Abstract
Biomedical event extraction is an important branch of biomedical information extraction. Trigger extraction is the most essential sub-task in event extraction, which has been widely concerned. Existing trigger extraction studies are mostly based on conventional machine learning or neural networks. But they neglect the ambiguity of word representations and the insufficient feature extraction by shallow hidden layers. In this paper, trigger extraction is treated as a sequence labeling problem. We introduce the language model to dynamically compute contextualized word representations and propose a multi-layer residual bidirectional long short-term memory (BiLSTM) architecture. First, we concatenate contextualized word embedding, pretrained word embedding and character-level embedding as the feature representations, which effectively solves the tokens’ ambiguity in biomedical corpora. Then, the designed BiLSTM block with residual connection and gated multi-layer perceptron is adopted to extract features iteratively. This architecture improves the ability of our model to capture information and avoids gradient exploding or vanishing. Finally, we combine the multi-layer residual BiLSTM with CRF layer to obtain more reasonable label sequences. Comparing with other state-of-the-art methods, the proposed model achieves the competitive performance (F1-score: 80.74%) on the biomedical multi-level event extraction (MLEE) corpus without any manual participation and feature engineering.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Event extraction is a high-level task of information extraction, which aims to extract complex and pre-defined structured events from large amounts of unstructured data. In biomedical field, biomedical event extraction is employed to mine detailed and complicated molecular activities, such as describing the intermolecular interactions.
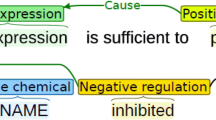
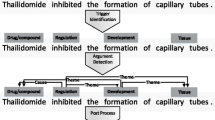
Usually a biomedical event consists of a trigger and several arguments. Each trigger corresponds to a biomedical event and determines the corresponding event type. Arguments refer to multiple participants in the events. As shown in Fig. 1 and Table 1, the example sentence contains multiple trigger-guided biomedical events. A representative biomedical event extraction process consists of three stages: trigger extraction, arguments extraction and post-processing. Among these stages, trigger extraction plays a vital role in the whole process. On one side, the trigger directs different types of events and related arguments. Moreover, triggers help to parse the association between multiple arguments in complex events. They are the basis and prerequisite for the following steps. Case studies show that more than 60% of event extraction errors are caused by incorrect identification of triggers [1]. Therefore, trigger extraction is not only the beginning and core of the biomedical event extraction, but also the key to the overall performance of the task. Early rule-based biomedical trigger extraction methods relied on manual design of rule templates, which were lack of flexibility and portability, and could not be applied to large-scale biomedical corpora. Recent biomedical trigger extraction studies can be roughly divided into two categories: conventional machine learning methods and neural networks. Pyysalo et al. [1] adopted more abundant contexts and dependencies as the representations. Based on support vector machine (SVM), they obtained 75.84% of the F1-score on MLEE corpus, which also became the baseline result of the corpus. Zhou et al. [2] developed a semi-supervised trigger extraction model, which identified triggers with unlabeled corpora and hidden information. This model achieved reasonable performance compared with the baseline result above. After combining feature engineering with domain knowledge, Zhou et al. [3] employed SVM to classify biomedical triggers. He et al. [4] divided trigger extraction into two stages: detection and classification. Based on this two-stage method, the F1-score of 79.75% was achieved on MLEE corpus. It was also the best performance in the current studies based on conventional machine learning. The conventional machine learning methods avoid manual participation to a certain degree, but they are still limited by feature engineering.
Recently, with the rapid development of artificial intelligence, deep learning technology is widely used in various fields [5,6,7,8,9]. Many biomedical trigger extraction researches are based on neural networks. Wang et al. [10] captured the semantic and syntactic information between tokens by regenerating dependency word embedding, and then identified triggers by multi-layer neural networks. Nie et al. [11] modelled the syntactic and semantic features of sequences based on the word embedding generated by Skip-gram model. This effectively assisted neural network in extracting biomedical triggers. Rahul et al. [12] regarded biomedical trigger extraction as sequence labeling problem. They combined word embedding with entity type embedding and adopted bidirectional recurrent neural network (BiRNN) to label triggers. He et al. [13] employed additional sentence vectors on dependency-based word embedding. They designed an attention-based bidirectional long short-term memory (BiLSTM) network. This model achieved performance close to the best conventional machine learning method [4]. Wang et al. [14] adopted the classical BiLSTM-CRF architecture to recognize biomedical triggers, and word-level embedding and character-level embedding were combined as input representations. Li et al. [15] concatenated dependency-based word embedding to entity type embedding as input representations. They proposed a contextual label sensitive gated network to extract triggers. The model obtained a competitive F1-score on MLEE corpus.
Previous work has achieved advanced performance in biomedical trigger extraction, but there are still several problems. First, the triggers in biomedical field are usually verbs or verb phrases with fixed collocation. It is well known that verbs usually represent multiple meanings according to context information. This ambiguity is more common in biomedical literatures than in the general field. The main reason is that the relatively monotonous verbs are used to trigger multiple biomedical events. Moreover, abbreviations and coreferences also interfere with the extraction of biomedical triggers. For example, the trigger “increase”, which belongs to the event “E1” in Fig. 1, is also treated as a non-trigger in one third of samples on the MLEE corpus. Furthermore, the token “activates” has only a third of the probability of being used as triggers in different corpora, and the rest are non-triggers. “play” can be serve as different types of triggers (Regulation, Positive_regulation, Negative_regulation) and non-trigger. Most of the current neural network models are based on conventional pretrained word embedding. They are limited by fixed feature representations and cannot effectively express the different semantics of vocabulary in complex contexts. Employing pretrained language model to dynamically compute contextualized representations is an effective avenue to improve the above problems. Second, tokens in the fixed collocation verb phrase are highly label dependent, all tokens in a phrase must be complete and in a fixed order. Third, most of the current representative trigger extraction models are based on conventional shallow neural networks, such as single-layer convolutional neural network (CNN) or recurrent neural network (RNN) and their improved structures. The single-layer structures greatly limit the ability of feature capturing in the encoding layer of the models, while the stacked deep network structures will cause training difficulties due to too many training parameters, which is more likely to cause gradient exploding or gradient vanishing. Therefore, how to properly enhance the depth of neural networks under the premise of avoiding information redundancy and gradient anomaly has become one of the solutions to this problem. Residual structure with recurrent trick has achieved great success in the field of computer vision [16, 17]. In the field of natural language processing (NLP), Huang et al. [18] designed a 9-layer CNN based on residual learning for distantly supervised noisy relation extraction in general field. Gui et al. [19] proposed a residual LSTM, which used dynamic skip connection based on reinforcement learning to simulate the variable distance dependence between words. These studies suggest that the residual structure has positive effects on NLP problems. However, the effect of residual structure on trigger extraction task is still not well understood, especially in biomedical field. The deep neural networks based on residual structure can effectively make up for the lack of feature extraction ability of shallow neural networks. Therefore, this paper considers the use of residual architecture to alleviate this problem.
In this paper, we propose a multi-layer residual BiLSTM-CRF architecture based on contextualized word embedding. First of all, embeddings from language models (ELMo) [20] is used to compute contextualized word embedding. The generated embedding is combined with pretrained word embedding and character embedding at the same time. The introduction of contextualized word embedding can solve the problem of triggers ambiguity in different contexts. Then, we treat the trigger extraction as a sequence labeling problem, considering the dependency of token labels in trigger phrases, CRF [21] is employed as the output layer to combine with BiLSTM. Finally, because the shallow neural networks cannot obtain the hidden valuable features of complex sequences effectively, we design a multi-layer residual BiLSTM block. The residual connection avoids the gradient exploding or vanishing during the training period. The introduction of gate mechanism in this block can dynamically control the transmission of information and avoid the information redundancy problem of multi encoding layers. In summary, the main contributions of this paper are as follows:
-
1.
We dynamically express the semantic information of tokens in different sequences through contextualized word representations.
-
2.
The multi-layer residual BiLSTM-CRF architecture is designed to capture hidden valuable features from complex biomedical text sequences. The residual BiLSTM block realizes the adaptive stacking of multi encoding layers and the CRF layer alleviates the problem of strong label dependence among tokens in trigger phrases.
-
3.
The experimental results show that our model obtains the advanced performance on the biomedical MLEE corpus.
The structure of this paper is as follows: Sect. 2 introduces the multi-layer residual BiLSTM architecture proposed in this paper. Section 3 describes the experimental corpus and settings. Section 4 reports the experimental results and analysis. Section 5 summarizes our work and draws conclusions.
2 Methods
This section first introduces the overall structure of our approach, and then describes each part of the model in detail.
2.1 Overall framework
The multi-layer residual BiLSTM architecture is shown in Fig. 2. It is divided into embedding layer, residual BiLSTM block and CRF layer. First, original sequences are transformed into multiple embedding at the embedding layer. They are standardized to be consistent with the dimension of residual BiLSTM block. Secondly, in multi-layer residual BiLSTM block, these features representations are captured by BiLSTM at first. Then residual connection and layer normalization strategies in residual architecture are performed between these features and the output of BiLSTM. They are sent to the gated multi-layer perceptron at last, and two strategies of residual architecture are implemented again. In order to fully extract hidden features, the above process is repeated 4 times. The output of each iteration serves as input for the next iteration. Finally, the output of the multi-layer residual BiLSTM block are fed into the CRF layer to parse labels.
The framework of multi-layer residual BiLSTM. The contextualized word embedding, pretrained word embedding and character embedding are generated by ELMo, \(PMC+PubMed\) pretrained vocabulary and BiLSTM, respectively. They are concatenated and input into the residual BiLSTM block. The green dots represent the output of BiLSTM. The residual connection operation is represented by dotted line. The blue striped dots represent the gated multi-layer perceptron. The output of the residual BiLSTM block after four iterations is fed into the CRF layer to predict the labels
2.2 Embedding layer
The embedding layer adopts three kinds of representations, including: pretrained word embedding, contextualized word embedding and character embedding. They are used to optimize the ambiguity of tokens and to parse the internal structure of tokens.
2.2.1 Pretrained word embedding
First, we introduce the pretrained word embedding. Word embedding is a kind of distributed representation, which is used to transform high-dimensional, sparse one-hot vectors into low-dimensional, dense representations. They map original tokens into vectors of fixed length, and distinguish the similarity between tokens according to the vectors’ distribution. Word embedding optimizes the performance of neural networks in NLP tasks. Common tools for computing distributed representations include Word2vec [22], Glove [23], etc.
In view of the fact that high-quality word embedding can better express potential semantic features and language patterns. The word embedding pretrained in large-scale biomedical corpora provided by Pyysalo et al. [24] is adopted as input of the model in this paper.
2.2.2 Contextualized word embedding
Previous pretrained word embedding is based on fixed vectors trained from large-scale corpora, which is context-independent and cannot reflect the proper semantics of tokens. On the basis of the pretrained word embedding, this paper additionally employs the 2-layer ELMo to compute the contextualized representations as one of our model’s input. Unlike other fine-tuning-based pretrained language models [25, 26], ELMo can flexibly adapt to the neural networks of various downstream tasks, and does not rely on high-performance devices and large-scale computing. In order to express the semantic features of tokens and the syntactic features of sequences dynamically, we directly transfer the pretrained ELMo language model proposed by Peters et al. [20] to generate the contextualized word representations. As shown in Fig. 3, the model includes two unidirectional LSTMs in opposite directions. Given the original embedding as input, the contextualized word representations are obtained as follows:
where \(x_{k}^{LM}\) represents the original word embedding of the k-th token, \(\overrightarrow{h_{k,j}^{LM}}\) and \(\overleftarrow{h_{k,j}^{LM}}\) represent the k-th output of two unidirectional LSTMs in opposite directions, w represents the softmax-normalized parameters and L represents the layer number. ELMo calculates the contextualized word representations through the above formulas. The representations is generated by performing addition operation on each layer’s output of the language model, or only adopts the output of the last layer. The former is adopted in this paper. The contextualized word representations can be directly concatenated to other input embedding as feature representations.
2.2.3 Character embedding
In addition to the above two kinds of coarse-grained representations, the lexical features are also beneficial for trigger extraction because triggers of biomedical events are usually words or phrases suffixed with various tenses. In order to make the model learn these fine-grained features, we adopt BiLSTM to compute character embedding which has advantages in capturing continuous features. As shown in Fig. 4. First, all characters of tokens are randomly initialized to the embedding. Then BiLSTM is employed to capture bidirectional global features of character matrix. Finally, these global features are integrated as the character-level embedding of tokens.
2.2.4 Multiple embedding integration
Let n denote the number of tokens, \(W_{elmo}\in R^{n\times d_e}\) denote the contextualized word embedding generated by ELMo, \(W_{word}\in R^{n\times d_w}\) denote the pretrained word embedding generated by \(PMC+PubMed\) pretrained vocabulary, \(W_{char}\in R^{n\times d_c}\) denote the character embedding generated by BiLSTM. In this step, the concatenate operation is performed to integrate the above three kinds of embedding. The final input representations of the model are as follows:
where \(\oplus\) denotes the concatenate operation. \(W_{f}\in R^{n\times d_{e+w+c}}\).
2.3 Residual BiLSTM block
2.3.1 Background
LSTM [27] is an evolutionary model of RNN. It is proposed to optimize the gradient vanishing or exploding problems of RNN when capturing long distance information. LSTM designs memory cell and gate mechanism based on RNN structure. The memory cell is designed for caching and propagating information. The gate mechanism can be seen as “valve”. There are three kinds of gates designed to control and update the input information (input gate i, forget gate f and output gate o). Due to the gate mechanism, LSTM is able to capture features from longer distance than RNN. The internal structure of LSTM unit is shown in the Fig. 5 and its updating process at time t is as follows:
where \(x_t\) is the original input of the unit. \(\sigma\) is the sigmoid function of gate mechanism. \(*\) is the dot product between vectors. \(W_i\), \(W_f\), \(W_o\) and \(W_c\) are weight parameter matrices. \(b_i\), \(b_f\), \(b_o\) and \(b_c\) are biases. \(i_t\), \(f_t\) and \(o_t\) denote the state of input gate, forget gate and output gate at time t, respectively. \(c_t\) and \(\widetilde{c_t}\) denote the final state and candidate state of the memory cell at time t, respectively. \(h_t\) is the output of the LSTM unit at time t.
In order to capture context information adequately, this paper employs bidirectional LSTM (BiLSTM) to extract features. First, BiLSTM captures the forward and backward features of the input sequence respectively. Then the concatenate operation is performed to merge two features as the global features. Finally, the global features are output to the residual architecture. The global features of BiLSTM are as follows:
where \(\oplus\) denotes the concatenate operation, \(\overrightarrow{h}\) and \(\overleftarrow{h}\) denote the forward and the backward features captured by BiLSTM, respectively.
The residual architecture is designed in two strategies, including residual connection and normalization. Residual connection is proposed by He et al. [28] in image recognition. It merges the current layer’s input and output as the input of the next layer. The residual connection avoids the problem of gradient exploding or vanishing with the continuous iteration.
The normalization is to optimize the convergence rate of parameters in complex neural networks. Because the conventional batch normalization relies on large batch size, it is difficult to apply in sequence labeling models with inconsistent sample lengths. So we employ layer normalization [29] to optimize neurons. It normalizes different neurons based on a single sample, which is more suitable for our task than batch normalization.
2.3.2 Residual BiLSTM block structure
The residual BiLSTM block is the most important part of our model. Its structure is shown in the middle shaded part of Fig. 2. We extract features iteratively by the residual BiLSTM block. The output of the current iteration serves as the input for the next iteration. The final output is fed into the CRF layer. The residual architecture guarantees the performance of feature extraction for our multi-layer BiLSTM block. We perform residual connection and layer normalization of the input and output of BiLSTM. The relevant formula is as follows:
where x denotes the input of BiLSTM, h denotes the output of BiLSTM, \(y_{B}\) denotes the final output of BiLSTM through the above strategies.
After residual connection and layer normalization, \(y_{B}\) is input to gated multi-layer perceptron (GMLP). GMLP is a feed-forward network with gate mechanism that maps a set of input vectors to a set of output vectors. It is proposed to integrate captured information while avoiding redundancy and controlling output. The structure of GMLP is shown in the Fig. 6.
The original formula is as follows:
where \(y_{B}\) and y denote the input and output of GMLP, respectively. \(M(y_{B})\) and \(M'(y_{B})\) denotes the identical MLP without activation function. \(\sigma\) denotes the sigmoid function, which is the gate mechanism. Because its interval is [0, 1], it can act as a “valve” to control output. We also adopt residual connections in GMLP:
Since \(M(y_{B})\) is a linear transform without activation function. The above formula is equivalent to:
where \(\times\) denotes element-wise multiplication, y is equivalent to \(y'\). The above formula can more intuitively explain the motivation of the designed GMLP: the input information is directly output with the probability of \(1-\sigma (M'(y_{B}))\), and output with the probability of \(\sigma (M'(y_{B})\) through the gate mechanism. Similar to GRU [30], this effectively optimizes information transmission and avoids gradient vanishing.
In this block, residual connection and layer normalization are performed again for the output of GMLP. The relevant formula is as follows:
y denotes the output of GMLP. \(y_{G}\) denotes the output of GMLP through the above two strategies, which is fed into the CRF layer.
2.4 CRF Layer
2.4.1 Background
In this paper, we treat biomedical trigger extraction as a sequence labeling task based on BIO label scheme. In detail, each token is classified into three boundary labels (label “B”, label “I” and label “O”). “B” denotes the first token of a trigger. “I” denotes the internal tokens of a trigger. “O” denotes the non-triggers. Although independent decision functions such as Softmax can generate prediction labels, they ignore the label dependency problem and cannot recognize illegal label sequences. For example, it is illogical for label “I” to follow behind label “O”. Therefore, our model employs CRF layer to solve this problem. CRF considers not only the initial probability generated by decision function, but also the transition probability between labels. It focuses on sentence-level label prediction, not limited to token-level. The effectiveness of adopting CRF layer has been demonstrated in previous biomedical trigger extraction studies [14].
2.4.2 CRF structure
Assuming that given an input sequence \(x=\{x_1,x_2,\ldots ,x_n\}\), there exists a corresponding label sequence \(l=\{l_1,l_2,\ldots ,l_n\}\), n represents the number of tokens. In addition, for a probability matrix P, \(P\in R^{n\times m}\), m represents the types of labels. First, let i and j denote the i-th token in the sequence and the j-th class in the label, respectively. The formula of P is as follows:
where T represents the transition matrix of labels. \(T_{ij}\) represents the score from the previous label to the current label. Let \(l_0\) and \(l_n\) be the head and end of the label sequence, \(T\in R^{(m+2)\times (m+2)}\).
Then, let \(l_x\) denote the predicted trigger labels of original sentence x. The prediction probability of all possible label sets l in CRF layer is expressed as \(P(l\mid x)\). Its formula is as follows:
where \(\widetilde{l}\) represents the actual labels of original samples.
Next, the following likelihood function monitors the distance between predicted and real results in order to optimize the model.
Finally, the CRF layer maximizes the likelihood function by adjusting the parameters during training, and generates the optimal labels \(l^*\) under this condition:
3 Experimental settings
This section introduces the relevant settings during model training, including dataset, label scheme, optimizer, regularization, parameters and evaluation measures.
3.1 Dataset and label scheme
In order to evaluate the performance, the trigger extraction experiments are carried out on MLEE [1], the mainstream corpus of biomedicine. MLEE corpus includes 4 categories of 19 biomedical event types. It covers molecular, cellular, organ and so on. The size of MLEE corpus adopted by us is consistent with Rahul et al. [12] and Li et al. [15]. There are 2608 samples in the corpus, of which 1728 are used for training and 880 are used as test sets. The trigger statistics for MLEE are shown in Table 2.
In this paper, trigger extraction is treated as a sequence labeling problem. Therefore, a label scheme is needed to re-label the samples containing triggers. We adopt BIO as the label scheme, which is one of the most common method in sequence labeling. B denotes the beginning of triggers, I denotes the middle part of trigger phrases, and O denotes that the token does not belong to a trigger. Taking sentence “matrix metalloproteinases are present at elevated levels during early wound healing when angiogenesis occurs and suggest that matrix metalloproteinase-9 may play a significant role” as an example, the result of BIO-based labeling is shown in Table 3.
3.2 Optimizer and regularization
Adam [31] is a first-order optimization algorithm which can replace the traditional stochastic gradient descent (SGD). It iteratively updates the parameters during training. Adam designs independent adaptive learning rates for parameters by computing the first-order and second-order moment estimations of gradients. It retains a learning rate for each parameter to improve the performance on the sparse gradient. Therefore, this paper employs Adam as the optimizer.
Dropout [32] is a conventional regularization method during model training. In each batch, the over-fitting can be significantly reduced by ignoring the fixed proportion of neurons in forward propagation. This method can reduce the interaction between neurons and make the model more generalized which does not depend too much on some local features in hidden layer.
3.3 Parameter settings
The specific parameter settings in our model are shown in Table 4. This paper sets the dimension of pretrained word embedding to 200, the contextualized word representations to 1024, and the character-level embedding to 50. The number of unidirectional LSTM units is 100. In addition, the dropout rate, learning rate and batch size are set to 0.5, 0.001 and 20, respectively.
3.4 Evaluation measures
In order to objectively and comprehensively compare with existing studies, the following three kinds of evaluation measures are employed in this paper: Precision, Recall and F1-score. Their definitions are described below.
where TP, FP, FN and TN respectively represent the true positive samples with correct prediction, the true negative samples with incorrect prediction, the true positive samples with incorrect prediction and the true negative samples with correct prediction.
4 Results and discussions
The performance comparisons are reported and discussed in this section. First, experiments 1 compares the performance of different pre-training word vectors (Sect. 4.1). The second experiment verifies the validity of each feature representation employed in this model (Sect. 4.2). In order to evaluate the effect of LSTM unit number on performance, we set up the third experiment (Sect. 4.3). Then we experiment on the strategies designed in the model (Sect. 4.4). Finally, we report and discuss the performance comparison between our model and existing studies (Sect. 4.5). In addition, we conduct case study and error analysis on the prediction results of the model (Sects. 4.6 and 4.7).
4.1 Performance comparison of pre-trained embedding
Experiments 1 evaluates the effects of different pretraining word embedding on the performance of our model. We provide 200-dimensional word vectors, which are randomly initialized, pretrained by GloVe in general large-scale corpus, and pretrained by Word2Vec in four kinds of biomedical corpora, namely PMC, PubMed, \(PubMed+Wiki\) and \(PMC+PubMed\). The experimental results are shown in Table 5. The experimental results show that the performance of the model using pretrained word embedding is better than that of randomly initialized word embedding. In addition, the performance of biomedical pretraining word embedding is better than that of general domain pretraining word embedding, among which \(PMC+PubMed\) has the best performance. Therefore, \(PMC+PubMed\) is adopted as the pretraining word embedding in our model.
4.2 Performance comparison of each representation
This paper combines contextualized word embedding, pretrained word embedding and character-level embedding as input feature representations. Experiment 2 compares the performance of each feature representation. The experimental results are shown in Table 6. The results show that contextualized word embedding can effectively improve performance. In the case study section, we provide an actual comparison result between our model and baseline model in dealing with typical complicated cases. The experimental results show that this model can effectively judge the actual meaning of triggers when they have multiple meanings, which also prove that the token ambiguity problem proposed in this paper can be effectively optimized. In addition, character-level embedding can deeply analyze the internal structure of tokens and better capture their semantic and lexical features. Therefore, the above 3 feature representations are input into our model after connecting.
4.3 Performance comparison of LSTM unit dimensions
The number of units in neural networks plays an important role in feature extraction. The third experiment compares the effects of different unit numbers on the performance. Four parameters, 50, 100, 150 and 200, are set in the parameter set of LSTM units. The experimental results are shown in Table 7. It can be concluded that the optimum number of units in LSTM is 100. Inadequate or excessive number of units will interfere with feature extraction, which may result in feature missing or feature redundancy.
4.4 Performance comparison of different strategies
Next, this paper verifies the effectiveness of various strategies designed in the model, including bidirectional LSTM, CRF layer, residual mechanism and multi-layer residual blocks. In this paper, unidirectional LSTM is used as the baseline model to evaluate the performance improvement of the model after employing the above strategies. The experimental results are shown in Table 8. After adopting the above strategies in turn, the performance of the model is significantly improved. From the table, the introduction of CRF layer can effectively improve the performance of the model. Moreover, with the adoption of the residual block we designed, the precision rate of the model has been significantly improved, which may be due to the residual structure providing more abundant hidden information for the model, and the gate mechanism strictly limits the transmission of low-quality features, which may lead to the reduction of recall rate to a certain extent. Furthermore, with the iteration of the residual block, the model captures more features, so its generalization begins to improve gradually, that is, the precision rate decreases slightly while the recall rate increases significantly. Finally, the model achieves the advanced overall performance. The number of layers of residual blocks is determined to be 4 after many adjustments. From the above results, we conclude that the strategies designed in this model have positive effects on biomedical trigger extraction.
4.5 Performance comparison with existing methods
Finally, as shown in Table 9, we report the performance comparison between our model and existing methods. The following is a brief introduction to the existing methods in the table.
Pyysalo et al. [1] proposed a SVM model that connected contexts and dependencies features.
Nie et al. [11] employed skip-gram model to generate word embedding to optimize the performance of neural networks.
Wang et al. [14] introduced a BiLSTM-CRF model and used word-level and character-level representations in the input layer.
Wang et al. [10] adopted artificial neural network (ANN) to identify triggers after regenerating raw dependency-based word embedding.
Zhou et al. [3] proposed a SVM model based on feature engineering and domain knowledge.
Rahul et al. [12] treated trigger extraction as a sequence labeling problem. They combined word embedding with entity type embedding and fed them into BiRNN to label triggers.
He et al. [13] merged dependency-based word embedding and sentence vectors. The attention-based BiLSTM was employed to extract triggers.
He et al. [4] proposed a SVM classifier based on two-stage method.
Fei et al. [33] proposed a RecurNN-CRF model, which combined the dependency-tree based RNN with CRF layer.
As can be seen from the table above, our model achieves the competitive performance with an F1-score of 80.74% on the premise of relying only on a simple and flexible architecture. Compared with the above methods based on BiLSTM such as He et al. [13] and Wang et al. [14], our model enhances the ability of feature extraction based on residual connection and gate mechanism. Meanwhile, the contextualized word embedding generated by ELMo can effectively alleviate the ambiguity and polysemy of biomedical triggers. He et al. [4] divide biomedical trigger extraction into two stages: detection and classification. They classify biomedical triggers by SVM based on two-stage method. This method achieves the best performance of conventional machine learning methods (F1-score: 79.75%). Compared with this method, although the precision rate of our model is reduced by 0.46%, the recall rate and F1-score are increased by 2.45% and 0.99%, respectively. More importantly, our model avoids any manual participation and feature engineering. This saves time and labor costs associated with data preprocessing and feature selection. In addition, compared with the recent advanced deep learning approach of Fei et al. [33], the F1-score of our model is increased by 0.46%. Moreover, our approach does not rely on special parsing tools to generate the dependency tree features, which makes our model more flexible.
4.6 Case study
The case study is shown in the Table 10. The sentence structure of this example is common in MLEE corpus. It contains multiple biomedical triggers where includes a complex phrase. We adopt LSTM as the baseline model to verify the effectiveness of strategies in this paper. The baseline model recognizes the single triggers “elevated” and “angiogenesis” correctly, but cannot fully extract the phrase “play a significant role”. Moreover, the single trigger “present” is also mispredicted. This is caused by the insufficient feature extraction and ambiguity of triggers. Our model correctly extracts all triggers, including a complex phrase. The proposed multi-layer residual BiLSTM architecture and the adopted of contextualized word representations accurately express the semantic information of triggers according to the context, and sufficiently capture the global features. The above design enables the biomedical trigger to be extracted correctly and completely.
4.7 Error analysis
We further analyze the error samples of the experimental results and conclude that the two factors most likely to cause the prediction errors are as follows.
Firstly, Table 11 lists the proportion of the categories with the most and the least samples in MLEE corpus. There is a serious sample imbalance between the categories, and the proportion exceeds 300:1. Due to the lack of training samples, our model cannot fully capture and extract the features of categories with fewer samples.
In addition, triggers in some samples are various and dense. Continuous triggers make our model more difficult to identify them completely. For example, in example sentences “endothelial cell proliferation plays a pivotal role in angiogenesis.” and “endothelial cell proliferation plays a pivotal role in angiogenesis.”, the number of triggers accounts for almost half of the whole sequence and belongs to different categories. These trigger categories include Cell_Proliferation, Localization, Positive_Regulation, Blood_Vessel_development and so on. Our model does not show reasonable performance when there are dense triggers distributed in short sequences.
5 Conclusion
This paper proposes a multi-layer residual BiLSTM-CRF architecture with contextualized word representations in biomedical trigger extraction. The trigger extraction task is treated as a sequence labeling problem. Through the contextualized word representations are generated by ELMo, our model can effectively solve the ambiguity problem in biomedical literature and make up for the deficiency of conventional word embedding. By multi-layer residual BiLSTM block, the model is able to improve the ability of capturing features and avoiding gradient exploding or vanishing. The experimental results show that the model obtains the competitive performance on the biomedical MLEE corpus. In the future, we plan to adopt multi-task learning to address the lack of training samples, so as to alleviate the imbalance of samples between categories. Moreover, we can employ additional linguistic features and multiple contextualized word embedding.
References
Pyysalo S, Ohta T, Miwa M, Cho H-C, Tsujii J, Ananiadou S (2012) Event extraction across multiple levels of biological organization. Bioinformatics 28(18):i575–i581
Zhou D, Zhong D (2015) A semi-supervised learning framework for biomedical event extraction based on hidden topics. Artif Intell Med 64(1):51–58
Zhou D, Zhong D, He Y (2014) Event trigger identification for biomedical events extraction using domain knowledge. Bioinformatics 30(11):1587–1594
He X, Li L, Liu Y, Yu X, Meng J (2017) A two-stage biomedical event trigger detection method integrating feature selection and word embeddings. IEEE/ACM Trans Comput Biol Bioinf 15(4):1325–1332
Zhou W, Zhu Z (2020) A novel bnmf-dnn based speech reconstruction method for speech quality evaluation under complex environments. Int J Mach Learn Cybern 12(11):1–14
Zhang T, Yang X, Wang X, Wang R (2020) Deep joint neural model for single image haze removal and color correction. Inf Sci 541:16–35
Xiao Y, Yin H, Duan T, Qi H, Zhang Y, Jolfaei A, Xia K (2020) An intelligent prediction model for ucg state based on dual-source LSTM. Int J Mach Learn Cybern. https://doi.org/10.1007/s13042-020-01210-7
Li Y, Xu Z, Wang X, Wang X (2020) A bibliometric analysis on deep learning during 2007–2019. Int J Mach Learn Cybern 11(12):2807–2826
Zhang Y, Lin H, Yang Z, Wang J, Sun Y, Xu B, Zhao Z (2019) Neural network-based approaches for biomedical relation classification: a review. J Biomed Inform 99:103294
Wang J, Zhang J, An Y, Lin H, Yang Z, Zhang Y, Sun Y (2016) Biomedical event trigger detection by dependency-based word embedding. BMC Med Genom 9(2):45
Nie Y, Rong W, Zhang Y, Ouyang Y, Xiong Z (2015) Embedding assisted prediction architecture for event trigger identification. J Bioinform Comput Biol 13(03):1541001
Rahul PV, Sahu SK, Anand A (2017) Biomedical event trigger identification using bidirectional recurrent neural network based models. BioNLP 2017:316–321
He X, Li L, Wan J, Song D, Meng J, Wang Z (2018) Biomedical event trigger detection based on bilstm integrating attention mechanism and sentence vector. In: 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). IEEE, pp 651–654
Wang Y, Wang J, Lin H, Tang X, Zhang S, Li L (2018) Bidirectional long short-term memory with crf for detecting biomedical event trigger in fasttext semantic space. BMC Bioinform 19(20):507
Li L, Huang M, Liu Y, Qian S, He X (2019) Contextual label sensitive gated network for biomedical event trigger extraction. J Biomed Inform:103221
Wang C, Zhou SK, Cheng Z (2020) First image then video: a two-stage network for spatiotemporal video denoising. arXiv:2001.00346
Claus M, van Gemert J (2019) Videnn: Deep blind video denoising. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition (CVPR) workshops, Long Beach, CA, pp 1843–1852
Huang YY, Wang WY (2017) Deep residual learning for weakly-supervised relation extraction. arXiv:1707.08866
Gui T, Zhang Q, Zhao L, Lin Y, Peng M, Gong J, Huang X (2019) Long short-term memory with dynamic skip connections. Proc AAAI Conf Artif Intell 33:6481–6488
Peters ME, Neumann M, Iyyer M, Gardner M, Clark C, Lee K, Zettlemoyer L (2018) Deep contextualized word representations. arXiv:1802.05365
Lample G, Ballesteros M, Subramanian S, Kawakami K, Dyer C (2016) Neural architectures for named entity recognition. arXiv:1603.01360
Mikolov T, Chen K, Corrado G, Dean J (2013) Efficient estimation of word representations in vector space. arXiv:1301.3781
Pennington J, Socher R, Manning C (2014) Glove: global vectors for word representation. In: Proceedings of the 2014 conference on empirical methods in natural language processing (EMNLP), Doha, Qatar, pp 1532–1543
Moen S, Ananiadou TSS (2013) Distributional semantics resources for biomedical text processing. In: Proceedings of the 5th international symposium on languages in biology and medicine (LBM), Tokyo, Japan, pp 39–44
Radford A, Narasimhan K, Salimans T, Sutskever I (2018) Improving language understanding by generative pre-training. https://s3-us-west-2.amazonaws.com/openai-assets/research-covers/language-unsupervised/language_understanding_paper.pdf
Devlin J, Chang M-W, Lee K, Toutanova K (2018) Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv:1810.04805
Hochreiter S, Schmidhuber J (1997) Long short-term memory. Neural Comput 9(8):1735–1780
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition (CVPR), Las Vegas, Nevada, pp 770–778
Ba JL, Kiros JR, Hinton GE (2016) Layer normalization. arXiv:1607.06450
Chung J, Gulcehre C, Cho K, Bengio Y (2014) Empirical evaluation of gated recurrent neural networks on sequence modeling. arXiv:1412.3555
Kingma DP, Ba J (2014) Adam: a method for stochastic optimization. arXiv:1412.6980
Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R (2014) Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 15(1):1929–1958
Fei H, Ren Y, Ji D (2020) A tree-based neural network model for biomedical event trigger detection. Inf Sci 512:175–185
Acknowledgements
This work was supported by National Natural Science Foundation of China (No. 61976124).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wei, H., Zhou, A., Zhang, Y. et al. Biomedical event trigger extraction based on multi-layer residual BiLSTM and contextualized word representations. Int. J. Mach. Learn. & Cyber. 13, 721–733 (2022). https://doi.org/10.1007/s13042-021-01315-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13042-021-01315-7