors, such as climate, ecological opportunity, and adaptive novelties (Ricklefs 2007; Yoder et al. 2010; Dumont et al. 2012; Givnish 2015). However, differences in diversity are expected because the processes that generate biodiversity (i.e., speciation and genetic differentiation) are stochastic to varying degree. Thus, to determine whether a group presents a significantly elevated (or reduced) diversification rate, it is necessary to use sophisticated statistical methods within a time-scaled, comprehensive phylogenetic framework (e.g., Chan and Moore 2002; Rabosky 2006; Rabosky and Glor 2010; Stadler 2011b; Rabosky et al. 2014).
We focus here in one group within bats (Chiroptera: Mammalia), an order that includes ca. 1400 currently recognized extant species classified into 21 families (Wilson and Mittermeier 2019; Simmons and Cirranello 2020). This remarkable diversity is, among the mammalian orders, only second to that of rodents. Bats have also evolved a notable variety of morphofunctional, ecological, and ethological adaptations. Remarkably, the number of species greatly varies among currently recognized families, from just 1 (in Craseonycteridae) or 2 (in Cistugidae, Myzopodidae, and Mystacinidae) to more than 400 in Vespertilionidae (Simmons and Cirranello 2020), prompting hypotheses of accelerated diversification rates in several families (e.g., Jones et al. 2005; Lack and Van Den Bussche 2010; Fenton 2010; Almeida et al. 2011; Monteiro and Nogueira 2011; Rojas et al. 2012). Nevertheless, these hypotheses have not been generally supported. In particular, Shi and Rabosky (2015) addressed diversification rates in bats using a supermatrix tree including 812 bat species, but they found only a single diversification rate shift located at the base of Stenodermatinae, a subfamily of New World leaf-nosed bats (Phyllostomidae) with frugivorous habits, thereby rejecting all other hypotheses of rapid diversification within Chiroptera. Shi and Rabosky (2015) concluded that speciation rates decelerated toward the present, and, more importantly, that diversification rates have been relatively homogeneous across the different chiropteran clades; this, in spite of the dramatic richness differences observed across chiropteran families (see above).
Pteropodidae (Old World fruit bats), with approximately 200 species (45 genera) and several peculiar morphofunctional traits, stands among the highly diverse bat families (Wilson and Mittermeier 2019; Simmons and Cirranello 2020). While a majority of bats feed on insects, Pteropodidae is the only exclusively phytophagous bat family. In fact, phytophagy has rarely evolved in bats, but intensely so within Phyllostomidae and in the ancestor of all pteropodids (Giannini 2019). Pteropodids are also outstanding among bats for lacking the laryngeal echolocation capability present in all other bat families (probably as a result of a secondary loss; see Teeling et al. 2012; Boonman et al. 2014; Wang et al. 2017; Thiagavel et al. 2018), and for their wide interspecific body size variance, including the by far largest bat species (Nowak 1994). Mean body size in species of Old World fruit bats ranges from 14 to ca. 1500 g, while in all other bat families, body size has a median value of ~ 16 g (never exceeding 250 g; see Moyers Arévalo et al. 2020 and citations therein). The evolution of very large sizes in pteropodids has been hypothesized to be a consequence of the functional release from physical constraints imposed by echolocation on body size (see Moyers Arévalo et al. 2020 and citations therein). Lack of sophisticated laryngeal echolocation made pteropodids dependent upon olfaction and vision to navigate in search for food, mates, and roost, although echolocation based on tongue-clicking (in Rousettus) and wing clicking (in several other genera) evolved secondarily in a few pteropodids allowing these forms to inhabit dark caves (Yovel et al. 2011; Boonman et al. 2014).
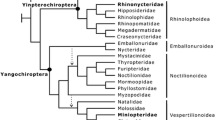
Pteropodids are restricted to the tropics of Africa, Asia, and Oceania, including many islands of the Indian and Pacific Oceans (Nowak 1994; Giannini 2019). This family composes the clade Yinpterochiroptera (Teeling et al. 2005; Amador et al. 2018) along with the superfamily Rhinolophoidea, which includes six families of insectivorous bats, also distributed in the Old World. The first comprehensive phylogeny of Pteropodidae included 56 terminals representing 42 genera, and it was based on an 8181 bp molecular matrix that combined mitochondrial and nuclear loci (Almeida et al. 2011). Despite recovering several suprageneric relationships with high support (including many of the subfamilies proposed by Bergmans 1997), the analyses were not able to resolve the family’s basal relationships. Both systematic bias and phylogenetic conflict were statistically dismissed as causes of the polytomy. This result led the authors to hypothesize that crown pteropodids experienced an explosive radiation, which would account for the blurred phylogenetic signal at the base of their phylogeny and for their high taxonomic and morphological diversity. More recently, several phylogenetic studies analyzed ever more comprehensive samplings of Pteropodidae at the species level and confirmed the difficulty in resolving its deep relationships (Shi and Rabosky 2015; Amador et al. 2018; Almeida et al. 2020). Nevertheless, the hypothesis of high diversification rate at the origin of Pteropodidae was not supported by a recent macroevolutionary study on bats (Shi and Rabosky 2015).
Diversification rates are intrinsically dependent on divergence time estimates. Yet, there are significant differences between the estimates of bat families ages obtained in different studies. In the particular case of Pteropodidae, the origin of the family has been dated, depending on the study, from ~ 25 million years ago (mya; Teeling et al. 2005, Jones et al. 2005, Amador et al. 2018) to ~ 40 mya (Shi and Rabosky 2015). These important differences in clade age estimates prompted us to readdress the hypothesis of an explosive radiation of Pteropodidae (Almeida et al. 2011) using macroevolutionary methods that allow for the detection of diversification rate shifts on a dated phylogeny. Our analyses were based on a recent, updated tree for the family (Almeida et al. 2020) that includes 75% of the currently recognized species. We used age estimates obtained in the study of Amador et al. (2018), which included 804 bat terminals and 44 fossil-based calibration points (the largest sample so far in bat studies), anchoring the origin of Pteropodidae at ca. 26 mya. We controlled for the effects of outgroup size using simulations and different datasets. The results showed a strong statistical support for an increased diversification rate in the origin of Pteropodidae, in agreement with the advanced hypothesis. Surprisingly, similar results were also obtained for other large bat clades related to Pteropodidae, thus questioning the general perception of bats evolving at uniform rates since the Eocene.
Methods
Phylogenetic tree
Our analyses used a phylogenetic hypothesis obtained from a matrix that includes 139 pteropodid species, representing all 45 currently recognized pteropodid genera, plus six outgroup species (Almeida et al. 2020). The majority of the ~ 47 pteropodid species missing from the matrix belong to the highly diverse genera Nyctimene (~ 15 species), Dobsonia (~ 14), and Pteropus (~ 63), which were represented in the matrix by 6, 8, and 51 species, or 40%, 57%, and 81% of genus richness, respectively. Species richness in Pteropodidae is only a rough estimate as several pteropodid genera are awaiting taxonomic revision, and genetic samples have been difficult to obtain for the missing species. The six outgroup species belong in the superfamily Rhinolophoidea, which has been consistently recovered as sister to Pteropodidae (e.g., Teeling et al. 2000; Lei and Dong 2016; Amador et al. 2018). Our supermatrix included eight loci; nuclear loci included exon 28 of the von Willebrand Factor gene (vWF, 1230 bp), partial recombination activating gene 1 (RAG1, 1084 bp), partial recombination activating gene 2 (RAG2, 760 bp), and partial breast cancer 1 gene (BRCA1, about 1370 bp); mitochondrial loci included complete cytochrome b gene (Cytb, 1140 bp), partial rRNA 12S gene (1069 bp), the valine tRNA gene (67 bp), and partial rRNA 16S gene (1330 bp). Phylogenetic relationships were inferred using both maximum likelihood and Bayesian methods and were the basis for a new classification of pteropodids (Almeida et al. 2020). Further details on the molecular methods, phylogenetic analysis, and taxonomic implications of the results are described in Almeida et al. (2020).
Dated phylogeny
The first step for the diversification analyses was to obtain a dated tree using Bayesian methods implemented in the program BEAST 1.8.4 (Drummond and Rambaut 2007). A drawback for divergence time estimation in Pteropodidae is the dearth of fossils assigned to this family in general and the lack of old fossils in particular. We therefore used divergence dates from Amador et al. (2018) as the most reliable estimates obtained so far because their analysis included a comprehensive sampling of bat diversity (~ 800 species from all families), and dating was based on multiple, taxonomically undisputed fossil calibration points (44 in total) across the entire order Chiroptera. From that analysis, we took seven time confidence intervals for cladogenetic events as soft bounds with normal distribution in order to calibrate our pteropodid phylogeny (Supporting Information Table S1). The sequence data were separated into the optimum partitioning scheme to which were applied the best substitution models according to PartitionFinder2 (Lanfear et al. 2017). The analysis was set to use an uncorrelated relaxed clock (Drummond et al. 2006) and a birth-death process with incomplete sampling as tree prior. A Markov Chain Monte Carlo (MCMC) was set in two runs of 15 million generations, sampling every 5000 generations, after which convergence was checked with Tracer. Results of the two runs were combined using LogCombiner and resumed in a consensus tree with TreeAnnotator (both from BEAST 1.8.4 package, Drummond and Rambaut 2007). Resulting divergence date estimates were very similar to the ones obtained in Amador et al. (2018), generally within ± 1 my.
Macroevolutionary analysis
As a first approach, we calculated the γ-statistic and its significance for the pteropodid tree using the package phytools for R (Revell 2012). The γ-statistics is a measure of diversification-rate trends along the phylogeny: positive values are associated with increasing rates, while negative values reflect higher rates at the clade origin with a subsequent decrease (Pybus and Harvey 2000).
To test the hypothesis of an explosive radiation of crown pteropodids, we used the Bayesian method introduced by Rabosky et al. (2014) and implemented in the program BAMM v. 2.5.0 (Bayesian Analysis of Macroevolutionary Mixtures, Rabosky et al. 2014). The method uses reversible jump to find shifts in evolutionary regimes (i.e., speciation and extinction rates) in a time-calibrated phylogeny. This Bayesian framework allows extracting the probability of different shift events that may have occurred in the phylogeny from their relative frequencies in the MCMC output, and the significance of differences in these probabilities can be assessed with Bayes factor. The analysis assumes that transitions between regimes occur under a Poisson process with a minimum number of shifts equal to zero (i.e., the entire phylogeny evolves under the same rate) and no upper limit. To generate priors for the analysis, we used the package BAMMTools v. 2.1.6 for R (Rabosky et al. 2014). We ran BAMM using these priors for 1 million generations, sampling every 1000 generations. The results were summarized and visualized with BAMMTools. Upon inspection of the MCMC log likelihoods, we discarded the first 10% generations as burn-in, resulting in effective sample sizes of 694 and 758 for the number of shifts and log likelihood, respectively. The analysis was rerun twice, increasing the number of expected shifts to 2 and 3, but the results did not change. We obtained from BAMM the number of shifts favored by Bayes factor, best shift configuration, 95% credibility set of shift configuration, and macroevolutionary cohorts (i.e., lineages with high pairwise probabilities of having originated under the same diversification processes; Rabosky et al. 2014).
Additionally, we calculated the species-level lineage diversification rate (DR; Jetz et al. 2012) to check for more recent shifts in diversification rates. The DR statistic is less conservative compared with BAMM, allowing for the detection of more sudden shifts, although this estimate presents a larger variance (Title and Rabosky 2017).
Effect of missing taxa
Diversification analysis should be ideally run on complete phylogenies (no missing taxon), but our dataset included only about 75% of known pteropodid species. To evaluate the effect of missing taxa in the BAMM analysis and the possibility of additional shifts within Pteropodidae, we simulated complete sampling phylogenies including 186 pteropodid species based on Simmons (2005; more recent estimates point to ~ 200 extant species). For that, we added missing pteropodid species to the matrix and scored missing data to all characters. We then set MrBayes v. 3.2.6 (Ronquist and Huelsenbeck 2003) searches constraining the monophyly of the genera with species with all missing data. We also set divergence time constraints to the speciose genera with many missing species (i.e., Dobsonia, Nyctimene, Pteropus, and Rousettus) setting the ages according to the Pteropodidae dated tree obtained herein. For Nyctimene, we set a uniform age prior varying from 9.3 to 14 million years (my), which were the mean crown Nyctimene age and the mean age of the split from Paranyctimene (sister genus), respectively. For Dobsonia, we used the same approach and applied the range 6.8 to 14 my, considering the divergence with respect to Aproteles. For Pteropus and Rousettus, for which we had more comprehensive samplings in previous phylogenies (e.g., Almeida et al. 2014; Amador et al. 2018; Tsang et al. 2020), we set fixed ages at 8.28 and 10.4 my (crown ages), respectively. We also fixed the stem and crown Pteropodidae ages at 55.4 and 25.3, respectively, following our previous estimates (Amador et al. 2018). A strict clock was applied with a rate normally distributed with mean 0.0041 substitutions per site per million year and standard deviation of 0.001 (as previously estimated; Amador et al. 2018). The resulting consensus tree is available as supporting information (Fig. S1). From the posterior distribution of the obtained trees, after discarding 20% samples as burn-in, we randomly selected 10 trees to run BAMM as described above.
Effects of divergence time estimates and reduced outgroup
To evaluate whether the use of a reduced outgroup (only six species) could be affecting the detection of a shift at the origin of Pteropodidae, we ran BAMM on trees simulated under two different time regimes. Using the TreeSim v.2.4 package for R (Stadler 2011b) and the sim.bd.age.taxa command, we obtained 20 trees containing 139 in-group terminals. In half of the trees, the in-group was constrained to have diverged in the last 25 million years (my), and in the other half, in the last 57 my (estimated date of the split between Pteropodidae and the other yinpterochiropteran families). We then grafted into those simulated trees the same outgroup clade from the original Pteropodidae tree (see above) in a way that the total time was similar to that of the real dated phylogeny. The simulated phylogenies were analyzed with BAMM to identify rate shifts and their branch location in case they were detected.
To overcome a possible bias caused by the use of a reduced outgroup in the Pteropodidae tree, we ran an additional analysis using the entire Yinpterochiroptera tree. For this analysis, we used the dated tree from Amador et al. (2018) and pruned off branches to leave only the Yinpterochiroptera clade (including Rhinolophoidea and Pteropodidae) plus eight Yangochiroptera species (representatives of most chiropteran families) as outgroup. This dataset includes a total of 227 in-group species (represented by 229 terminals): 114 Pteropodidae (out of 186), 45 Hipposideridae (81), 55 Rhinolophidae (83), 6 Rhinonycteridae (9), 3 Rhinopomatidae (4), 3 Megadermatidae (5), and 1 Craseonycteridae (1). Then, we ran BAMM as previously described for 20 million generations, keeping the results every 2000 generations. The first 2000 samples (20%) were discarded as burn-in, and the effective sample sizes of the number of shifts and log likelihood were 1284 and 537, respectively. Again, the analysis was run several times using different priors on the numbers of expected shifts, without significant changes in the results.
Alternative diversification analysis with TreePar
As an alternative to BAMM, we applied the method proposed by Stadler (2011a) to detect diversification rate shifts through time. This method uses the birth-death-shift model to derive the likelihood of different number of shifts, given estimated diversification rates (speciation rate minus extinction rate) and turnover (extinction/speciation rates) for fixed time intervals. In contrast to BAMM, the birth-death-shift method does not identify clades where rate shifts occurred, but instead time periods that experienced rate shifts in the history of a group. Derived likelihoods are then compared with a likelihood ratio test to determine which number of shifts is best supported. This method is implemented in the R package TreePar v. 3.3 (Stadler 2011c) and was applied to both the novel Pteropodidae tree and the Yinpterochiroptera tree pruned from Amador et al. (2018). The analysis was carried out without outgroup and accounting for incomplete sampling (75% of pteropodids, 60% of yinpterochiropterans), with a grid of 1 million years.
Results
The γ-statistic was not significant for the Pteropodidae dataset analyzed (γ = 1.14, p = 0.26), indicating that no general trend in diversification rate can be detected for the group using this sampling and phylogenetic hypothesis. Nevertheless, the γ-statistic was significantly positive for 9 of the 10 “taxonomically complete” simulated trees analyzed (ranging from1.4 to 2.75, Supporting Information Table S2), suggesting an increase in the diversification rate toward their tips. A detectable increase in diversification rates in the recent history of pteropodids is likely due to the explosive diversification of the genus Pteropus and possibly other genera (see below).
Results of BAMM applied to Pteropodidae indicated that two shifts occurred with the highest posterior probability (pp = 49%, Bayes factor = 517), while three shifts had the second highest pp (31%, Fig. 1a). The best shift configuration placed one shift at the origin of crown Pteropodidae and the other shift in the origin of Pteropus as suggested by the marginal posterior probabilities (Fig. 1 b and c). Three macroevolutionary cohorts were recovered, i.e., three sets of taxa that differ among each other in macroevolutionary parameters, which is in agreement with two rate shifts (Supporting Information Fig. S2). The 95% credible shift set also included, albeit with a much lower probability, a third shift in Epomophorinae (Supporting Information Fig. S3). Rate-through-time plots clearly captured the initial increase of speciation rate, followed by stability, and a secondary increase in Pteropus (Fig. 2). These two shifts may cancel out across the entire family and thus explain the lack of a general trend pointed out by the γ-statistics (see previous paragraph).
Results of the BAMM applied to the pteropodid phylogeny. a Posterior distribution of the number of diversification shifts in all replicates. b Tree in which branch lengths are proportional to the marginal probability of a shift occurring along that branch. c Time-calibrated pteropodid tree showing the best shift configuration and colored according to estimated net diversification rates. Below the tree is a time axis in million years before present
When BAMM was applied to the taxonomically complete trees that included all pteropodid species (see “Methods”), the results were very similar to those obtained with the “real data” tree, but with a third shift in Epomophorinae appearing in the best shift configuration of 8 out of 10 analyzed trees (Supporting Information Table S1, Fig. S4). In one of those trees, a fourth shift was placed in the genus Dobsonia. These extra shifts revealed in the analysis of recreated, taxonomically complete trees were hypothetical in the sense that missing species were introduced assuming their phylogenetic positions (i.e., placed in the corresponding genus) and need to be tested with a real data tree for all pteropodid species. Nevertheless, these results were supported by the DR statistic, which had higher values in the same three pteropodid genera: Pteropus, Dobsonia, and Epomophorus (Supporting Information Fig. S5).
The results of BAMM applied to the Pteropodidae tree were challenged, however, by the analysis of simulated trees. In all 20 trees, obtained with both age schemes, a diversification shift was detected at the origin of the in-group clade (simulating Pteropodidae) (Supplementary Information, Figs. S6 and S7). This is an important result that shows how BAMM is affected by the outgroup choice if the hypothesis to test involves a basal tree branch. Although the different dating schemes did not affect the number of shifts detected in simulated trees, they directly affected the diversification rates, as expected (Supplementary Information, Figs. S6 and S7): the 57-my scheme resulted in lower rates that kept relatively stable or slightly increased from origin to tips, whereas the 25-my scheme resulted in slightly higher rates at the in-group origin that decreased through time, more similar to what we obtained with the real Pteropodidae data.
Because the results obtained from the analyses of simulated trees suggested that the BAMM results for the Pteropodidae tree might be spurious, we applied BAMM to a real data tree including a larger outgroup set, i.e., Rhinolophoidea (114 terminals), and a few yangochiropteran species (6 terminals). In this tree, Pteropodidae was represented by 118 species making the ratio between in-group and outgroup more even. BAMM favored four diversification rate shifts (Bayes factor = 76), with five shifts being the second best scheme. It confirmed the shifts observed in the first analysis, i.e., at crown Pteropodidae and Pteropus, and surprisingly suggested that both Hipposideridae (minus Rhinonycteridae; see Foley et al. 2015, Amador et al. 2018) and Rhinolophidae also had increased diversification rates at their origins (Fig. 3); this result was supported by both marginal posterior probabilities (Supporting Information, Fig. S8) and cohort analysis (Fig. 4). The three most probable shift configurations of the 95% credibility set also supported the four shifts observed in the best shift configuration (Supporting Information, Fig. S9). Rate estimates revealed that such pattern might be due to strong and moderate increases in speciation rates in Pteropus and Rhinolophidae, respectively, while Pteropodidae and Hipposideridae had slight increases in speciation rates coupled with a reduction in extinction rates (Supporting Information, Fig. S10).
Best shift configuration obtained with BAMM for a time-calibrated Yinpterochiroptera tree, showing branches where shifts have occurred (red circles) and colored according to net diversification rates. Below the tree is a time axis in million years before present and a temperature through time curve based on the δ18O isotope as estimated by Zachos et al. (2001)
Applying the likelihood-based method implemented in TreePar, the best fitting model for the Pteropodidae phylogeny included a single rate shift (LRT, p = 0.97). This shift represents a fivefold decrease in the diversification rate at 21 mya, from the high rate (0.48 s/my) estimated for the first ~ 4 million years of pteropodid evolution (Fig. 5a), mainly due to a substantial decrease in the speciation rate (although the extinction rate also experienced a decrease). When the method was applied to the Yinpterochiroptera dataset, a single shift was detected (LRT, p = 1): an ~ 2.7 times increase in the diversification rate at ~ 25 mya (from 0.05 to 0.14 s/my); this shift is coincident with the origin of both crown Pteropodidae and Hipposideridae (Fig. 5b). The shifts revealed by BAMM at the origins of Rhinolophidae and Pteropus were not detected by this method.
Discussion
Explosive diversification in Pteropodidae
The hypothesis that Pteropodidae went through an explosive diversification at about 25 million years ago was raised by Almeida et al. (2011) to explain the apparent hard polytomy observed at the origin of the crown group (following Whitfield and Lockhart 2007), after statistically discarding several potential biases in the data. This hypothesis was put to test in the broad phylogenetic framework of Chiroptera by Shi and Rabosky (2015), but their results did not show evidence supporting it. Here, we readdressed this hypothesis using divergence dates estimated in a comprehensive, fossil-calibrated analysis (Amador et al. 2018) and a dataset that included additional pteropodid sequences. The results of both methods applied herein supported the explosive diversification hypothesis recovering elevated rates at the origin of Pteropodidae. Our analyses also recovered an increased diversification rate at the origin of Pteropus, as previously suggested by Shi and Rabosky (2015) and long suspected by many systematists (e.g., Almeida et al. 2014).
We argue that the main reason for the disagreement between our results and those of a previous study where the same method did not detect shifts at the base of Pteropodidae and other Yinpterochiroptera families (Shi and Rabosky 2015) lies in differences in node ages between dated trees. As expected and empirically demonstrated, node age estimates have a significant effect on results of macroevolutionary analysis (Title and Rabosky 2017). The other possible reason for the disagreement was sampling size, as we were able to include 139 Pteropodidae species, as compared with the 117 species included by Shi and Rabosky (2015). However, our results (increased diversification and the origin of Pteropodidae) did not change when we used a reduced dataset of 118 pteropodid species (Yinpterochiroptera tree). On the other hand, the dated phylogeny used by Shi and Rabosky (2015) in their BAMM had node ages significantly older; in particular, the Pteropodidae crown age was estimated at 40 million years. The same node was about 15 million years younger in our dated tree. There were some differences between studies in taxonomic sampling, choice of genes, and more importantly, in the calibration of the molecular clock, which resulted in clade-age estimates generally older or much older by Shi and Rabosky (2015) than the ones obtained by Amador et al. (2018) as well as previous studies (e.g., Teeling et al. 2005). For instance, and only among clades analyzed herein, ages for crown Rhinolophidae were 8.7, 6.5, and 14.3 my in Teeling et al. (2005), Jones et al. (2005), and Amador et al. (2018), respectively, whereas it was 37.2 my (2.7-fold) in Shi and Rabosky (2015). For crown Pteropodidae, age estimates were 24.8, 36.1, and 25.9 my, respectively, as above, and 40.2 in Shi and Rabosky (2015); in fact, the latter authors acknowledged the uncertainty of age estimates for Pteropodidae as a result of a dearth of old fossil pteropodids (see Giannini 2019).
Explosive radiations may be driven by different factors, such as colonization of a new habitat, origin of a key innovation, demography, and high intrinsic evolutionary rates (Schluter 2000; Gavrilets and Losos 2009; Yoder et al. 2010; Givnish 2015; Stroud and Losos 2016). Pteropodids possess several characteristics that are expected in clades that experienced explosive radiations (Almeida et al. 2011). Among those features are key innovations that allowed them to explore a radically different niche: pteropodids are primarily phytophagous that evolved from strictly insectivorous ancestors and likely have lost sophisticated laryngeal echolocation capabilities. Loss of echolocation has released pteropodids from constraints imposed on body size upper limit by a high-frequency sonar system (Jones 1999; Moyers Arévalo et al. 2020). Consequently, body size in pteropodids remains constrained exclusively by physical limitations related to powered flight (Norberg and Norberg 2012). Nevertheless, maximum theoretical size (as calculated by Norberg and Norberg 2012) has not yet been attained in living species, which appear to have rapidly evolved large sizes in the past few million years (Moyers Arévalo et al. 2020). These notable changes in diet, sensory system, and size altogether suggest adaptive factors underpinning their explosive diversification (Gavrilets and Vose 2005).
Demographic factors may also play a role in the diversification of pteropodids, especially in the diversification of island taxa, particularly important in Pteropus. Most Pteropus species are island specialists (see Giannini 2019 and citations therein), and the isolation and small sizes of their populations favor genetic differentiation and allopatric speciation (Losos and Ricklefs 2009). Moreover, Pteropus presents the highest size variation in bats with species weighting from mean ca. 120 g (e.g., P. molossinus) to ca. 1 kg (e.g., P. vampyrus; Moyers Arévalo et al. 2020). Possibly, this notable size span, and the aerodynamic consequences of it (see Amador et al. 2019), would have allowed Pteropus species to exploit a variety of plant resources and to reach remote-island environments (see Giannini 2019), thus enhancing niche partition and, ultimately, geographic speciation. Other speciose pteropodid genera like Dobsonia and Nyctimene are also mostly distributed on islands (Giannini 2019).
Diversification dynamics in Chiroptera
Aside from Pteropodidae, two additional diversification rate shifts at the origin of Rhinolophidae and Hipposideridae were detected by BAMM using a more comprehensive sampling. The evolution of a high duty-cycle echolocation system that has allowed these bats to detect insects in highly cluttered spaces (e.g., inside rainforest vegetation) was hypothesized as a novel trait capable of driving an adaptive radiation (Simmons and Conway 2003). These results are not in agreement with those obtained by Shi and Rabosky (2015), whose study did not find diversification rate shifts associated with the origin of any bat family, suggesting that “the global radiation of bats is characterized by a remarkably homogeneous diversification process.” Conversely, Jones et al. (2005), by means of a different methodological approach, reconstructed significant diversification rate shifts within Phyllostomidae and Molossidae, as well as some evidence for rate shifts within Pteropodidae, Emballonuridae, Rhinolophidae, Hipposideridae, and Vespertilionidae (although their significance depended on polytomy resolution). It is important to point out, though, that Jones et al. (2005) used a very different approach from the one employed both here and in Shi and Rabosky (2015).
Besides the increased diversification rates recovered herein in three bat families, hypotheses of explosive or adaptive radiation have also been raised in relation to some Yangochiroptera families (not included in our analysis). An adaptive radiation within phyllostomids has been proposed based on their use of a large diversity of dietary resources, particularly fruits, along with accompanying morphological adaptations (Dumont et al. 2012; Rojas et al. 2012; Shi and Rabosky 2015; Rossoni et al. 2019). Regarding molossids, it has been shown that this group has evolved high-performance wing morphology and flight style among bats, which allow them to catch flying insects high above the canopy (Amador et al. 2019). These examples show that not only is Chiroptera a distinctive mammalian order, capable of powered flight and sophisticated echolocation, but several clades within Chiroptera have evolved key innovations that might account for shifts in their diversification rates. Here, we contribute evidence for shifts in diversification rates at the origin of three families and also within one of them, reflecting the spectacular diversification of an island taxon, Pteropus.
Tempo of Yinpterochiroptera diversification
According to estimates by Amador et al. (2018), the age of pteropodid origin and first diversification burst used in our analysis was 22.4–27.7 mya (95% confidence interval), corresponding to the late Oligocene. Interestingly, the age of crown Hipposideridae was estimated within roughly the same time interval (21.3–27.9). This interval overlaps with a period of marked temperature rise known as the Late Oligocene warming (Fig. 3) (Zachos et al. 2001; Zhang et al. 2013). These two bat families share most of their geographic distributions, which encompass the entire Old World tropics, although they would not compete for resources as they exhibit very different ecologies: pteropodids are phytophagous, whereas hipposiderids are specialized aerial-hawking insectivores (Nowak 1994). Palynological data from SE Asia, probable region of pteropodid origin (Almeida et al. 2011), suggest that the Oligocene-Miocene boundary was characterized by a super-wet rain forest climate that followed a time of drier, seasonal climate (Lelono and Morley 2011; Hall 2012). Therefore, the environmental conditions were optimal for tropical taxa at the time the two families started to diversify. In addition, the region was experiencing substantial tectonic activity at that time, with the collision of the Australian and Asian Plates (Morley 2011; Hall 2012), therefore favoring allopatric speciation across islands and land connections.
After the late Oligocene warming, in the Early Miocene, temperatures remained relatively stable (despite brief glaciation periods) until the Mid Miocene Climatic Optimum (MMCO) at approximately 15 mya (Fig. 3) (Zachos et al. 2001; Zhang et al. 2013). The origin of crown rhinolophids, estimated at between 12 and 16 mya, occurred during the MMCO, a time when rainforest resources were likely more abundant. It is important to notice here that rhinolophids are also present in temperate areas, unlike pteropodids. The MMCO was followed by a gradual cooling and establishment of polar ice sheets, and this cooling trend persisted until the early Pliocene (~ 6 mya), when temperatures rose subtly until the onset of the Plio-Pleistocene Northern Hemisphere Glaciations at 3.2 mya (Zachos et al. 2001; Zhang et al. 2013). The fourth clade that appears to have experienced an explosive radiation, Pteropus, originated at the Late Miocene, around 8 mya, but diversified more intensely from 5 mya to the recent.
In conclusion, our results reject the hypothesis of homogeneous diversification processes in bats and point to complex diversification dynamics in the evolution of bat families linked to evolutionary innovation and environmental opportunity. Moreover, it serves as an example of why the estimation of divergence dates should be the focus of diversification studies, as the outcome of the latter is absolutely dependent on clade age estimates.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Alfaro, M. E., Santini, F., Brock, C., Alamillo, H., Dornburg, A., Rabosky, D. L., Carnevale, G., & Harmon, L. J. (2009). Nine exceptional radiations plus high turnover explain species diversity in jawed vertebrates. Proccedings of the National Academy of Sciences USA, 106, 13410–13414. https://doi.org/10.1073/pnas.0811087106
Almeida, F. C., Giannini, N. P., DeSalle, R., & Simmons, N. B. (2011). Evolutionary relationships of the old world fruit bats (Chiroptera, Pteropodidae): another star phylogeny? BMC Evolutionary Biology, 11(1), 281. https://doi.org/10.1186/1471-2148-11-281.
Almeida, F. C., Giannini, N. P., Simmons, N. B., & Helgen, K. M. (2014). Each flying fox on its own branch: a phylogenetic tree for Pteropus and related genera (Chiroptera: Pteropodidae). Molecular Phylogenetics and Evolution, 77, 83–95.
Almeida, F. C., Simmons, N. B., & Giannini, N. P. (2020). A species-level phylogeny of Old World fruit bats with a new higher-level classification of the Family Pteropodidae. American Museum Novitates, 2020(3950), 1–24. https://doi.org/10.1206/3950.1.
Amador, L. I., Moyers Arévalo, R. L., Almeida, F. C., Catalano, S. A., & Giannini, N. P. (2018). Bat Systematics in the light of unconstrained analyses of a comprehensive molecular supermatrix. Journal of Mammalian Evolution, 25(1), 37–70. https://doi.org/10.1007/s10914-016-9363-8.
Amador, L. I., Almeida, F. C., & Giannini, N. P. (2019). Evolution of traditional aerodynamic variables in bats (Mammalia: Chiroptera) within a comprehensive phylogenetic framework. Journal of Mammalian Evolution. https://doi.org/10.1007/s10914-019-09475-8.
Bergmans, W. (1997). Taxonomy and biogeography of African fruit bats (Mammalia, Megachiroptera). 5. The genera Lissonycteris Andersen, 1912, Myonycteris Matschie, 1899, and Megaloglossus Pagenstecher, 1885. General remarks and conclusions. Beaufortia, 47, 11–90.
Boonman, A., Bumrungsri, S., & Yovel, Y. (2014). Nonecholocating fruit bats produce biosonar clicks with their wings. Current Biology, 24(24), 2962–2967. https://doi.org/10.1016/j.cub.2014.10.077.
Chan, K. M., & Moore, B. R. (2002). Whole-tree methods for detecting differential diversification rates. Systematic Biology, 51, 855–865. https://doi.org/10.1080/10635150290102555.
Cornwell, W., & Nakagawa, S. (2017). Phylogenetic comparative methods. Current Biology, 27, R327–R338.
Drummond, A. J., & Rambaut, A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology, 7(1), 214. https://doi.org/10.1186/1471-2148-7-214.
Drummond, A. J., Ho, S. Y. W., Phillips, M. J., & Rambaut, A. (2006). Relaxed phylogenetics and dating with confidence. PLoS Biology, 4(5), e88. https://doi.org/10.1371/journal.pbio.0040088.
Dumont, E. R., Dávalos, L. M., Goldberg, A., Santana, S. E., Rex, K., & Voigt, C. C. (2012). Morphological innovation, diversification and invasion of a new adaptive zone. Proceedings of the Royal Society B: Biological Sciences, 279(1734), 1797–1805. https://doi.org/10.1098/rspb.2011.2005.
Fenton, M. B. (2010). Convergences in the diversification of bats. Current Zoology, 56(4), 454–468. https://doi.org/10.1093/czoolo/56.4.454.
Foley, N. M., Thong, V. D., Soisook, P., Goodman, S. M., Armstrong, K. N., Jacobs, D. S., et al. (2015). How and why overcome the impediments to resolution: lessons from rhinolophid and hipposiderid bats. Molecular Biology and Evolution, 32(2), 313–333. https://doi.org/10.1093/molbev/msu329.
Gavrilets, S., & Losos, J. B. (2009). Adaptive radiation: contrasting theory with data. Science, 323(5915), 732–737. https://doi.org/10.1126/science.1157966.
Gavrilets, S., & Vose, A. (2005). Dynamic patterns of adaptive radiation. Proceedings of the National Academy of Sciences USA, 102(50), 18040–18045.
Giannini, N. P. (2019). Pteropodidae. In D. E. Wilson & R. Mittermeier (Eds.), Handbook of the Mammals of the World. 9. Bats (pp. 16–61). Lynx Edicions: Barcelona.
Givnish, T. J. (2015). Adaptive radiation versus ‘radiation’ and ‘explosive diversification’: why conceptual distinctions are fundamental to understanding evolution. New Phytologist, 207(2), 297–303. https://doi.org/10.1111/nph.13482.
Hall, R. (2012). Sundaland and Wallacea: geology, plate tectonics and palaeogeography. In D. J. Gower, K. G. Johnson, J. E. Richardson, B. R. Rosen, L. Rüber, & S. T. Williams (Eds.), Biotic evolution and environmental change in Southeast Asia (pp. 32–78). Cambridge: Cambridge University Press.
Jetz, W., Thomas, G. H., Joy, J. B., Hartmann, K., & Mooers, A. O. (2012). The global diversity of birds in space and time. Nature, 491(7424), 444–448.
Jones, G. (1999). Scaling of echolocation call parameters in bats. The Journal of Experimental Biology, 202, 3359–3367.
Jones, K. E., Bininda-Emonds, O. R. P., & Gittleman, J. L. (2005). Bats, clocks, and rocks: diversification patterns in Chiroptera. Evolution, 59(10), 2243–2255. https://doi.org/10.1111/j.0014-3820.2005.tb00932.x.
Lack, J. B., & Van Den Bussche, R. A. (2010). Identifying the confounding factors in resolving phylogenetic relationships in Vespertilionidae. Journal of Mammalogy, 91(6), 1435–1448. https://doi.org/10.1644/09-MAMM-A-354.1.
Lanfear, R., Frandsen, P. B., Wright, A. M., Senfeld, T., & Calcott, B. (2017). PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution, 34(3), 772–773.
Lei, M., & Dong, D. (2016). Phylogenomic analyses of bat subordinal relationships based on transcriptome data. Scientific Reports, 6(1), 27726. https://doi.org/10.1038/srep27726.
Lelono, E. B., & Morley, R. J. (2011). Oligocene palynological succession from the East Java Sea. Geological Society, London, Special Publications, 355(1), 333–345. https://doi.org/10.1144/SP355.17.
Losos, J., & Ricklefs, R. (2009). Adaptation and diversification on islands. Nature, 457, 830–836. https://doi.org/10.1038/nature07893.
Monteiro, L. R., & Nogueira, M. R. (2011). Evolutionary patterns and processes in the radiation of phyllostomid bats. BMC Evolutionary Biology, 11(1), 137. https://doi.org/10.1186/1471-2148-11-137.
Morley, R. J. (2011). Cretaceous and Tertiary climate change and the past distribution of megathermal rainforests. In M. Bush, J. Flenley, & W. Gosling (Eds.), Tropical Rainforest Responses to Climatic Change (pp. 1–34). Berlin: Springer.
Moyers Arévalo, R. L., Amador, L. I., Almeida, F. C., & Giannini, N. P. (2020). Evolution of body mass in bats: insights from a large supermatrix phylogeny. Journal of Mammalian Evolution, 27(1), 123–138. https://doi.org/10.1007/s10914-018-9447-8.
Mullen, S. P., Savage, W. K., Wahlberg, N., & Willmott, K. R. (2011). Rapid diversification and not clade age explains high diversity in neotropical Adelpha butterflies. Procceeding of the Royal Society B: Biological Sciences, 278, 1777–1785. https://doi.org/10.1098/rspb.2010.2140.
Norberg, U. M. L., & Norberg, R. Å. (2012). Scaling of wingbeat frequency with body mass in bats and limits to maximum bat size. Journal of Experimental Biology, 215(5), 711–722. https://doi.org/10.1242/jeb.059865.
Nowak, R. M. (1994). Walker’s bats of the world. Baltimore: Johns Hopkins University Press.
Pybus, O. G., & Harvey, P. H. (2000). Testing macro–evolutionary models using incomplete molecular phylogenies. Proceedings of the Royal Society of London. Series B: Biological Sciences, 267(1459), 2267–2272. https://doi.org/10.1098/rspb.2000.1278.
Rabosky, D. L. (2006). Likelihood methods for detecting temporal shifts in diversification rates. Evolution, 60, 1152–1164. https://doi.org/10.1111/j.0014-3820.2006.tb01194.x.
Rabosky, D. L., & Glor, R. E. (2010). Equilibrium speciation dynamics in a model adaptive radiation of island lizards. Procceedings of the National Academy of Sciences USA, 107, 22178–22183. https://doi.org/10.1073/pnas.1007606107.
Rabosky, D. L., Grundler, M., Anderson, C., Title, P., Shi, J. J., Brown, J. W., et al. (2014). BAMMtools: an R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods in Ecology and Evolution, 5(7), 701–707. https://doi.org/10.1111/2041-210X.12199.
Revell, L. J. (2012). phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution, 3(2), 217–223. https://doi.org/10.1111/j.2041-210X.2011.00169.x.
Ricklefs, R. E. (2007). Estimating diversification rates from phylogenetic information. Trends in Ecology and Evolution, 22, 601–610. https://doi.org/10.1016/j.tree.2007.06.013.
Rojas, D., Vale, Á., Ferrero, V., & Navarro, L. (2012). The role of frugivory in the diversification of bats in the Neotropics. Journal of Biogeography, 39(11), 1948–1960. https://doi.org/10.1111/j.1365-2699.2012.02709.x.
Ronquist, F., & Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19, 1572–1574.
Rossoni, D. M., Costa, B. M. A., Giannini, N. P., & Marroig, G. (2019). A multiple peak adaptive landscape based on feeding strategies and roosting ecology shaped the evolution of cranial covariance structure and morphological differentiation in phyllostomid bats. Evolution, 73(5), 961–981. https://doi.org/10.1111/evo.13715.
Schluter, D. (2000). The ecology of adaptive radiation. New York: Oxford University Press.
Shi, J. J., & Rabosky, D. L. (2015). Speciation dynamics during the global radiation of extant bats. Evolution, 69(6), 1528–1545. https://doi.org/10.1111/evo.12681.
Simmons, N. B. (2005). Order Chiroptera. In D. E. Wilson & D. M. Reeder (Eds.), Mammal species of the world: a taxonomic and geographic reference (pp. 312–529). Baltimore: The Johns Hopkins University Press.
Simmons, N. B., & Cirranello, A. L. (2020). Bat species of the world: a taxonomic and geographic database. http://batnames.org. Accessed 15 April 2020.
Simmons, N. B., & Conway, T. M. (2003). Evolution of ecological diversity in bats. In T. H. Kunz & M. B. Fenton (Eds.), Bat Ecology (pp. 493–535). Chicago: The University of Chicago Press.
Stadler, T. (2011a). Mammalian phylogeny reveals recent diversification rate shifts. Proceedings of the National Academy of Sciences, 108(15), 6187–6192. https://doi.org/10.1073/pnas.1016876108.
Stadler, T. (2011b). Simulating trees on a fixed number of extant species. Systematic Biology, 60, 676–684.
Stadler, T. (2011c). TreePar in R-estimating diversification rates in phylogenies. http://cran.r-project.org/web/packages/TreePar/index.html. Accessed 20 January 2020.
Stroud, J. T., & Losos, J. B. (2016). Ecological opportunity and adaptive radiation. Annual Review of Ecology, Evolution, and Systematics, 47(1), 507–532. https://doi.org/10.1146/annurev-ecolsys-121415-032254.
Teeling, E. C., Springer, M. S., Madsen, O., Bates, P., & O’brien, S. J., & Murphy, W. J. (2005). A molecular phylogeny for bats illuminates biogeography and the fossil record. Science, 307(5709), 580–584.
Teeling, E. C., Scally, M., Kao, D. J., Romagnoli, M. L., Springer, M. S., & Stanhope, M. J. (2000). Molecular evidence regarding the origin of echolocation and flight in bats. Nature, 403(6766), 188–192. https://doi.org/10.1038/35003188.
Teeling, E. C., Dool, S., & Springer, M. S. (2012). Phylogenies, fossils and functional genes: the evolution of echolocation in bats. In G. F. Gunnell & N. B. Simmons (Eds.), Evolutionary history of bats: fossils, molecules, and morphology (pp. 1–22). Cambridge: Cambridge University Press.
Thiagavel, J., Cechetto, C., Santana, S. E., Jakobsen, L., Warrant, E. J., & Ratcliffe, J. M. (2018). Auditory opportunity and visual constraint enabled the evolution of echolocation in bats. Nature Communications, 9(1), 98. https://doi.org/10.1038/s41467-017-02532-x.
Title, P. O., & Rabosky, D. L. (2017). Do macrophylogenies yield stable macroevolutionary inferences? An example from squamate reptiles. Systematic Biology, 66(5), 843–856.
Tsang, S. M., Wiantoro, S., Veluz, M. J., Sugita, N., Nguyen, Y. L., Simmons, N. B., & Lohman, D. J. (2020). Dispersal out of Wallacea spurs diversification of Pteropus flying foxes, the world’s largest bats (Mammalia: Chiroptera). Journal of Biogeography, 47(2), 527–537.
Wang, Z., Zhu, T., Xue, H., Fang, N., Zhang, J., Zhang, L., et al. (2017). Prenatal development supports a single origin of laryngeal echolocation in bats. Nature Ecology & Evolution, 0021(1), 1–5. https://doi.org/10.1038/s41559-016-0021.
Whitfield, J. B., & Lockhart, P. J. (2007). Deciphering ancient rapid radiations. Trends in Ecology & Evolution, 22(5), 258–265. https://doi.org/10.1016/j.tree.2007.01.012.
Wilson, D. E., & Mittermeier, R. A. (2019). Handbook of the Mammal Species of the World – Volume 9: Bats. Barcelona: Lynx Edicions.
Yoder, J. B., Clancey, E., Des Roches, S., Eastman, J. M., Gentry, L., Godsoe, W., et al. (2010). Ecological opportunity and the origin of adaptive radiations: ecological opportunity and origin of adaptive radiations. Journal of Evolutionary Biology, 23(8), 1581–1596. https://doi.org/10.1111/j.1420-9101.2010.02029.x.
Yovel, Y., Geva-Sagiv, M., & Ulanovsky, N. (2011). Click-based echolocation in bats: not so primitive after all. Journal of Comparative Physiology A, 197, 515–530. https://doi.org/10.1007/s00359-011-0639-4.
Zachos, J., Pagani, M., Sloan, L., Thomas, E., & Billups, K. (2001). Trends, rhythms, and aberrations in global climate 65 Ma to present. Science, 292(5517), 686–693. https://doi.org/10.1126/science.1059412.
Zhang, Y. G., Pagani, M., Liu, Z., Bohaty, S. M., & DeConto, R. (2013). A 40-million-year history of atmospheric CO 2. Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences, 371, 20130096. https://doi.org/10.1098/rsta.2013.0096.
Acknowledgments
We thank the two anonymous reviewers for their helpful comments.
Funding
This work was supported by Grants PICT 2015-2389 and PICT 2016-3682 to NPG.
Author information
Authors and Affiliations
Contributions
FCA conceived the idea of the paper and ran the analyses; NPG and LIA discussed methods and results. All three authors contributed to the writing
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethics approval
Not applicable.
Consent to participate
Inform consent was obtained from all individual participants included in this study.
Consent for publication
All authors approved the final version to be published.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Almeida, F.C., Amador, L.I. & Giannini, N.P. Explosive radiation at the origin of Old World fruit bats (Chiroptera, Pteropodidae). Org Divers Evol 21, 231–243 (2021). https://doi.org/10.1007/s13127-021-00480-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13127-021-00480-5