Abstract
Quantification of mammalian skull development has received much attention in the recent literature. Previous results in different lineages have shown an effect of historical legacy on patterns of skull growth. In marsupials, the skull of adults exhibits high variation across species, principally along a size axis. The development keys of the marsupial skull are fundamental to understanding the evolution of skull function in this clade. Its generally well-resolved phylogeny makes the group ideal for studying macroevolution of skull ontogeny. Here, we tested the hypothesis that ontogenetic similarity is correlated with phylogeny in New World marsupials, so that developmental patterns are expected to be conserved from ancestral opossums. We concatenated our previously published ontogenetic cranial data from several opossum species with new ontogenetic sequences and constructed an allometric space on the basis of a set of comparable cranial linear measurements. In this ontogenetic space, we determined the degree of correspondence of developmental patterns and the phylogeny of the group. In addition, we mapped ontogenetic trajectories onto the opossum phylogeny, treating the trajectories as composite, continuously varying characters. Didelphids differed widely in the magnitude of skull allometry across species. Splanchnocranial components exhibited all possible patterns of inter-specific variation, whereas mandibular variables were predominantly allometrically “positive” and neurocranial components were predominantly allometrically “negative.” The distribution of species in allometric space reflected the compounded effect of phylogeny and size variation characteristic of didelphids. The terminal morphology of related species differed in shape, so their ontogenetic trajectories deviated with respect to that of reconstructed common ancestors in varying degree. Phylogeny was the main factor structuring the allometric space of New World marsupials. Didelphids inherited an ancestral constellation of allometry coefficients without change and retained much of it throughout their lineage history. Conserved allometric values on the nodes splitting placental outgroups and marsupials suggest a developmental basis common to all therians.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The ontogeny of the mammalian skull has been a rich subject of comparative work relevant to understand the relationship between form and function in a developmental and evolutionary context (Segura and Prevosti 2012). Quantitative studies include advances in comparative allometry (e.g., Weston 2003; Giannini et al. 2004, 2010; Flores et al. 2015), allometric disparity (Wilson and Sánchez-Villagra 2010; Wilson 2013), direct ontogenetic mapping onto phylogenies (Flores et al. 2013, 2015; Giannini 2014; Wilson and Sánchez-Villagra 2010; Wilson 2018), and phylomorphospace construction from ontogenetic trajectories (e.g., Tavares et al. 2016; Esquerré et al. 2017). The quantification of postnatal ontogenetic patterns of the mammalian skull has identified adaptive changes associated with the acquisition of full adult performance (e.g., Abdala et al. 2001; Tanner et al. 2010; Segura et al. 2013, 2016; Tarnawski et al. 2014), from both phenetic (e.g., Klingenberg and Froese 1991; Gerber et al. 2007; Wilson 2013) and phylogenetic (e.g., Giannini 2014; Flores et al. 2015; Tavares et al. 2016) perspectives. The phylogenetic approach demonstrated a considerable level of historical variation in cranial growth via the detection of specific allometric changes associated with clades of marsupials, rodents, and carnivores (e.g., Flores et al. 2013, 2015; Giannini 2014; Segura 2014) or through the discovery of patterns in clades or functional groups of rodents in ontogenetic morpho-spaces (e.g., Wilson and Sánchez-Villagra 2010; Wilson 2013; Tavares et al. 2016). Because simultaneous allometry estimation of skull components is a multivariate concept, and as such it is quantified considering the major axis of covariance (Jolicoeur 1963), allometric space studies are directly comparable to covariance matrix evolution (Wilson 2013). Thus, quantification of ontogeny in multivariate terms implies the comparative analysis of covariance matrices (e.g., Marroig and Cheverud 2005, 2010). According to Wilson (2013), most of previous studies on covariance matrix evolution have focused on variation in the adult stage (static allometry), therefore considering only the endpoints of individual ontogenetic trajectories. However, understanding the evolution of phenotypes in a given group also requires a quantification of the evolution of development (Hall 2000; Raff 2000), as the terminal size and shape of an organism depend on its ontogenetic trajectory understood as an orderly developmental sequence, i.e., changing across intermediate morphologies of the developing organism. Recent empirical approaches dealing with continuous developmental characters in phylogenies (e.g., Giannini 2014; Segura 2014; Flores et al. 2015) allowed detection of changes in the rate of growth in internal nodes of a given phylogeny, and thus to infer ancestral allometries and their evolutionary context.
When compared with any clade of placental mammals, marsupials as a group exhibit low morphological disparity and high integration of skull components (Shirai and Marroig 2010; Goswami et al. 2012; Porto et al. 2013). These patterns have been attributed to intrinsic factors such as reproduction and development (e.g., Goswami et al. 2012; Porto et al. 2013) and/or extrinsic factors such as the history of diversification and biogeography in marsupials (Sánchez-Villagra 2013). Nonetheless, the variation of skull shape in marsupials is compelling, even among the apparently homogeneous carnivorous marsupials (e.g., Horovitz and Sánchez-Villagra 2003; Wroe and Milne 2007). As a consequence, the study of skull ontogeny in carnivorous marsupials is highly relevant not only from a purely descriptive perspective but also in terms of the peculiar evolution of the group.
The New World opossum Didelphidae comprises the most diverse living radiation of marsupials outside Australia (Voss and Jansa 2009). Didelphimorphia, the order containing opossums, split from the remaining marsupials in the Upper Cretaceous (Astúa 2015). However, the living opossums are morphologically derived from most Cretaceous and Paleogene metatherians. The well-resolved phylogeny of Didelphidae (Amador and Giannini 2016) and the considerable size variation across species of the didelphid crown group (e.g., Astúa 2015; Voss and Jansa 2009; Amador and Giannini 2016) make this diverse Neotropical group ideal for an integral study of evolution of skull ontogeny. Here, we review previous works on post-weaning skull ontogeny in didelphids, and concatenate data from these studies with new ontogenetic sequences that consistently cover extant didelphid diversity. We specifically test the hypothesis that evolutionary history (phylogeny) is correlated with ontogenetic similarity, and so the clade-level diversity of New World opossums is established in ancestrally shared developmental patterns. To this end, we report the skull ontogenetic patterns of representative didelphids and relevant marsupial outgroups (caenolestids, microbiotheriids, and dasyurids), as well as more distant placental outgroups in which we root the reconstructed ontogenetic changes. We map elements of ontogenetic trajectories onto the recent phylogeny of didelphid marsupials from Amador and Giannini (2016), treating these elements as continuously varying characters (Giannini 2014). This method was successfully applied recently in other mammals (Flores et al. 2015; Segura 2014). In addition, following Gerber et al. (2008), we constructed the ontogenetic space of didelphid marsupials on the basis of species-specific ontogenetic trajectories, and described the correspondence of patterns in this space with the phylogeny of didelphids. This contribution represents the more speciose taxonomic sample of carnivorous marsupials analyzed in the context of ontogeny optimization and allometric disparity. We reveal the strong phylogenetic component in patterns of skull development in didelphid marsupials and discuss the historical context of ontogenetic evolution in New World marsupials.
Material and methods
The group
Living didelphids comprise 103 currently recognized species distributed from Canada to Patagonia (Astúa 2015). This diversity is classified into well-supported monophyletic groups (see Amador and Giannini 2016 and citations therein). In the last decades, the knowledge of didelphid taxonomy, phylogenetic relationships, and distribution patterns has grown enormously with the aid of new molecular data (e.g., Voss and Jansa 2009), morphology (e.g., Reig et al. 1987; Voss and Jansa 2003; Flores 2009), and new distributional and ecological data (Vieira and Astúa 2003; Birney and Monjeau 2003). Three living subfamilies—Caluromyinae (Kirsch 1977), the highly diverse Didelphinae (Gray 1821), and the monotypic Hyladelphinae (Voss and Jansa 2009)—compose this speciose family (Gardner 2007; Voss and Jansa 2009; Astúa 2015). The systematics of living didelphines were widely discussed in past decades; most topologies recently recovered from morphological and molecular data split this group into three monophyletic tribes: Didelphini, including the large opossums (Didelphis, Philander, Lutreolina, Chironectes, and Metachirus); Thylamyini, including the mouse opossums Cryptonanus, Gracilinanus, Marmosops, Thylamys, and Lestodelphys; and Marmosini, including the mouse opossums Marmosa, the monotypic Tlacuatzin, and the short-tailed opossums Monodelphis (Voss and Jansa 2009). Most of the genera were recovered as natural groups in the taxonomic arrangement considered at present (e.g., Gardner 2007; Astúa 2015; Amador and Giannini 2016). The didelphid radiation occupied a diversity of ecological niches (Astúa 2015); the observed morphological variation reveals a mosaic of some highly conserved features (both within the family and shared with earlier metatherian lineages), such as the dental formula and postcranial general morphology (Horovitz and Sánchez-Villagra 2003; Flores 2009; Voss and Jansa 2009), whereas other characters vary considerably within the family, such as molar morphology (Chemisquy et al. 2015) or body size (Amador and Giannini 2016). For instance, adult individuals of North American opossum Didelphis virginiana weigh up to 5 kg, whereas the pygmy opossum Monodelphis kunsi is only 10 g (Astúa 2015). Although didelphids were considered as models of “living fossils” when compared with mesozoic and paleogene metatherians (e.g., Crompton 1989), also called “basal ameridelphians” (Goin et al. 2016), some characters in living forms exhibit derived conditions with respect to early metatherians (e.g., Reig et al. 1987; Rougier et al. 1998; Horovitz and Sánchez-Villagra 2003; Luo et al. 2003; Horovitz et al. 2009). Recent estimations of time-calibrated phylogenies inferred an Early Oligocene (26.3 Ma) origin of crown-clade Didelphidae (Meredith et al. 2008; Jansa et al. 2014; Goin et al. 2016), with a radiation of living forms and extinction processes occurring from the middle-late Miocene in consonance with orogenic pulses and the rising of the Andes (e.g., Patton and da Silva 1997; Jansa et al. 2014; Palma et al. 2014; Díaz-Nieto et al. 2016).
Sample
We included in our study previously published ontogenetic samples from four placental outgroups, seven marsupial outgroups including the New World caenolestids and microbiotheriids and Australian dasyurids and peramelids, and three didelphids (Caluromys, Didelphis, and Lutreolina). To this set, we added eight newly generated ontogenetic series of didelphid species measured from museum collections (Appendix SI.1): Chironectes minimus, Philander opossum, Metachirus nudicaudatus, Thylamys sponsorius, Marmosops incanus, Marmosa murina, M. demerarae, and Monodelphis brevicaudata. This dataset comprised 383 new specimens examined (Appendix SI.1) and composed a total set of 1155 specimens representing 22 species (Table 1). The dataset represented ontogenetic sequences of 11 didelphids, thus including 10 living genera out of the 17 represented in the family and certainly members from all suprageneric natural groups. All series included young specimens with only the first molar erupted (except for Caenolestidae, see Flores et al. 2015). In general, the condylobasal length of the youngest specimen was approximately 50% or less than that of the largest adult specimen of the conspecific series (Table 1).
Cranial measurements
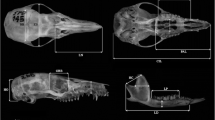
We took 15 cranial measurements that represent splachnochranial and neurochranial components described in detail in previous work (e.g., Abdala et al. 2001; Giannini et al. 2004; Flores et al. 2015) and illustrated in Fig. 1. For statistical comparisons, we considered exactly the same set of measurements in our 22 ontogenetic series.
Skull and mandible measurements considered in this study. Abbreviations: CIL, condyloincisive length; BB, breadth of the braincase; BP, breadth of the palate; HC, height of the coronoid process; HD, height of the dentary; HO, height of the occipital plate; LC, length of the coronoid process; LD, length of the dentary; LN, length of the nasals; LP, lower postcanine toothrow; MH, muzzle height; ORB, orbit length; PAL, palate length; UP, upper postcanine toothrow; ZB, Zygomatic breadth
Multivariate analysis of allometry
Multivariate coefficients of allometry were obtained from the unit-scaled, first eigenvector of a principal component analysis (PCA) based on a variance–covariance matrix of log10-transformed data (see founding paper by Jolicoeur 1963). For all multivariate coefficients of allometry, the expected value of isometry depends on the number of variables (p), being calculated as 1/p0.5 (0.258 for our set of 15 variables). We tested the significance of the coefficients of allometry (i.e., statistical departures from isometry) with a resampling strategy based on the jackknife, in which each specimen was removed from the sample at a time, generating n pseudovalues in the process that were used to calculate confidence intervals (CIs) for the original coefficients (see Giannini et al. 2004 and Flores et al. 2006 for details on this method). If CIs excluded the expected value of isometry, the variable was considered positively or negatively allometric. However, our comparisons were based on the coefficients as continuous values. Trimming the m largest and smallest m pseudovalues (with m = 1) for each variable may decrease the standard errors calculated under jackknife and allow more accurate allometric estimations (Manly 1997; Giannini et al. 2004). Here, we report untrimmed and trimmed calculations, but opting for the results with lower average bias (i.e., the difference between the observed and resampled allometry coefficients; Giannini et al. 2004). For the multivariate statistical analyses (PCA + jackknife resampling), we used the R script of Giannini et al. (2010; Appendix SI. 2).
Allometric disparity
We followed Wilson and Sánchez-Villagra (2010) and Wilson (2013) in applying the concept of allometric disparity (formalized by Gerber et al. 2007, 2008, 2011) to our sample, so an allometric morpho-space was expanded on the basis of the intraspecific ontogenetic trajectories included in this study. Following the multivariate generalization of allometry described above (Jolicoeur 1963), the first principal component of the ontogenetic series represents the vector of multivariate allometric coefficients, in which size is taken as a latent variable that affects all the original variables simultaneously. Hence, 22 intraspecific PCAs (one for each species studied) were calculated as above, and the first normalized eigenvector of each analysis was included in a matrix that was submitted to another PCA. The result is interpreted as the allometric space of the sample, in which each species is represented by a coordinate. The principal components for the first and second axes were plotted to examine species position in this space. Given that each point in allometric space reflects a normalized vector (PC1, i.e., species’ trajectories), it is possible to quantify the allometric distance between any two species as inter-trajectory angles in the allometric space (e.g., Solignac et al. 1990; Wilson and Sánchez-Villagra 2010). Total variance can be quantified using an average angle between trajectories, which provides an indication of spacing in state space (see Wilson 2013). When two PC1 vectors x and y are normalized (x′, y′), then the angle α between the two trajectories is computed as α = arcos (x′·y′) or the arccosine of the dot product of two normalized vectors (Klingenberg 1996; Wilson and Sánchez-Villagra 2010; Wilson 2013). A vector with all coefficients equal to 1/p0.5 (i.e., 0.258) defines the isometric vector, which was used to assess allometric trends for given trajectories. Angles between species or trajectories of monophyletic groups (e.g., Didelphini, Marmosini, Thylamini) and the isometric vector were also calculated. The Kruskal–Wallis test was used in order to evaluate the statistical significance of comparisons and detect significant intertrajectory angle values between species and for each species with the isometric vector. Kruskal–Wallis tests were coupled with Mann–Whitney pairwise tests, resulting in P values which were corrected using the Bonferroni method (Zar 1974). Our analysis of the allometric space was computed using a marsupial subsample with a more complete ontogenetic coverage. For instance, caenolestids were excluded because they lack the earliest age stages represented in the remaining marsupials (Flores et al. 2015). The exclusion of the earliest ontogenetic stages could impact the multivariate slope, and thus, the angular comparisons in allometric space (and the relative position of those taxa) would be prone to error (see below). Therefore, our analysis of the multivariate allometric space included species of Didelphidae, Dromiciops, Dasyurus, and two peramelids (i.e., 15 species). To estimate the standard error of the PC1 coefficients for each species, a bootstrap with replacement was performed for 1000 iterations.
Evolution of ontogeny
Macroevolutionary patterns, such as interspecific variation in body mass or growth rate, can only be discerned in an explicit phylogenetic framework (e.g., Gould and MacFadden 2004; Butler and Goswami 2008; Flores et al. 2015; Giannini 2014; Amador and Giannini 2016). The quantification of ontogeny in a phylogenetic framework (e.g., Weisbecker et al. 2008; Wilson and Sánchez-Villagra 2010; Bennett and Goswami 2013; Koyabu et al. 2011; Goswami et al. 2012; Segura 2014; Giannini 2014; Flores et al. 2015) has added quantitative rigor to the studies of developmental bases in mammalian evolution. Here, we applied the methods of character optimization and phylogenetic signal as in Flores et al. (2015). Optimization, or character mapping, is a fundamental tool used to determine the cost of a character state on a given tree topology and the representation of ancestral character states in internal nodes and the root. Giannini (2014) demonstrated that allometric coefficients should be mapped onto phylogenies as continuous characters (see also Segura 2014 and Flores et al. 2015 for empirical applications). Optimization of continuous characters (using either point estimates or intervals for terminal values) is implemented in the computer program TNT (Goloboff et al. 2008) as an extension of Farris’ (1970) multistate character optimization (Goloboff et al. 2006). Allometric similarities can be explained by common ancestry, and ancestral allometry vectors can be determined (Giannini 2014 and citations therein). We reconstructed the allometric vector at all internal nodes, calculated the optimal cost of reconstruction (steps on the tree), and examined the evolutionary changes (increases and decreases in individual allometric coefficients) as implied by the tree topology, to uncover the general phylogenetic pattern of cranial ontogeny evolution and its functional implications.
For this analysis, we included our sample of marsupials and placentals (see above) arranged in the more accepted current topology, considering Ameridelphia as paraphyletic and Australidelphia as monophyletic (i.e., successively Caenolestidae, Didelphidae, Microbiotheriidae, and the remainder of Australidelphia; see Horovitz and Sánchez-Villagra 2003; Asher et al. 2004; Nilsson et al. 2004; Beck 2008; Meredith et al. 2008). In the didelphid clade, Marmosini (Monodelphis–Marmosa) is sister of the Thylamyini (Thylamys–Marmosops)–Didelphini (Metachirus–Chironectes–Lutreonina–Philander–Didelphis) clade, a relationship supported by both genetic and molecular evidence (see Voss and Jansa 2009; Amador and Giannini 2016).
Phylogenetic signal was estimated using random swapping of characters (allometric coefficients) in terminal branches (Laurin 2004). The distribution of character states (i.e., resampled allometric coefficients) on the observed phylogenies was compared with the distribution of states on 20,000 randomly generated trees. The number of steps of each character on the randomly generated datasets was also compared with the number of steps of that character in the observed phylogenies. We considered a character to have significant phylogenetic signal if the observed number of steps was smaller than the steps calculated for at least 95% of the randomly generated trees. For phylogenetic signal calculation, we used the R script of Prevosti et al. (2010, Appendix SI. 3), computed with the program TNT v. 1.1 (Goloboff et al. 2008).
Results and discussion
Multivariate allometric trends
We report in Table SI.1(Supplementary Information) the detailed multivariate results of the seven additional didelphid species considered in this study; we choose to interpret the confidence intervals provided by untrimmed analyses given their generally lower average bias, except for Lestoros inca, in which trimmed analysis showed lower bias (Table SI.2, Supplementary Information). We summarized the allometric trends of all species considered in this study, i.e., the observed allometric trends in Table 2. The variance summarized by each intraspecific PCA (used a posteriori to construct the allometric space) ranged from 85 to 97% (variance of PC2 between 1.7 and 16.4% across species; Table SI.3, Supplementary Information).
As a first step in interpreting our results, here we highlight trends in the data on a per-variable basis. Condyloincisive length (CIL) was predominantly isometric in our marsupial sample; however, there were cases of both positive and negative allometry, and these cases were disconnected (i.e., not clearly associated to either taxonomic or functional groupings). Rostral, or splanchnocranial, components exhibited a highly complex pattern of growth across our sample. The length and width of the palate (PAL and BP) showed either isometry or negative allometry across the majority of the sample, including placental outgroups; PAL was positive only in Echymipera, whereas BP was negative in the whole sample. The growth of nasal length (LN) reflected the characteristically long rostrum in most opossums, so it was a “positive” variable in most cases; exceptions were Lutreolina and the australidelphian Dromiciops that feature a typical short rostrum. The height of the rostrum (MH) experienced isometric growth in all groups, with both positive and negative exceptions in particular species. As reported in previous analyses (e.g., Abdala et al. 2001), the growth of both upper (UP) and lower (LP) tooth rows is coordinated, but with the lower tooth row outstripping the upper tooth row by one element in their respective eruption sequences. In most cases (Table 2), the upper and lower tooth rows shared the same allometric trend (isometry or negative allometry), with Chironectes minimus being the only species showing positive allometry in both variables. The zygomatic breadth (ZB) showed positive allometry in relatively large or more carnivorous marsupials, such as Dasyurus, Lutreolina, Didelphis, Marmosa demerarae, and Monodelphis. This growth pattern expands the zygoma laterally and generates the inner space for a larger temporal muscle that is accommodated between the braincase (negative allometry) and the zygoma (positive allometry). All neurocranial variables exhibited negative allometry, with some isometric exceptions and a considerable variation in the actual value of allometry coefficients (see below). The allometry of the braincase was negative in all placentals and marsupials included in our sample, except Metachirus, in which it was isometric.
The mandible, a complex structure part of the splanchnocranium and mostly affected to trophic functions, grew with positive allometry, or isometry, in most of its component variables across the sample. These variables include length of the mandible, height of the mandibular body, and height of the coronoid process, indicating general increasing size and strength of mandibular components. The only exception was L. inca, which showed the length of the coronoid process (LC) with (marginal) negative allometry.
Allometric disparity
The multivariate generalization of allometry (Jolicoeur 1963) provides an accurate reflection of growth, with PC1 axes representing the direction of growth in the multivariate space of the log-transformed dataset. Allometric space was constructed from 15 allometric trajectories. All the unbiased multivariate coefficients obtained for 15 variables ranged between 0.011 and 0.807 across marsupial species (Table 3).
In the allometric space, PC1 and 2 summarized 32.6 and 19.7% of total variation, respectively, and the variance of measurements along PC1 and PC2 is shown in Table 4 and Fig. 2. The normalized vectors on the plane of the allometric space (Fig. 2) exhibited phylogenetic structuring. First, generalized, substantial allometric growth was indicated in the fact that all taxa are removed from the position of the hypothetical isometric taxon (coded Hyp in Fig. 2). This is in clear agreement with the large inter-trajectory angles observed in the comparison with the isometric vector (SI 4, Supplementary Information; Fig. 3). Second, there was overlap in allometric space of some australidelphians (Dasyurus and peramelids) with large didelphids (Didelphini), which is also evidenced in the non-significant differences in the average angle among these groups (SI 4, Supplementary Information). Third, Dromiciops appeared isolated on the negative side of PC1, largely due to the influence of the variable LC, although a close inter-trajectory angle was detected with Thylamys (see below). Fourth, all didelphid marsupials grouped in close correspondence with clade membership: Caluromys appeared separate as the only Caluromyinae included; the five genera of large didelphids (Didelphis, Chironectes, Metachirus, Lutreolina, and Philander, tribe Didelphini) were relatively disparate although always on the positive side of PC2, maintaining a phylogenetic arrangement; so were Marmosops and Thylamys (Thylamini), and Marmosa (Micoureus), Marmosa (Marmosa), and Monodelphis (Marmosini), on the negative side of PC2. Finally, australidelphian outgroups also showed recognizable patterns, particularly the distant position of Isoodon with respect to Echymipera, which in principle reflected the divergent growth pattern in peramelids suggested by Flores et al. (2013). However, both peramelids show a non-significant difference in the inter-trajectory average angle (χ2 = 0.119, p = 0.747). Species with positive scores on PC1 had a lower positive allometric coefficient for the length of the coronoid process (LC; being Dromiciops the taxon with the higher allometric coefficient for this sample, Table 3), two neurocranial variables (BB and OH), breadth of palate (BP), and lower postcanine (LP, Table 4). Change along PC2 reflected deviations in growth trends for orbit length (ORB) principally, upper postcanine (UP), height and length of the coronoid process (HC and LC), and height of the dentary (HD, Table 4).
Allometric space for 15 marsupial species, comprising representatives from the major lineages of carnivorous marsupials (didelphids, microbiotheriids, dasyurids, and peramelids). Each species is represented by a single point in allometric space, describing its allometric trajectory. Squares: Didelphinae; vertical ellipse: Caluromyinae; cross: Microbiotheriidae; circle: Peramelidae; pentagon: Dasyuridae; horizontal ellipse: hypothetic isometric taxon. Lines contacting didelphids indicate monophyletic groups defined by Voss and Jansa (2009). Abbreviations: Dg, Dromiciops gliroides; Da, Didelphis albiventris; Po, Philander opossum; Cm, Chironectes minimus; Lc, Lutreolina crassicaudata; Mn, Metachirus nudicaudatus; Mb, Monodelphis brevicaudata; Md, Marmosa demerarae; Mm, Marmosa murina; Mi, Marmosops incanus; Ts, Thylamys sponsorius; Cp, Caluromys philander; Im, Isoodon macrourus; Ec, Echymipera kalubu; Dm, Dasyurus albopunctatus; Hyp, hypothetic isometric taxa. PC1 = 32.6%; PC2 = 19.7% of sample variance. Red: Didelphini; blue: Marmosini; green: Thylamyni. Abbreviations of variables as in Fig. 1
Boxplots showing the distribution of inter-trajectory angle comparisons (whiskers, minima and maxima; horizontal bar, median value; boxes, middle two quartiles). Comparisons, from left to right, are within members of a clade (colors as in Fig. 2), between members of a clade and the isometric vector (coefficient value of 0.258), and between members of different clades (tribes)
The occupation of allometric space for the didelphid species studied appeared concentrated, as indicated by a low average angle of 10.65° between PC1 components. The closest pair of trajectories was Thylamys and Dromiciops (1.39°) whereas the widest inter-trajectory angle was 28.99° between Didelphis and Chironectes (SI 4, Supplementary Information). The large inter-trajectory angle observed for Didelphis–Chironectes could be an artifact caused by the sample completeness of taxa, where the size of the largest specimen is almost three times the size of the smallest, which is a size variation comparatively larger than the other ontogenetic series (see Table 1). The average angle among PC1 components for tribe Didelphini was 16.7°, while the average angles for Marmosini and Thyamini clades were 6.49 and 3.56° respectively. The average angle of the species in tribe Didelphini was significantly different from that observed for Marmosini (χ2 = 6.42, p = 0.014) and Thylamini species (χ2 = 4.61, p = 0.041; Fig. 4), whereas differences in the average angle between the species of Marmosini and Thylamini were non-significant (χ2 = 3, p = 0.149; Fig. 3). The angles between species and the isometric vector were high (average 20.1°), being only minor in didelphids (average 19.9°), in which it ranges between 15.3° in Thylamys and 32.7° in Didelphis (Fig. 3; SI 4, Supplementary Information). When examining all inter-trajectory angles in Didelphidae, comparisons between tribes were lower than the greatest difference between Didelphini trajectories (Fig. 3). The average inter-trajectory angles between Didelphini–Marmosini and Didelphini–Thylamyini were statistically similar (χ2 = 1.23, p = 0.279), whereas the average angles between Thylamyini–Marmosini and Didelphini–Marmosini were statistically different (χ2 = 12.27, p = 0.0004); so were the average angles between Thylamyini–Marmosini and Didelphini–Thylamyini (χ2 = 5.19, p = 0.026; Fig. 3).
Optimization of confidence intervals obtained from multivariate analysis of allometry on the hypothesis supported by Voss and Jansa (2009) and Amador and Giannini (2016). Numbers indicate the difference between confidence interval of allometry between ancestor and descendent node (or terminal). Sign + or − indicate increase or decrease of the confidence interval. Arrow indicates the node defining Didelphidae, showing the confidence intervals of the multivariate coefficient of allometry of didelphid ancestor. The allometric trend is indicated in parentheses. Colors in monophyletic groups as indicated in Fig. 2. Abbreviations on internal nodes and terminals indicate variables as in Fig. 1
Ontogenetic character optimization and phylogenetic signal
We detected changes in the rate of growth of several variables at nodes which support monophyletic groups. Seven out 20 internal nodes were assigned changes in the rate of growth of at least one skull variable. Caenolestidae was diagnosed by a decreasing rate of growth of the zygomatic width. The clade containing australidelphians and Didelphidae was diagnosed by two changes: increasing rate of growth of the length of the upper postcanine tooth row and height of the dentary at the horizontal ramus (=mandibular body; Fig. 4). Changes in the height of the occipital plate support two clades: increased rate in Isoodon + Echymipera (i.e., Peramelidae) and decreasing rate support the didelphid clade Marmosini (Monodelphis + Marmosa). Finally, within Didelphini, the clade Didelphis–Philander was supported by a decreasing rate of growth of the breadth of the palate (Fig. 4).
Our analysis exhibited a degree of phylogenetic signal, although statistically significant in just four skull variables (Table 5). A significant phylogenetic signal was present in two braincase variables (breadth of the braincase, BB, and height of the occipital plate, HO) and two variables related with the trophic apparatus (height of the dentary, HD, and length of the upper postcanine tooth row, UP).
How the didelphid skulls grow
Species of didelphids grew each in a particular fashion, but an analysis of allometry across variables reveals inter-specific ontogenetic patterns that we discuss in the following. Condyloincisive length (CIL) is one measure of overall skull size widely used even as proxy of the entire body size (see Emerson and Bramble 1993). However, multivariate analyses, which do not assume a single size proxy in regression analysis, but allow perceiving size as a latent factor affecting all variables at once, revealed that the allometry of CIL varied across species and was characterized by no common ontogenetic pattern. Both the placental outgroup and the marsupial sample offered examples of isometry, “negative” allometry, and “positive” allometry. In fact, CIL was one skull dimension that varied much inter-specifically. Similarly to CIL, the zygomatic breadth (ZB) also presented a mixed pattern of variation. Remarkably, CIL and ZB varied in opposition to a great extent among marsupials, with some intermediate cases. Allometries resulting in short, wide skulls (“negative” or “isometric” CIL and “positive” ZB) corresponded to more animalivorous species (Dasyurus, Lutreolina, Chironectes, Monodelphis; Flannery 1990; Vieira and Astúa 2003), whereas long and narrow skulls corresponded with more insectivorous species (Marmosa; Marmosops; e.g., Vieira and Astúa 2003). The skull of Didelphis albiventris, a large species that is considered a dietary generalist (e.g., Vieira and Astúa 2003), grew with “positive” allometry in both CIL and ZB, showing also a strongly divergent trajectory regarding other species (see Table SI 4, Supplementary Information). The evolutionary flexibility of the mammalian face allows it to be remodeled simply by modifying postnatal growth. A general pattern in mammals consists of an apparent rule in which face development is in close positive relation with cranial size (Cardini and Polly 2013). Although we analyzed the rate of growth of cranial measurements (including indirectly rostrum through nasal and/or palate lengths), but not the final size of those, our observations on the proportional size of the face and skull in adult didelphids, rate seemed to be more related with food habits than with size. For instance, the small insectivore Marmosops showed a proportionally longer rostrum than the larger carnivore Lutreolina.
The three neurocranial variables (braincase breadth BB, occipital height OH, and orbital length ORB) tended to grow with negative allometry in all species studied. These variables remained strictly “negative” in the placental outgroup; by contrast, we detected some variability in the marsupial sample. Cases of isometry were common in ORB, including two cases of marginally “positive” allometry in caenolestids (see also Flores et al. 2015). The isometry of the braincase (BB) in the highly terrestrial Metachirus is a trend not observed in any other member of our sample. The value of the BB multivariate coefficient is negative but represented by high in Didelphini and Thylamyni (except Didelphis, Table 3; Supplementary Information SI.1). Splanchnocranial components (ZB being one of them) exhibited all patterns of inter-specific variation, from predominantly “positive” allometry in the nasal length (LN) to almost uniformly “negative” in the breadth of the palate (BP). The length of the upper tooth row (UP) was also predominantly “negative” whereas the length of the palate (PAL) was predominantly isometric across species. The remarkable “positive” exception in the latter is in the bandicoot Echymipera kalabu (Peramelidae), discussed in Flores et al. (2013), which stands out as a singularity in the context of a wider sample of marsupials presented here. Echymipera exhibits a particularly long rostrum originated in the development of clear diastemata on the premolar series, which is not present in Isoodon (see Figs. 2 and 3 in Flores et al. 2013).
The pattern of growth in mandibular variables was predominantly “positive” (four variables of the body and coronoid process) whereas the length of the lower postcanine row was “negative” in most cases (Table 2). As expected, the latter was coordinated with the growth pattern of the upper tooth row although it may show a slight out-of-phase pattern in the actual level of “negative” allometry of upper versus lower tooth row (first noted in marsupials in Abdala et al. 2001). Teeth are discrete units, but their emergence is gradual, so the length of the tooth row increases in a discontinuous manner (Abdala et al. 2001). The last element is added belatedly (depending on the species), but in general, the entire skull continues to grow well beyond this event. This particular condition of a measurement composed of the addition of discrete units likely causes the generalized negative allometry (or isometry) of the postcanine series, also suggesting that growth rate may not be strictly constant (see Abdala et al. 2001). In adult didelphids, lengths of both postcanine series are nearly the same, but during ontogeny, the emergence of the lower postcanines is usually advanced in one element with respect to the upper postcanines (Astúa and Leiner 2008). This sequence of emergence can explain the allometric differences between upper and lower rows in species such as Metachirus nudicaudatus and Marmosa murina (Table 2), with the coefficient of the lower postcanine row being smaller than that of the upper row. However, the upper and lower rows exhibit similar multivariate allometric coefficients in most species, in both cases “negative,” where the lower and upper tooth rows are equally involved in the mutual adjustment during growth. According to Radinsky (1983, 1984), “negative” allometry of tooth series is not an unusual condition in mammals. The one “positive” exception in the didelphid Chironectes is replicated in the upper tooth row only in this taxon; the biological basis of these contrasting “positive” allometries remains elusive.
Taking into consideration this inter-specific variation, the didelphid skull grows, across species, with a variable and generally opposing pattern in overall length (CIL) and zygomatic width (ZB), with a negative neurocranium and a more variable orbit, variable muzzle height, positive nasals (rostral length), a combination of isometry in palate length and “negative” palate breadth, and also “negative” upper tooth row length, which was coordinated with the lower tooth row in acute contrast with the predominantly “positive” variables of the mandibular body and coronoid. Opposing trends shape the trophic function of the skull in the already mentioned CIL–ZB opposition, but also in the abundantly documented opposition of ZB and BB in marsupials, as well as in palate length versus width (see Abdala et al. 2001; Giannini et al. 2004; Flores et al. 2015). Braincase breadth (BB) was always “negative” although to varying extent, whereas ZB ranged from highly “positive” to slightly “negative” across species. The space for the temporal muscle is shaped between the zygoma and the braincase, so the more “positive” the ZB, the more space is generated to develop a large temporal muscle and the potential for a stronger bite increases. In turn, the palate length was more frequently isometric whereas the palate width was typically “negative,” meaning that the palate started as a short, posteriorly wide platform that supported the tongue during permanent, pump suckling, an essential function during the early life of marsupials (Clark and Smith 1993; Maier 1993), and eventually developed the more elongated palate of marsupials (see Abdala et al. 2001; Flores et al. 2003, 2010, 2015). The early development and integration of the palate were also related with the breathing capacity of young during the fixation period (e.g., Shirai and Marroig 2010; Goswami et al. 2012). Finally, orbit allometry might be positively associated with habits (nocturnality, arboreality); Pilatti and Astúa (2017) described for Didelphidae that elongated rostra implied orbits more convergent and more dorsally positioned, whereas the widening of the rostrum implied a more vertical position and a little divergent orbit orientation. According to Rasmussen (1990), the rostrum and the convergence of the orbits of Caluromys are suited to capture arthropod prey. This marsupial also makes extensive use of the terminal branch feeding, as observed in prosimians. Although we do not include variables related to the position of the orbit, the short face of Caluromys (expressed in an isometric length of nasals, LN) is coincident with that in Lutreolina that shows a negative trend for this variable (Table 2). Both taxa show similar orbital orientation (see Pilatti and Astúa 2017). However, it is clear that the use of the substrate in Caluromys (arboreal) and Lutreolina (mostly terrestrial) is not similar (Nowak 2005).
From the perspective of the didelphid species themselves, they widely differed in the level of allometry expressed in the developing skull. Species ranged from having just one isometric variable, as in Monodelphis, to species such as Thylamys (of comparable size) with as many as 11 isometric variables (Table 2). Inter-specific differences and similarities were reflected with greater precision in allometric space (see above), where disparity was associated to major didelphid clades (Figs. 2 and 3; Table S4 Supplementary Information). As a general principle, adults of related species may differ in morphology because their ontogenetic trajectories changed with respect to that of a common ancestor (Arthur 2011). Disparity as measured in allometric space expresses these differences and allows the recognition of shared versus convergent patterns of ontogenetic evolution (Wilson and Sánchez-Villagra 2010). In rodents, major clades differed in the level of disparity and convergence (e.g., sciurids being more conserved), and trophic ecology (diet) played an important role in the evolution of allometric trajectories (Wilson and Sánchez-Villagra 2010; Wilson 2013). Such result is not replicated in our species, perhaps because didelphids (and Neotropical marsupials in general) are taxonomically diverse but quite homogeneous ecologically; most didelphids can be seen as rather generalist mammals with similar trophic, locomotive, and reproductive functions across species (see Vieira and Astúa 2003). Living Neotropical marsupials possess similar body form and do not exhibit great niche diversity; for instance, Chemisquy et al. (2015) showed that a highly conserved molar morphology exists in all didelphids, with few exceptions (e.g., Caluromys, a mostly frugivore; Charles-Dominique et al. 1981, Leite et al. 1996).
Here, we show that phylogeny was the main factor structuring allometric space in didelphid marsupials and hence explaining much of their ontogenetic disparity: species included in this study grouped in correspondence with major clade membership in allometric space (Fig. 2), meaning that each of them grow in a way similar to its closest relatives. This has the potential implication that growth patterns were inherited from founders of major groups (see below). These clades also reflect much of the body size variation in the group. For instance, the inter- and intra-trajectory angles of ontogenetic vectors showed lower differences in small opossums (i.e., Marmosini–Thylamyini) in comparison with large opossums (Didelphini; Fig. 3; Table SI 4, Supplementary Information). Body size is the only parameter of major ecological importance with high inter-specific variation in didelphids (Voss and Jansa 2009; Amador and Giannini 2016). Morphometric studies on the skull have shown that didelphids are among the most morphologically integrated (i.e., least modular) groups of mammals (Porto et al. 2009; Shirai and Marroig 2010). Here, body size is key because high integration arises chiefly when most morphological variation is highly correlated (aligned) with size variation (Astúa 2009; Marroig et al. 2009; Porto et al. 2009; Shirai and Marroig 2010). Thus, the distribution of species in allometric space likely reflected the compound phylogenetic and size variation characteristic of didelphids.
However, our results offer in Fig. 2 a glimpse into a larger pattern of disparity at the level of marsupials, by which certain didelphids resemble more the ontogenetic patterns of some australidelphians rather than the patterns of other didelphids. This is the case for large didelphids (the Didelphini) overlapped with dasyurids and one peramelid, showing that their growth patterns are convergently similar to some significant extent. Thus, convergence in allometric space appears at a higher level in marsupials just as in major rodent groups (Wilson 2013), strongly suggesting that a study including a denser taxonomic sample in the whole marsupial clade is likely to uncover other patterns of this kind.
How the ontogenies of didelphid skulls evolved
Considering the strong phylogenetic imprint of the allometric space (Fig. 2), we confidently traced the evolution of allometry in the didelphid (+ outgroup) phylogenetic tree (Fig. 4). The rationale for using coefficients of allometry as phylogenetic characters has been clarified in Giannini (2014) who noted that the allometric sampling is repeatable, the coefficients reliably express patterns of a heritable growth program (new generations demonstrably will grow in the same fashion), and that the genetic network that defines the growth program evolves (see Pavlicev et al. 2008). Thus, coefficients of allometry that are similar between closely related species can be safely attributed to inherited variation (common ancestry), and allometries that differ may thus be attributed to diverging evolution, thereby modifying the ontogenetic trajectories in the descendants (see Arthur 2011). The conservative nature of skull morphology in didelphids reveals few changes in the rate of growth in internal nodes, but strong phylogenetic signal. Size variation in the group is consistent with a strong evolutionary component, from the ancestor’s developmental program. As previously indicated (e.g., Abdala et al. 2001; Flores et al. 2003), larger didelphids (i.e., Didelphini) grow acquiring characters (e.g., sagittal crest, lambdoidea crest) absent in adults of the small Marmosini and Thylamyini, which maintained a size similar to that of the didelphid ancestor. The ancestor of Didelphidae would show a size similar to that of Dromiciops or some mouse opossums (Amador and Giannini 2016), with a highly conservative ontogenetic pattern (described below; see Fig. 4). In our case, with didelphids and other marsupial and placental outgroups, a phylogenetic signal was detected in four skull variables (Table 5), two neurocranial (height of the occipital plate and breadth of the braincase) and two splanchnocranial (height of the dentary and upper postcanine tooth row), and nearly half of the nodes were assigned evolutionary changes in allometric coefficients (Fig. 4). Significantly, no changes were detected on the nodes defining placental outgroups versus marsupials, strongly suggesting the existence of a common therian basis that can be reconstructed in this way with more samples (see Flores et al. 2013, 2015). This suggests that the divergent life history strategies between these groups (in organogenesis, integration, development, and reproduction; e.g., Clark and Smith 1993; Shirai and Marroig 2010; Goswami et al. 2012) were not directly reflected in ontogenetic changes in postnatal skull development at the marsupial ancestor. Marsupials exhibited many highly relevant evolutionary changes, which can be taken as potential synapomorphies of corresponding clades, but didelphids in particular inherited the ancestral constellation of coefficients without change. This pattern passed onto descendants with little alteration, and here lies the phylogenetic basis of the conservative structure of the didelphid skull, which from a modularity perspective is among the most highly integrated mammalian skulls, and, concomitantly, the less evolutionarily flexible (Shirai and Marroig 2010). In this sense, the low average of intertrajectory angle differences observed between the species of the Didelphidae (see above) is in agreement with the conservative nature of the group. The didelphid allometric pattern that produced the ancestral didelphid skull (Fig. 4) can be reconstructed as follows. The variables that define the overall shape of the skull, condyloincisive length, and zygomatic width were slightly negative and isometric, respectively; remarkably, this combination is present only in Dromiciops among extant forms. The ancestral didelphid was estimated to weigh 22–33 g (Amador and Giannini 2016), which overlaps with the 22 g which is the average body weight of Dromiciops. The three neurocranial variables, including the orbit, were strongly negative. The rostrum modeled via the combination of a strongly positive allometry of nasals and the slightly negative allometry of muzzle height. Palate and upper tooth row were all negative, more strongly so in the latter, with a correspondingly negative trend for the lower tooth row (coefficients overlap in the upper and lower tooth rows). All other mandible variables were strongly positive. This ancestral pattern resembles much the way extant didelphids in general grow (see previous section), thereby reinforcing the conservative nature of the didelphid skull―this time from a purely ontogenetic perspective.
Changes were reconstructed only in the nodes of origin of major subclades (Caluromyinae, Marmosini, Didelphini), which in turn were characterized by ontogenetic changes of their own, as well as some parallelisms. For instance, the height of the occipital plate, HO, shows a lower rate of growth in Marmosini, and increasing it in Peramelidae (Fig. 4).
Conclusions
With a coverage of major opossum groups, and comparative outgroups selected among ameridelphians (caenolestids), australidelphians, and placentals, we reconstructed the evolutionary patterns of skull growth in Didelphidae. We recognize a common therian pattern of growth, and more into the details, specific patterns of skull variables are shared with basal australidelphians, but more forms, particularly herbivores, should be added to the sample to test this hypothesis at the marsupial subclass level. We note, however, that herbivorous marsupials are largely derived forms in the Diprotodontia, so the base of the phylogeny is not misrepresented in our sample and we should expect changes toward the derived Australasian groups and less so toward the root of the marsupial tree. The growth of the ancestral didelphid skull resembles the growth of a generalized didelphid: trends correspond to most species when exceptions in different terminals are set aside. In turn, the skull patterns differ across species and groups within didelphids because the sharing of the therian ontogenetic basis is subtly altered in some variables in specific nodes, whose combination appears to result in the bounded diversity of morphologies that we recognize across didelphid clades. Analysis of allometric disparity also reflected a phylogenetic structuring around the major didelphid clades.
References
Abdala, F., Flores, D., & Giannini, N. (2001). Postweaning ontogeny of the skull of Didelphis albiventris. Journal of Mammalogy, 82, 190–200.
Amador, L., & Giannini, N. (2016). Phylogeny and evolution of body mass in didelphid marsupials (Marsupialia: Didelphimorphia: Didelphidae). Organisms, Diversity and Evolution. https://doi.org/10.1007/s13127-015-0259-x.
Arthur, W. (2011). Evolution: a developmental approach. Oxford: Wiley Blackwell.
Asher, R., Horovitz, I., & Sánchez-Villagra, M. (2004). First combined cladistic analysis of marsupial mammal interrelationships. Molecular Phylogenetics and Evolution, 33, 240–250.
Astúa, D. (2009). Evolution of scapula size and shape in didelphid marsupials (Didelphimorphia: Didelphidae). Evolution, 63, 2438–2456.
Astúa, D. (2015). Order Didelphimorphia. In D. Wilson & R. Mittermeier (Eds.), Handbook of the mammals of the world (Vol. 5. Monotremes and marsupials, pp. 70–187). Barcelona: Lynx Editions.
Astúa, D., & Leiner, O. (2008). Tooth eruption sequence and replacement pattern in woolly opossums, genus Caluromys (Didelphimorphia: Didelphidae). Journal of Mammalogy, 89, 244–251.
Beck, R. (2008). A dated phylogeny of marsupials using a molecular supermatrix and multiple fossil constraints. Journal of Mammalogy, 89, 175–189.
Bennett, C. V., & Goswami, A. (2013). Statistical support for the hypothesis of developmental constraint in marsupial skull evolution. BMC Biology, 11, 52. https://doi.org/10.1186/1741-7007-11-52.
Birney, E., & Monjeau, A. (2003). Latitudinal variation in South American marsupial biology. In M. Jones, C. Dickman, & M. Archer (Eds.), Predators with pouches: the biology of carnivorous marsupials (pp. 297–317). Collingwood: CSIRO Publishing.
Butler, R. J., & Goswami, A. (2008). Body size evolution in Mesozoic birds: little evidence for Cope’s rule. Journal of Evolutionary Biology, 21, 1673–1682.
Cardini, A., & Polly, P. D. (2013). Larger mammals have longer faces because of size-related constraints on skull form. Nature Communications, 4, 2458.
Charles-Dominique, P., Atramentowicz, M., Charles-Dominique, M., Gerard, H., Hladik, C. M., & Prevost, M. F. (1981). Les mammiferes frugivores arboricoles nocturnes d’une foret Guyannaise: inter-relations plantes–animaux. Revue d’Ecologie (La Terre et La Vie), 35, 341–435.
Chemisquy, M. A., Prevosti, F. J., Martin, G., & Flores, D. (2015). Evolution of molar shape in didelphid marsupials (Marsupialia: Didelphidae): analysis of the influence of ecological factors and phylogenetic legacy. Zoological Journal of the Linnean Society, 173, 217–235.
Clark, C. T., & Smith, K. K. (1993). Cranial osteogenesis in Monodelphis domestica (Didelphidae) and Macropus eugenii (Macropodidae). Journal of Morphology, 215, 119–149.
Crompton, A. W. (1989). The evolution of mammalian mastication. In D. B. Wake & J. Roth (Eds.), Complex organismal function: integration and evolution in vertebrates. New York: John Wiley and Sons Ltd..
Díaz-Nieto, J. F., Jansa, S. A., & Voss, R. S. (2016). DNA sequencing reveals unexpected recent diversity and an ancient dichotomy in the American marsupial genus Marmosops (Didelphidae: Thylamyini). Zoological Journal of the Linnean Society, 176, 914–940.
Emerson, S. B., & Bramble, D. M. (1993). Scaling, allometry and skull design. In J. Hanken & B. K. Hall (Eds.), The skull (pp. 384–416). Chicago: The University of Chicago Press.
Esquerré, D., Sherratt, E., & Keogh, J. S. (2017). Evolution of extreme ontogenetic allometric diversity and heterochrony in pythons, a clade of giant and dwarf snakes. Evolution, 71, 2829–2844.
Farris, J. (1970). Methods for computing Wagner trees. Systematic Zoology, 19, 83–92.
Flannery, T. (1990). Mammals of New Guinea. Australia: Robert Brown & Associates.
Flores, D. A. (2009). Phylogenetic analyses of postcranial skeletal morphology in didelphid marsupials. Bulletin of the American Museum of Natural History, 320, 1–81.
Flores, D. A., & Casinos, A. (2011). Cranial ontogeny and sexual dimorphism in two New World monkeys: Alouatta caraya (Atelidae) and Cebusapella (Cebidae). Journal of Morphology, 272, 744–757.
Flores, D., Giannini, N., & Abdala, F. (2003). Cranial ontogeny of Lutreolina crassicaudata (Didelphidae): a comparison with Didelphis albiventris. Acta Theriologica, 48, 1–9.
Flores, D., Giannini, N., & Abdala, F. (2006). Comparative postnatal ontogenyof the skull in the australidelphian metatherian Dasyurus albopunctatus (Marsupialia: Dasyuromorpha: Dasyuridae). Journal of Morphology, 267, 426–440.
Flores, D. A., Abdala, F., & Giannini, N. (2010). Cranial ontogeny of Caluromys philander (Didelphidae, Caluromyinae): a qualitative and quantitative approach. Journal of Mammalogy, 91, 539–550.
Flores, D. A., Abdala, F., & Giannini, N. (2013). Post-weaning cranial ontogeny in two bandicoots (Mammalia, Peramelomorphia, Peramelidae) and comparison with carnivorous marsupials. Zoology, 116, 372–384.
Flores, D. A., Abdala, F., Martin, G., Giannini, N., & Martinez, J. (2015). Post-weaning cranial growth in shrew opossums (Caenolestidae): a comparison with bandicoots (Peramelidae) and carnivorous marsupials. Journal of Mammalian Evolution, 22, 285–303.
Gardner, A. L. (Ed.). (2007). Mammals of South America. Vol. 1: Marsupials, xenarthrans, shrews, and bats. Chicago: University of Chicago Press.
Gerber, S., Neige, P., & Eble, G. J. (2007). Combining ontogenetic and evolutionary scales of morphological disparity: a study of early Jurassic ammonites. Evolutionary Development, 9, 472–482.
Gerber, S., Eble, G. J., & Neige, P. (2008). Allometric space andallometric disparity: a developmental perspective in the macroevolutionary analysis of morphological disparity. Evolution, 62, 1450–1457.
Gerber, S., Eble, G. J., & Neige, P. (2011). Developmental aspects of morphological disparity dynamics: a simple analytical exploration. Paleobiology, 37, 237–251.
Giannini, N. (2014). Quantitative developmental data in a phylogenetic framework. Journal of Experimental Zoology, 322, 558–566.
Giannini, N., Abdala, F., & Flores, D. A. (2004). Comparative postnatal ontogeny of the skull in Dromiciops gliroides (Marsupialia: Microbiotheriidae). American Museum Novitates, 3460, 1–17.
Giannini, N., Segura, V., Giannini, M. I., & Flores, D. (2010). A quantitative approach to the cranial ontogeny of the puma. Mammalian Biology, 75, 547–554.
Goin, F. J., Woodburne, M. O., Zimicz, A. N., Martin, G. M., & Chornogubsky, L. (2016). A brief history of South American metatherians: evolutionary contexts and intercontinental dispersals. New York: Springer.
Goloboff, P. A., Mattoni, C. I., & Quinteros, A. S. (2006). Continuous characters analyzed as such. Cladistics, 22, 589–601.
Goloboff, P. A., Farris, J. S., & Nixon, K. C. (2008). TNT, a free program for phylogenetic analysis. Cladistics, 24, 774–786.
Goswami, A., Polly, P. D., Mock, O. B., & Sánchez-Villagra, M. R. (2012). Shape, variance and integration during craniogenesis: contrasting marsupial and placental mammals. Journal of Evolutionary Biology, 25, 862–872.
Gould, G. C., & MacFadden, B. J. (2004). Gigantism, dwarfism, and Cope’s rule: “nothing in evolution makes sense without a phylogeny”. Bulletin of the American Museum of Natural History, 285, 219–237.
Hall, B. K. (2000). Evo-devo or devo-evo—does it matter? Evolution and Development, 2, 177–178.
Horovitz, I., & Sánchez-Villagra, M. R. (2003). A morphological analysis of marsupial mammal higher-level phylogenetic relationships. Cladistics, 19, 181–212.
Horovitz, I., Martin, T., Bloch, J., Ladevèze, S., Kurz, C., & Sánchez-Villagra, M. R. (2009). Cranial anatomy of the earliest marsupials and the origin of opossums. Plos One, 4(12), e8278.
Jansa, S. A., Barker, F. K., & Voss, R. S. (2014). The early diversification history of didelphid marsupials: a window into South America’s “Splendid Isolation”. Evolution, 68, 684–695.
Jolicoeur, P. (1963). The multivariate generalization of the allometry equation. Biometrics, 19, 497–499.
Kirsch, J.A.W. (1977). The comparative serology of Marsupialia, and a classification of marsupials. Australian Journal of Zoology Supplementary series, 25, 1–152.
Klingenberg, C. P. (1996). Multivariate allometry. In L. F. Marcus, M. Corti, A. Loy, G. J. P. Naylor, & D. E. Slice (Eds.), Advances in morphometrics (pp. 23–49). New York: Plenum Press.
Klingenberg, C. P., & Froese, R. (1991). A multivariate comparison of allometric growth patterns. Systematic Zoology, 40, 410–419.
Koyabu, D., Endo, H., Mitgutsch, C., Suwa, G., Catania, K. C., Zollikofer, C. P. E., Oda, S., Koyasu, K., Ando, M., & Sánchez-Villagra, M. R. (2011). Heterochrony and developmental modularity of cranial osteogenesis in lipotyphlan mammals. Evo Devo, 2, 21.
Laurin, M. (2004). The evolution of body size, Cope’s rule and the origin of amniotes. Systematic Biology, 53, 594–622.
Leite, Y. L. R., Costa, L. P., & Stallings, J. R. (1996). Diet and vertical space use of three sympatric opossums in a Brazilian Atlantic forest reserve. Journal of Tropical Ecology, 12, 435–440.
Luo, Z. X., Ji, Q., Wible, J. R., & Yuan, C. X. (2003). An Early Cretaceous tribosphenic mammal and metatherian evolution. Science, 302, 1934–1940.
Maier, W. (1993). Cranial morphology of the therian common ancestor, as suggested by the adaptations of neonate marsupials. In F. S. Szalay, M. J. Novacek, & M. C. McKenna (Eds.), Mammal phylogeny: mesozoic differentiation, multituberculates, monotremes, early therians, and marsupials (pp. 166–181). Berlin: Springer-Verlag.
Manly, B. F. J. (1997). Randomization, bootstrap, and Monte Carlo methods in biology (2nd ed.). London: Chapman and Hall.
Marroig, G., & Cheverud, J. M. (2005). Size as a line of least evolutionary resistance: diet and adaptive morphological radiation in New World monkeys. Evolution, 59, 1128–1142.
Marroig, G., & Cheverud, J. M. (2010). Size as a line of least resistance II: direct selection on size or correlated response due to constraints? Evolution, 64, 1470–1488.
Marroig, G., Shirai, L. T., Porto, A., Oliveira, F. B., & De Conto, V. (2009). The evolution of modularity in the mammalian skull II: evolutionary consequences. Evolutionary Biology, 36, 136–148.
Meredith, R. W., Westerman, M., & Springer, M. S. (2008). A timescale and phylogeny for “bandicoots” (Peramelemorphia: Marsupialia) based on sequences for five nuclear genes. Molecular Phylogenetics and Evolution, 47, 1–20.
Nilsson, M. A., Arnason, U., Spencer, P. B. S., & Janke, A. (2004). Marsupial relationships and a timeline for marsupial radiation in south Gondwana. Gene, 340, 189–196.
Nowak, R. M. (2005). Walker’s marsupials of the world. Baltimore: The John Hopkins University Press.
Palma, R. E., Boric-Bargetto, D., Jayat, J. P., Flores, D. A., Zeballos, H., Pacheco, V., Cancino, R. A., Alfaro, F. A., Rodríguez-Serrano, E., & Pardiñas, U. F. J. (2014). Molecular phylogenetics of mouse opossums: new findings on the phylogeny of Thylamys (Didelphimorphia, Didelphidae). Zoologica Scripta, 43, 217–234.
Patton, J. L., & da Silva, M. N. F. (1997). Definition of species of pouched four-eyed opossums (Didelphidae, Philander). Journal of Mammalogy, 78, 90–102.
Pavlicev, M., Kenney Hunt, J. P., Norgard, E. A., Roseman, C. C., Wolf, J. B., & Cheverud, J. M. (2008). Genetic variation in pleiotropy: differential epistasis as a source of variation in the allometric relationship between long bone lengths and body weight. Evolution, 62, 199–213.
Pilatti, P. & Astúa, D. (2017). Orbit orientation in didelphid marsupials (Didelphimorphia: Didelphidae). Current Zoology, 63, 403–415.
Porto, A., Oliveira, F. B., Shirai, L. T., De Conto, V., & Marroig, G. (2009). The evolution of modularity in the mammalian skull I: morphological integration patterns and magnitudes. Evolutionary Biology, 36, 118–135.
Porto, A., Shirai, L. T., de Oliveira, F. B., & Marroig, G. (2013). Size variation, growth strategies, and the evolution of modularity in the mammalian skull. Evolution, 67, 3305–3322.
Prevosti, F., Turazzini, C. F., & Chemisquy, M. A. (2010). Morfología craneana en tigres dientes de sable: alometría. función y filogenia. Ameghiniana, 47, 239–256.
Radinsky, L. B. (1983). Allometry and reorganizationin horse skull proportions. Science, 221, 1189–1191.
Radinsky, L. B. (1984). Ontogeny and phylogeny in horse skull evolution. Evolution, 38, 1–15.
Raff, R. A. (2000). Evo-devo: the evolution of a new discipline. Nature Reviews Genetics, 1, 74–79.
Rasmussen, D. T. (1990). Primate origins: lessons from a Neotropical marsupial. American Journal of Primatology, 22, 263–277.
Reig, O. A., Kirsch, J. A. W., & Marshall, L. G. (1987). Systematic relationships of the living and neocenozoic American “opossum-like” marsupials (suborder Didelphimorphia), with comments on the classification of these and of the Cretaceous and Paleogene New World and European metatherians. In M. Archer (Ed.), Possums and opossums: studies in evolution (Vol. 1, pp. 1–89). Sydney: Surrey Beatty.
Rougier, G. W., Wible, J. R., & Novacek, M. J. (1998). New specimens of Deltatheridium, implications for the early history of marsupials. Nature, 396, 459–463.
Sánchez-Villagra, M. R. (2013). Why are there fewer marsupials than placentals? On the relevance of geography and physiology to evolutionary patterns of mammalian diversity and disparity. Journal of Mammalian Evolution, 20, 279–290.
Segura, V. (2014). Ontogenia craneana postnatal en cánidos y félidos neotropicales: funcionalidad y patrones evolutivos. PhD thesis, Universidad Nacional de La Plata, La Plata, Buenos Aires, Argentina.
Segura, V., & Prevosti, F. (2012). A quantitative approach to the cranial ontogeny of Lycalopex culpaeus (Carnivora: Canidae). Zoomorphology, 131, 79–92.
Segura, V., Prevosti, F., & Cassini, G. (2013). Cranial ontogeny in the Puma lineage, Puma concolor, Herpailurus yagouaroundi, and Acinonyx jubatus (Carnivora: Felidae): a three dimensional geometric morphometric approach. Zoological Journal of the Linnean Society, 169, 235–250.
Segura, V., Prevosti, F., & Cassini, G. (2016). Three-dimensional cranial ontogeny in pantherines (Panthera leo, P. onca, P. pardus, P. tigris; Carnivora, Felidae). Biological Journal of the Linnaean Society. https://doi.org/10.1111/bij.12888.
Shirai, L., & Marroig, G. (2010). Skull modularity in neotropical marsupials and monkeys: size variation and evolutionary constraint and flexibility. Journal Experimental Zoology, 314B, 663–683.
Solignac, M., Cariou, M. L., & Wimitzki, M. (1990). Variability, specificity and evolution of growth gradients in the species complex Jaera albifrons (Isopoda, Asellota). Crustaceana, 59, 121–145.
Tanner, J. B., Zelditch, M. L., Lundrigan, B. L., & Holekamp, K. E. (2010). Ontogenetic change in skull morphology and mechanical advantage in the spotted hyena (Crocuta crocuta). Journal of Morphology, 271, 353–365.
Tarnawski, B. A., Cassini, G. H., & Flores, D. (2014). Allometry of the postnatal cranial ontogeny and sexual dimorphism in Otaria byronia (Otariidae). Acta Theriologica, 59, 81.
Tavares, W. C., Pessoa, L. M., & Seuánez, H. N. (2016). Phylogenetic and size constrains on cranial ontogenetic allometryof spiny rats (Echimyidae, Rodentia). Journal of Evolutionary Biology, 29, 1752–1765.
Vieira, E. M., & Astúa, D. (2003). Carnivory and insectivory in Neotropical marsupials. In M. Jones, C. Dickman, & M. Archer (Eds.), Predators with pouches: the biology of carnivorousmarsupials (pp. 271–284). Collingwood: CSIRO.
Voss, R. S., & Jansa, S. (2003). Phylogeneticstudies in didelphid marsupials II. Nonmolecular data and new IRBP sequences: separate and combined analyses of didelphine relationships with denser taxon sampling. Bulletin of the American Museum of Natural History, 276, 1–82.
Voss, R. S., & Jansa, S. A. (2009). Phylogenetic relationships and classification of didelphid marsupials, an extant radiation of New World metatherian mammals. Bulletin of the AmericanMuseum of Natural History, 322, 1–177.
Weston E.M. (2003). Evolution of ontogeny in the hipoppotamus skull: using allometry to dissect developmental change. Biological Journal of the Linnean Society, 80, 625–638.
Weisbecker, V., Goswami, A., Wroe, S., & Sánchez-Villagra, M. R. (2008). Ossification heterochrony in the therian postcranial skeleton and the marsupial–placental dichotomy. Evolution, 62, 2027–2041.
Wilson, L. (2013). Allometric disparity in rodent evolution. Ecology and Evolution, 3, 971–984.
Wilson, L. (2018). The evolution of ontogenetic allometric trajectories in mammalian domestication. Evolution. https://doi.org/10.1111/evo.13464.
Wilson, L., & Sánchez-Villagra, M. R. (2010). Diversity trends and their ontogenetic basis: an exploration of allometric disparity in rodents. Proceedings of the Royal Society B, 277, 1227–1234.
Wroe, S., & Milne, N. (2007). Convergence and remarkably consistent constraint in the evolution of carnivore skull shape. Evolution, 61, 1251–1260.
Zar J.H. (1974). Biostatistical analysis. Upper Saddle River, NJ: Prentice Hall.
Acknowledgments
We thank the curators who allowed the examination of the material under their care: Sergio Lucero of the Museo Argentino de Ciencias Naturales (MACN, Buenos Aires); Bruce Patterson and Bill Stanley of the Field Museum of Natural History (FMNH, Chicago); Kris Helgen, Darrin Lunde, and Linda Gordon of the Smithsonian Institution (USMNH, Washington, D.C.); Rob Voss and Eileen Westwig of the American Museum of Natural History (AMNH, New York); Joao Alves de Oliveira of the Museu Nacional Universidad Federal do Rio de Janeiro (Brazil); and Mario de Vivo of the Museu do Zoología Universidad do Sao Paulo (Brazil). Two anonymous reviewers improved the quality of this manuscript. This work was supported by the Consejo Nacional de Investigaciones Cientificas y Técnicas of Argentina (CONICET); the projects PICT2008-1798, PICT2012-1583, and PICT 2015-2389 of the Agencia Nacional de Promoción Científica y Tecnológica, Argentina (ANPCyT); and the National Research Foundation of South Africa.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Appendix SI.1
Specimens examined in this study and sample size. Acronyms of collections: AMNH, American Museum of Natural History, New York; FMNH, Field Museum of Natural History, Chicago; MACN, Museo Argentino de Ciencias Naturales “Bernardino Rivadavia”, Buenos Aires, Argentina; MZUSM, Museu de Zoologia Universidade do Sao Paulo, Brazil; UFRJ; Universidade Federal do Rio de Janeiro, Brazil; USNHM, United Sates Natural History Museum, Smithsonian Institution, Washington DC. (DOCX 13 kb)
Appendix SI.2
R script performed by Giannini et al. (2010) for multivariate statistical analyses (PCA and jackknife resampling). (DOCX 5 kb)
ESM 1
(XLSX 53 kb)
ESM 2
(XLSX 23 kb)
ESM 3
(XLSX 13 kb)
ESM 4
(XLSX 11 kb)
Rights and permissions
About this article
Cite this article
Flores, D.A., Giannini, N. & Abdala, F. Evolution of post-weaning skull ontogeny in New World opossums (Didelphidae). Org Divers Evol 18, 367–382 (2018). https://doi.org/10.1007/s13127-018-0369-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13127-018-0369-3