Abstract
Stroke remains a leading cause of mortality; however, available therapeutics are limited. The study of ischemic tolerance, in paradigms such as resveratrol preconditioning (RPC), provides promise for the development of novel prophylactic therapies. The heavily oxidative environment following stroke promotes poly-ADP-ribose polymerase 1 (PARP1)-overactivation and parthanatos, both of which are major contributors to neuronal injury. In this study, we tested the hypothesis that RPC instills ischemic tolerance through decreasing PARP1 overexpression and parthanatos following in vitro and in vivo cerebral ischemia. To test this hypothesis, we utilized rat primary neuronal cultures (PNCs) and middle cerebral artery occlusion (MCAO) in the rat as in vitro and in vivo models, respectively. RPC was administered 2 days preceding ischemic insults. RPC protected PNCs against oxygen and glucose deprivation (OGD)–induced neuronal loss, as well as increases in total PARP1 protein, implying protection against PARP1-overactivation. Twelve hours following OGD, we observed reductions in NAD+/NADH as well as an increase in AIF nuclear translocation, but RPC ameliorated NAD+/NADH loss and blocked AIF nuclear translocation. MCAO in the rat induced AIF nuclear translocation in the ischemic penumbra after 24 h, which was ameliorated with RPC. We tested the hypothesis that RPC’s neuroprotection was instilled through long-term downregulation of nuclear PARP1 protein. RPC downregulated nuclear PARP1 protein for at least 6 days in PNCs, likely contributing to RPC’s ischemic tolerance. This study describes a novel mechanism by which RPC instills prophylaxis against ischemia-induced PARP1 overexpression and parthanatos, through a long-term reduction of nuclear PARP1 protein.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Reactive oxygen species (ROS) overproduction during and following cerebral ischemia leads to oxidative stress (OS) [1, 2]. High levels of ROS can result in DNA oxidation and eventual oxidative genotoxic stress (OGS) [3]. The DNA damage response is a collection of cellular mechanisms responsible for the repair of DNA lesions. These pathways are rapid and efficient. However, select mechanisms can induce cell death. For example, in the presence of overabundant single-strand breaks (SSB), the enzyme poly-ADP-ribose polymerase 1 (PARP1) drives the cell towards PARP1-dependent cell death [4].
PARP1-mediated cell death, also known as parthanatos, has been shown to contribute to cerebral ischemic injury both in vitro and in vivo [5,6,7,8]. PARP1 utilizes large amounts of nicotinamide adenine dinucleotide (NAD+) to produce poly-ADP-ribose (PAR). PARP1 itself is a common recipient of PAR and this molecular flag attracts SSB repair proteins [9,10,11]. During and following cerebral ischemia, PARP1 can be overactivated. Increases in PARP1 protein have been reported following in vitro cerebral ischemia in primary neuronal cultures (PNCs), suggesting the potential for overactivation [12]. PARP1-overactivation rapidly depletes nuclear NAD+ and allows for the diffusion of PAR [13]. PAR then binds and releases apoptosis-inducing factor (AIF) from the mitochondria [7]. In the cytosol of cortical neurons, AIF binds to macrophage migration inhibitory factor (MIF), which induces translocation to the nucleus and MIF-mediated DNA fragmentation [14, 15]. Targeting PARP1 through either pharmacological inhibition or genetic knockout has shown protection against middle cerebral artery occlusion (MCAO) [4, 16, 17].
Ischemic preconditioning (IPC) induces ischemic tolerance to protect against cerebral ischemia [18]. In the context of PARP1-mediated injury, IPC was shown to reduce PARP1 activity in astro-neuronal cultures, implying that ischemic tolerance may be achieved through targeting PARP1 [19]. Our group has studied mechanisms of IPC as well as a mimetic to IPC called resveratrol (RSV) [20,21,22,23,24]. Resveratrol preconditioning (RPC) instills ischemic tolerance in multiple windows and through various means [21,22,23,24,25]. Previously, we have shown that RPC induces nuclear erythroid 2-related factor 2 (NRF2)–dependent neuroprotection against MCAO in the mouse, suggesting that RPC may protect against OS [26]. Our current study sought to further expound on RPC’s mechanisms of neuroprotection, specifically in the context of OGS.
This study aimed to determine whether RPC promotes ischemic tolerance by targeting PARP1. We hypothesized that RPC initiates PARP1 downregulation to prevent PARP1 overexpression and parthanatos. To investigate this hypothesis, we assessed RPC for neuroprotection and prevention of PARP1 overexpression in vitro. We then assessed protection against parthanatos both in vitro and in vivo. Additionally, we characterized RPC’s downregulation of PARP1 in PNCs. Overall, this study describes a neuroprotective mechanism of RPC through the downregulation of PARP1.
Methods
Animals
Animal use in this study was approved by the University of Miami Institutional Animal Care and Use Committee. Experiments using animals followed the ARRIVE Guidelines and the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals. All animals were housed in the University of Miami Department of Veterinary Resources facilities under a 12-h light–dark cycle with food and water ad libitum.
Primary Neuronal Cultures
PNCs were prepared as previously described [27]. Briefly, primary neurons were sourced from embryonic day E18-20 rat cerebral cortex. Pregnant Sprague–Dawley rats were obtained from Charles River (Wilmington, MA). Rats were anesthetized with 3% isoflurane and decapitated. Embryos were extracted, decapitated, and cerebral cortices were isolated. Cortices were kept in ice-cold dissection medium and diced before trypsinization, with DNase, for 15 min at 37 °C. Trypsin was inactivated by the plating medium (MEM, 1% Glutamax, 5% fetal bovine serum, 15 mmol/L glucose) and brain bits were then dissociated to produce single cells. Homogenates were passed through a 70-μm cell strainer and then seeded at 2.5 \(\times\) 105 cells/cm2 on poly-d-lysine pre-coated sterile plates. The media was replaced with Neuronal Culture Medium (Neurobasal Medium, 1X B-27 Supplement, 0.5 mM Glutamax) following adherence. PNCs were maintained with half-media replacement every 2–3 days.
Resveratrol Preconditioning
For in vitro RPC, RSV (Sigma-Aldrich) was dissolved in DMSO at the concentration of 65 mg/mL and stored at − 20 °C. For in vivo RPC, RSV was dissolved in 0.5% carboxymethyl cellulose (CMC) (50 mg of CMC dissolved in 10 mL sterile water) at a concentration of 65 mg/mL and stored at − 20 °C. In vitro RPC on DIV8 began with expanding RSV in saline to 1 mg/mL, followed by vigorous mixing in the dark. Media was half-changed 1 and 2 h following RPC, with no protection from light. In vivo RPC took place 2 days before MCAO or sham. Aliquots were expanded with saline to 16.25 mg/mL and mixed vigorously immediately before use. A total of 50 mg/kg RPC or vehicle was injected intraperitoneally in the dark.
Middle Cerebral Artery Occlusion
Eight- to 12-week-old male rats were doubly housed and allowed to acclimate 8–14 days before being injected with RPC or vehicle, 2 days before MCAO or sham. MCAO was conducted as previously described [28]. Briefly, MCAO was implemented via an intraluminal suture blocking the right middle cerebral artery (MCA) for 90 min. Animals were first treated with atropine sulfate (0.5 mg/kg IP) and then anesthetized with 3.5% isoflurane, mixed in 70% N2O and 30% O2. Rats were then intubated, paralyzed, and mechanically ventilated. Blood gases, blood pressure, pH, and glucose were monitored periodically during the procedure. Blood flow was constantly monitored through laser Doppler perfusion monitoring (Perimed Inc.) via a fiberoptic probe at a burr hole made over the frontoparietal cerebral cortex. The right common carotid artery (CCA) was then exposed and the external carotid artery (ECA) isolated and coagulated. The ECA was then divided and an intraluminal 20-mm silicone-coated suture (Doccol, 403,956) was advanced along the internal carotid artery (ICA) to the origin of the MCA to occlude. After 90 min, the suture was removed and the CCA and ECA were inspected for patency. The animals were awakened and allowed to recover for 24 h. Rats were then anesthetized and perfused with ice-cold saline for 2 min, at 100–200 mmHg, before decapitation and brain removal. Brains were sectioned in a 2-mm matrix. The MCA penumbra was estimated as 2- to 0-mm anterior to bregma and − 6- to − 8-mm posterior to bregma of the cerebral cortex. Samples of both the ipsilateral and contralateral hemispheres were snap-frozen in liquid nitrogen before storage at − 80 °C.
This experiment had one exclusion criterion: ischemic phase blood flow drops below 30% of baseline. Three MCAO animals failed to reach this threshold (two RPC, one vehicle). Three animals did not survive to sample collection (two vehicle, one RPC). n = 5 for both MCAO groups and n = 6 for both sham groups.
Oxygen and Glucose Deprivation
Oxygen and glucose deprivation (OGD) experiments were performed as previously described [23, 27]. Briefly, cells were exposed to OGD medium and then transferred to an anoxia chamber (COY Lab Products, Grass Lake MI, USA) for 3.5 h at 37 °C. Following OGD, the culture media was replaced, and the plates were returned to the incubator. The OGD medium contained 1.26 mM CaCl2, 5.37 mM KCl, 0.44 mM KH2PO3, 0.49 mM MgCl2, 0.41 mM MgSO4, 136.9 mM NaCl, 4.17 mM NaHCO3, 0.34 mM Na2HPO4·7H2O, 20 mM sucrose, and 10 mM HEPES, adjusted to pH 7.4—all in distilled water and passed through a 0.2-μm sterile filter. Sham groups received media changes to account for cell death due to aspiration but did not receive OGD medium or anoxia.
Lactate Dehydrogenase Assay
Lactate dehydrogenase (LDH) cytotoxicity assessments were performed as previously described [23]. Twenty-four hours following OGD or sham, 100-μL aliquots of the culture supernatant were collected to conduct the colorimetric assay according to the manufacturer’s instructions (Cytotoxicity Detection Kit, Roche). All steps, aside from reaction incubation, were conducted on ice. Data were represented as the fold-change in LDH activity compared to sham.
Propidium Iodide and Hoechst Staining
Propidium iodide (PI) cytotoxicity assessments were performed as previously described [23]. Cell death was quantified through the ratio of PI-positive to Hoechst-positive cells. Twenty-four hours following OGD, PI at 2 μg/mL and two drops of NucBlue™ (Live ReadyProbes™) reagent were added to the cells and incubated for 10 min at 37 °C. Images were acquired on an EVOS™ inverted microscope (CKX41/CKX31) and analyzed via ImageJ for PI/Hoechst ratio.
Alkaline Comet Assay
DNA damage was assessed through the Alkaline Comet Assay. These experiments followed the manufacturer’s instructions (CometAssay® Kit, R&D System, Minneapolis, MN). Briefly, PNCs were collected in 1X PBS 1 h following OGD, sham, or H2O2 treatment and then mixed in agarose. Cell-agarose slurries were then added to glass slides and incubated in lysis buffer. DNA was denatured in an alkaline solution and then cells were subjected to electrophoresis. The DNA was then stained with SYBR™ Gold (Invitrogen) for 30 min at room temperature. Comets were imaged on a Leica Stellaris 5 Confocal Microscope and quantified for Head DNA, Tail DNA, and Tail Moment through the ImageJ plugin OpenComet.
NAD+/NADH Measurement
The levels of NAD+ and NADH were measured with the NAD+/NADH Quantification Colorimetric Kit (K337, Biovision. Waltham, MA). The procedure was conducted based on the manufacturer’s instructions. Briefly, 12 h following OGD or sham, PNCs were washed and collected in 1X PBS, pelleted, and resuspended in NADH/NAD+ Extraction buffer. Samples were lysed with freeze–thaw cycles and centrifuged to collect Total-NADH/NAD+ supernatant. Aliquots were taken and heated to 60 °C for NADH measurement. All samples and standards were transferred to a 96-well plate, treated with a reaction mixture, and developed. The optical density of 450 nm was measured every 40 min for 4 intervals via spectrophotometry. At lysis, aliquots were taken for protein quantification. Data were quantified as ng of NAD+ or NADH per microgram of protein.
Subcellular Fractionation
Cells
Cellular protein fractions were collected via the Subcellular Protein Fractionation Kit for Cultured Cells (Cat. # 78,840, Thermo Scientific. Waltham, MA). This experiment was conducted according to the manufacturer’s instructions. Twelve hours following OGD or sham, 7.5 \(\times\) 106 cells were washed and then collected in ice-cold 1X PBS. Cells were then pelletized to determine pellet size for buffer volume. Incubations with cytosolic, membrane, or nuclear extraction buffers produced all three protein fractions. Samples were stored at − 80 °C.
Tissue
Snap frozen cerebral cortex tissue samples underwent protein fractionation via the Subcellular Protein Fractionation Kit for Tissues (Cat. # 87,790, Thermo Scientific. Waltham, MA). Volumes of cytosolic, membrane, and nuclear extraction buffers were based on mass. Samples were diced into small bits and then homogenized in a Dounce Homogenizer 9 times in the cytosolic extraction buffer. The homogenate was then pelletized to yield the cytosolic fraction. Incubation with membrane and nuclear fraction buffers yielded these protein fractions. Samples were stored at − 80 °C.
RNA Extraction, cDNA Synthesis, and Real-Time PCR Analysis
RNA experiments were performed as previously described [23]. Briefly, RNA was extracted from PNCs with TRIzol reagent and QIAGEN RNeasy column (Venlo, The Netherlands) isolation. RNA was then converted to cDNA with the use of qScript cDNA Synthesis Kit (Quantabio). cDNA was used for real-time PCR with the Power SYBR Green PCR Master Mix on the CFX96™ Real-Time System (BioRad). All groups were assessed in triplicate and changes in expression were determined via the Delta-Delta-Ct Method. Primers were designed using Primer3 (version 0.4.0). Genomic DNA amplification was avoided with exon-exon covering primer design. The two sets of primers used are listed as follows:
-
Rat PARP1 forward: 5′-GCCAACCGTGAAAAGATGAC
-
Rat PARP1 reverse: 5′-CATCGGTAGGTCCGACACAA
-
Rat β-actin forward: 5′-ACAAACTGCAACTTCTGGAGTC
-
Rat β-actin reverse: 5′-CGATAACTGTGCCCACTCG
Western Blotting
Western Blotting was conducted as previously described [23, 27, 29]. Whole-cell culture lysates were homogenized using RIPA buffer and then passed through a 27-gauge needle 8 times. Cell and tissue subcellular fractionation samples were used in Western Blotting. Protein was loaded into a 4–20% SDS polyacrylamide gel (BioRad, Hercules, CA) and transferred onto a nitrocellulose membrane (Biorad), then blocked for 1 h at room temperature. Primary antibodies were incubated overnight at 4 °C. HRP-conjugated secondary antibodies (1:5000 or 1:10,000, GE Healthcare, Little Chalfont, UK) were incubated for 1 h at room temperature, and blots were developed with ECL reagents (ThermoFisher Scientific, Waltham, MA), and visualized with X-ray film (Denville Scientific, Holliston, MA) or Chemidoc™ MP Imaging System (BioRad). Blots were then digitalized and analyzed in ImageJ by densitometry. Antibodies used were as follows: rabbit anti-PARP1 (46D11, Cell Signaling Technologies CST), mouse anti-α-Tubulin (DM1A, CST), mouse anti-β-Actin (8H10D10, CST), rabbit anti-AIF (D39D2, CST), mouse anti-Lamin A/C (4C11, CST), rabbit anti-Na+/K+-ATPase (ab76020, abcam), rabbit anti-COX IV (3E11, CST).
Statistical Analysis
Rats were randomly assigned to groups for in vivo experiments. Grouping assignment, data acquisition, and data analysis were conducted blindly whenever possible. Tests for data normality and statistical analysis were conducted with the use of Prism9.2.0 Software (GraphPad, San Diego, CA). For all representations, data were presented with mean and error bars ± standard deviation. Statistical tests included one-way ANOVA, two-way ANOVA, and Welch’s T-test with appropriate post hoc testing. Welch’s T-test is used in place of Student’s T-test to assume unequal variances between groups, as well as to better control for type 1 error. p-values of p < 0.05 were considered significant. Sample sizes were determined from pilot studies or previous experiments conducted in the lab. When needed, power analysis determined effect sizes using G*power 3.1 software. Power analysis was conducted for MCAO animals. We used an α value of 0.05 and a β value of 0.8, with an effect size of 1.1, based on previous experiments. This yielded a total sample size of 22 rats.
Results
Resveratrol Preconditioning Protects Against Oxygen and Glucose Deprivation in a Two-Day Window
Our group has shown that RPC protects against ischemic damage, using both in vitro and in vivo models, in a 2-day window of ischemic tolerance [21, 23, 30]. Here we utilize the PNCs model in a 2-day window of RPC to determine its ability to protect against OGD. PNCs were exposed to a transient, 1-h, incubation with four concentrations of RPC (10 μM, 25 μM, 50 μM, 75 μM), 2 days before 3.5 h of OGD. Twenty-four hours following OGD, cytotoxicity was assessed via the LDH Assay (Fig. 1). Each group was first normalized as the percent increase in LDH activity compared to sham. Then, each concentration of RPC was normalized to its respective vehicle for fold change. The 50 μM RPC treatment showed significantly reduced LDH activity compared to its vehicle group (Veh 1 ± 0.5 vs RPC-50 μM 0.68 ± 0.39, p < 0.05, Fig. 1a). To confirm 50 μM RPC neuroprotection, we then conducted PI staining 24 h following OGD. The 50 μM RPC treatment reduced cell death by 42.8% compared to the vehicle (Veh 55.59 ± 6.3 vs RPC 31.8 ± 4.4, p < 0.001, Fig. 1c). With 50 μM RPC’s neuroprotection established, we next tested the hypothesis that RPC protects against PARP1 overexpression following OGD.
RPC protects against OGD in a two-day window. a Bar graph showing LDH levels 24 h following 3.5 h of OGD. A total of 50 μM RPC shows a significant reduction in LDH activity compared to the vehicle control (n = 9–11, (*) = p < 0.05, matched two-way ANOVA followed by Bonferroni’s post-test). b Representative images of Hoechst and PI staining for Sham, Veh-OGD, and RPC-OGD, 24 h following OGD. c Quantification of PI/Hoechst ratio in Veh-OGD and RPC-OGD groups showing RPC-OGD reduces PI-indicated death by 42.8%. (n = 6, (****) = p < 0.0001, Welch’s T-test)
RPC Protects Against OGD-Induced PARP1 Overexpression in PNCs
Following cerebral ischemia, PARP1 overactivation has been observed through increases in PARP1 protein and activity [31, 32]. RSV has been associated with reduced PARP1 activity in cardiomyocytes, suggesting that RPC may regulate PARP1 in neurons as well [33]. Based on these reports, we hypothesized that OGD induces PARP1 overexpression in PNCs, while RPC prevents this increase. To test this hypothesis, we measured total PARP1 protein levels, in a pilot titration, of immediately following OGD as well as 1 and 2 h following the insult. Total PARP1 protein levels from whole-cell lysates were assessed at these times, while sham protein was collected 1 h following media change (Fig. 2). OGD induced PARP1 overexpression, as seen through significant increases in total PARP1 protein levels compared to sham (Sham 1 ± 0.37 vs Immediate 3.38 ± 1 and vs 1-h 3.49 ± 0.86, p < 0.05, Fig. 2b). We also assessed apurinic/apyrimidinic endonuclease 1 (APE1) with no observed changes. Power analysis for APE1 produced a predicted sample size of 10 (n = 5) for a Welch’s T-test between Sham and 1-h groups (G*Power Version 3.1.9.3. α-value = 0.05, β-power = 0.8, effect size = 2.003). A separate experiment to assess APE1, between Sham and 1-h groups (n = 5), showed no differences in APE1, which was omitted from the study. We used this increased sample size experiment to assess PARP1 and confirm our previous observation. Additionally, a previous report indicated that 1 h following MCAO, penumbra regions show receding numbers of SSBs. However, core regions of injury demonstrate further accumulation of SSBs [34]. Thus, we selected 1-h post-OGD as a critical timepoint to test PARP1 overactivation. Again, we observed a significant induction of total PARP1 protein 1 h following OGD compared to sham (Sham 1 ± 0.51 vs 1-h post-OGD 5.2 ± 0.725, p < 0.0001, Fig. 2d).
RPC protects against OGD-induced PARP1 overexpression. a Representative Western Blot images of PARP1 (top) and Tubulin (bottom), left to right (Sham, Immediate, 1-h, 2-h) showing protein levels following OGD. b Bar graph showing quantification of total PARP1 protein normalized to sham. (n = 3, (*) = p < 0.05, one-way ANOVA followed by Tukey’s post-test). c Representative Western Blot images of PARP1 and Tubulin protein 1-h following OGD or sham. d Bar graph showing total PARP1 protein quantification normalized to sham. (n = 5, (****) = p < 0.001, Welch’s T-test). e Representative Western Blot images showing PARP1 and Tubulin protein levels. RPC or vehicle was administered on DIV8 with OGD on DIV10 and protein was collected 1 h post-OGD or sham. Left to right; Sham, Veh-OGD, RPC-OGD. f Bar graph of total PARP1 protein levels normalized to sham. RPC protects against PARP1 protein overexpression compared to the vehicle. (n = 7, (*) = p < 0.05, (***) = p < 0.005, one-way ANOVA followed by Tukey’s post-test)
We next determined whether RPC was able to mitigate OGD’s PARP1 induction. PNCs received 50 μM RPC, or vehicle, 2 days before OGD. Then whole-cell lysates were collected 1 h following OGD. OGD induced a significant increase in total PARP1 protein compared to sham. However, RPC prevented this increase (Sham 1 ± 0.48 vs Veh-OGD 2.6 ± 0.835, p < 0.005, Sham vs RPC-OGD 1.64 ± 0.55, p = 0.176). RPC showed significantly lower total PARP1 protein compared to the Veh-OGD group (p < 0.05). Thus, the propensity for PARP1 overactivation is likely reduced by RPC. Parthanatos has been reported following cerebral ischemia, suggesting that OGD’s induction of PARP1 may translate to this process [7, 35]. Based on RPC’s blocking of PARP1 overexpression, we next hypothesized that RPC protects against parthanatos following OGD.
Two-Day RPC Protects Against Features of Parthanatos Following OGD in PNCs
NAD+ depletion occurs in PARP1 overactivation and persists through parthanatos in rat hippocampal neurons [36]. Additionally, NAD+ repletion has been shown to protect against OGD [37]. Moreover, our lab has demonstrated that RSV bolsters mitochondrial NAD+ levels in mixed cortical neurons [38]. Based on these reports, we hypothesized that RPC protects against NAD+ and NADH depletion. To investigate, we measured NAD+ and NADH levels 12 h post-OGD (Fig. 3a, b). NAD+ levels were significantly reduced in both post-OGD groups compared to sham (Sham 2.945 ± 0.283 vs Veh-OGD 0.988 ± 0.255, Sham vs RPC-OGD 1.459 ± 0.12, p < 0.001, Fig. 3a). However, RPC significantly increased NAD+ levels compared to the vehicle group (p < 0.01, Fig. 3a), suggesting some capacity to retain post-OGD NAD+. Veh-OGD showed a significant reduction in NADH levels compared to both sham and RPC-OGD (Sham 0.1078 ± 0.018 vs Veh-OGD 0.0544 ± 0.012, p < 0.005, Veh-OGD vs RPC-OGD 0.085 ± 0.026, p < 0.05, Fig. 3b). There was no significant difference between sham and RPC-OGD (p = 0.131), showing RPC’s ability to normalize NADH levels 12 h following OGD.
RPC protects against features of parthanatos 12 h following OGD in PNCs. a Bar graph of NAD+ levels as ng-NAD+/μg-protein of starting material. OGD significantly reduced NAD+ levels compared to sham. However, RPC shows a significant increase over Veh-OGD. (n = 6, (**) = p < 0.01, (****) = p < 0.001, one-way ANOVA followed by Tukey’s post-test). b Bar graph of NADH levels as ng-NADH/μg-protein of starting material. OGD significantly reduced NADH levels compared to sham and RPC-OGD. (n = 6, (*) = p < 0.05, (***) = p < 0.005). c Representative Western Blot images. Top to bottom AIF, PARP1, Lamin C, Na+/K+-ATPase. Lamin C is the nuclear loading control and Na+/K.+-ATPase is the fraction purity control. Left to Right: Sham, Veh-OGD, RPC-OGD with fractions cytosolic, membrane, and nuclear. d Bar graph of nuclear AIF normalized first to Lamin C and then sham levels. AIF translocation was significantly increased in Veh-OGD compared to both sham and RPC-OGD. (n = 5, (**) = p < 0.01, (*) = p < 0.05). e Bar graph of nuclear PARP1 protein normalized first to Lamin C and then sham levels. Nuclear PARP1 is significantly reduced in the RPC-OGD group compared to both sham and Veh-OGD (n = 6, (*) = p < 0.05, (***) = p < 0.005)
Blocking PARP1-overactivation has been shown to prevent parthanatos [6, 12]. The execution of parthanatos occurs upon the translocation of AIF to the nucleus [7]. To test AIF translocation, we used subcellular fractionation followed by Western Blot, 12 h following OGD (Fig. 3c, d). Homeostatic AIF is observed in the membrane fraction, while pathological AIF is localized to the nuclear fraction. Twelve hours following OGD, Veh-OGD showed an increase in nuclear AIF compared to both sham and RPC-OGD (Sham 1 ± 0.68 vs Veh-OGD 4.171 ± 1.9, p < 0.01, Veh-OGD vs RPC-OGD 1.85 ± 0.71, Fig. 3d). There was no significant difference between the sham and RPC-OGD groups (p = 0.56). We next determined whether OGD’s induction of PARP1 continued out to 12 h following OGD (Fig. 3c, e). Twelve hours following OGD, nuclear PARP1 protein was increased compared to the sham group (Sham 1 ± 0.427 vs Veh-OGD 1.59 ± 0.35, p < 0.05). Interestingly, RPC significantly reduced nuclear PARP1 when compared to both Veh-OGD and sham (RPC-OGD 0.31 ± 0.23 vs Sham, p < 0.05, RPC-OGD vs Veh-OGD, p < 0.005). The difference between RPC-OGD and sham suggests that this effect on nuclear PARP1 protein is independent of OGD. RPC’s impact on NAD+/NADH levels, AIF translocation to the nucleus, and nuclear PARP1 suggest protection against parthanatos 12 h following OGD. With protection against PARP1-mediated injury established, we next sought to determine whether RPC induces changes in PARP1 expression independently of OGD.
RPC Instills a Long-Term Downregulation of Nuclear PARP1 Protein In Vitro
Our lab has shown downstream effectors of RSV that operate independently of cerebral ischemic insult [23, 24, 26]. Thus, RPC may regulate PARP1 independently of cerebral ischemia to produce protection against PARP1 overexpression and parthanatos. We hypothesize that RPC downregulates PARP1 expression independently of cerebral ischemia. To investigate this hypothesis, we assessed total PARP1 protein, as well as PARP mRNA levels, in PNCs 2 days following 50 μM RPC or vehicle (Fig. 4a–c). This time was selected to assess the 2-day window. We observed a significant reduction in total PARP1 protein in the RPC group compared to the vehicle (Veh 1 ± 0.214 vs RPC 0.69 ± 0.25, p < 0.05, Fig. 4b). However, we did not observe any changes in PARP1 mRNA levels following RPC (Veh 1 ± 0.14 vs RPC 1.1 ± 0.15, p = 0.227, Fig. 4c). Interestingly, RPC is sufficient to downregulate total PARP1 protein, but not mRNA expression. We next sought to determine the duration of RPC’s downregulation of PARP1. Based on our data in Fig. 3e, nuclear PARP1 is downregulated for at least 36 h following RPC. Our lab has previously shown that RPC protects astro-neuronal mixed cultures against OGD in a 6-day window [23]. Thus, we hypothesized that RPC’s downregulation of nuclear PARP1 protein lasts at least 6 days. We used subcellular fractionation to assess nuclear PARP1 levels 2, 4, and 6 days following RPC or vehicle (Fig. 4d). Nuclear PARP1 protein was then assessed via Western Blotting. At all three time points, RPC significantly reduced nuclear PARP1 protein compared to the respective vehicle group (2-day Veh 1 ± 0.35 vs 2-day RPC 0.583 ± 0.273, p < 0.05. 4-day Veh 1 ± 0.423 vs 4-day RPC 0.38 ± 0.37, p < 0.005, 6-day Veh 1 ± 0.57 vs 6-day RPC 0.33 ± 0.285, p < 0.005). The largest magnitude downregulation of nuclear PARP1 protein was observed 6 days following RPC (67%), as opposed to 4 days (62%) and 2 days (41.7%). This effect suggests a strong, long-term, prophylactic potential of RPC for the prevention of PARP1 overexpression and parthanatos in vitro. To determine whether RPC impacts parthanatos in vivo, we next tested the hypothesis that RPC protects against AIF translocation in the ischemic penumbra following MCAO in the rat.
RPC instills long-term downregulation of nuclear PARP1 protein in vitro. a Representative Western Blot images of PARP1 (top) and Actin (bottom) for Veh (left) and RPC (right) groups, showing total PARP1 protein levels after 2 days. b Bar graph of total PARP1 protein showing 50 μM RPC significantly reduced total PARP1 protein compared to the vehicle. Data represented as fold change from the vehicle. (n = 7, (*) = p < 0.05, Welch’s T-test). c Bar graph of PARP1 mRNA expression 2 days following 50 μM RPC or vehicle showing no significant difference. Data represented as fold change from the vehicle. (n = 6, p = 0.227, Welch’s T-test). d Representative Western Blot images following subcellular fractionation of PARP1 (top) and Lamin C (bottom) with Veh (left) and RPC (right). Left to right; 2 days, 4 days, and 6 days. e Bar graph showing nuclear PARP1 levels in 2-, 4-, and 6-day groups. Data represented as fold change from the respective vehicle. RPC significantly reduced nuclear PARP1 compared to the vehicle at all time points. (n = 6, (*) = p < 0.05, (***) = p < 0.005, two-way ANOVA followed by Bonferroni’s post-test)
Two-Day RPC Protects Against Nuclear Translocation of AIF 24 h Following MCAO In Vivo
Our lab has shown RPC’s ability to protect the brain against both MCAO and cardiac arrest (CA) in vivo [22, 24, 26, 30]. Additionally, the translocation of AIF has been shown to increase in the ischemic penumbra following MCAO in rats [39, 40]. Thus, we hypothesized that RPC protects against parthanatos in the ischemic penumbra following MCAO in the rat (Fig. 5). To investigate this hypothesis, male rats were injected intraperitoneally with either 50 mg/kg RSV or vehicle, 2 days before 90-min MCAO. In this experiment, there were no significant differences observed in physiological parameters for MCAO animals (Online Resource Supplemental Table.1). Twenty-four hours following reperfusion, penumbra tissue regions were collected from the ipsilateral cerebral cortex. As an internal control, the same regions of the cerebral cortex were collected on the contralateral hemisphere for each rat. The groups of this experiment were Veh-Sham, Veh-MCAO, RPC-Sham, and RPC-MCAO. The ratio of nuclear AIF to membrane AIF was determined for both the ipsilateral and contralateral samples (Fig. 6). There were no significant differences observed in translocated AIF from either hemisphere when assessed alone (Fig. 5c, d). However, the ratio of ipsilateral to contralateral translocated AIF showed a significant increase in the Veh-MCAO group compared to all other groups (Veh-MCAO 1.436 ± 0.44 vs Veh-Sham 0.77 ± 0.22, p < 0.01. Veh-MCAO vs RPC-Sham 0.95 ± 0.16, p < 0.05. Veh-MCAO vs RPC-MCAO 0.843 ± 0.17, p < 0.05, Fig. 5b). There was no statistical difference between the RPC-Sham and RPC-MCAO groups (p = 0.99). This data shows that RPC prevents AIF nuclear translocation in the MCA penumbra region 24 h following MCAO. These results further support our findings in vitro and suggest that RPC can block parthanatos in the peri-infarct regions of the cerebral cortex following focal cerebral ischemia.
RPC protects against AIF translocation in the ischemic penumbra of rat cerebral cortex following MCAO. a Representative Western Blot images of (top to bottom) AIF, Lamin C, and COX IV in all four groups (left to right) Veh-Sham, Veh-MCAO, RPC-Sham, and RPC-MCAO. Lamin C is the nuclear loading control and COX IV is the membrane loading control. b Bar graph of the ipsilateral to contralateral translocated AIF ratio (ipsilateral-nuclear/membrane/contralateral-nuclear/membrane). The Veh-MCAO group showed a significant increase in translocated AIF compared to all other groups. All other groups were statistically similar. (n = 5–6, (*) = p < 0.05, (**) = p < 0.01, two-way ANOVA followed by Bonferroni’s post-test). c, d Bar graphs representing the ipsilateral (c) and contralateral (d) translocated AIF values. No significant differences were observed
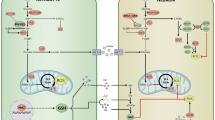
RPC-induced changes in PARP1 and parthanatos. (Left panel) the depiction of RPC’s effect on nuclear PARP1 in the absence of ischemia. (Middle panel) ischemia’s effect on PARP1 induction, AIF nuclear translocation, and reductions in NAD+/NADH levels. (Right panel) RPC’s reversal of ischemia-induced deleterious effects. Adapted from “How Does the Environment Determine Sex?,” by BioRender.com (2022). Retrieved from https://app.biorender.com/biorender-templates
Discussion
RPC has the potential to delineate effective therapies and prophylactics for individuals with a higher proclivity for a stroke, such as those with diabetes, cardiovascular disease, and hypertension [18, 38, 41, 42]. It is imperative to determine novel downstream targets of RPC to better exploit the ischemic tolerance provided. PARP1 overactivation and parthanatos contribute substantially to ischemic injury and targeting this pathway may prevent neuronal loss [43]. In the current study, we demonstrated that the 2-day window of 50 μM RPC protects PNCs against OGD, in part, by blocking OGD-induced PARP1 upregulation and eventual parthanatos. RPC protected against features of parthanatos by ameliorating NAD+/NADH loss and preventing AIF nuclear translocation. RPC’s ischemic tolerance was likely the result of long-term nuclear downregulation of PARP1 protein. To our knowledge, this is the first report depicting RSV’s ability to induce long-term downregulation of PARP1 in neurons. Moreover, the use of RPC to specifically protect against PARP1 overexpression, as well as parthanatos, has yet to be described in the brain.
As previously mentioned, SSB formation and PARP1 upregulation have been reported 1 h following MCAO [32, 34]. Thus, targeting 1 h following OGD allows for an approximate assessment of PARP1 overactivation. Increases in PARP1 may compensate for increased SSB formation. However, in PARP1−/− mice, the loss of PARP1 significantly reduced infarct volume following MCAO but did not compromise genome integrity [44]. Additionally, the use of PARP1 inhibitors in a wide range of experimental contexts has shown protection against cerebral ischemia [45]. Here we observe the capacity of RPC to block OGD-induced increases in total PARP1 protein (Fig. 2). We also show that RPC’s effect on PARP1 does not exacerbate DNA damage following OGD (Online Resource Supplemental Fig. 1). PARP1 overactivation drives metabolic dysfunction, ATP deficiency, and neuronal death [46, 47]. RPC’s attenuation of PARP1 overexpression likely reduces the risk of PARP1 overactivation, without compromising genomic integrity, thus providing effective prophylaxis against cerebral ischemia.
PARP1 overactivation progresses to parthanatos to induce cell death. NAD+ depletion contributes to this pathology, and in this report, we show RPC’s ability to lessen NAD+ loss and restore NADH following OGD (Fig. 3a, b). Due to diffusion, depletion of nuclear NAD+ levels may reduce cytosolic NAD+ levels, potentially affecting glycolysis [48, 49]. Additionally, NAD+ levels were normalized due to the inhibition of PARP1 with JPI-289 treatment before and after OGD [50]. Thus, the effect RPC has on NAD+ levels following OGD may be the result of reduced PARP1 utilization of NAD+. Previous studies in our lab have shown that RPC increases glycolysis in vitro [23, 30]. RPC was able to prevent the loss of NADH levels 12 h following OGD (Fig. 3b), potentially suggesting a change in glycolysis. The increase in NAD+ levels and normalization of NADH levels implies RPC may improve metabolic recovery following OGD, where increases in NAD+ may be due to reduced PARP1 activity.
Our data shows that RPC blocks the translocation of AIF to the nucleus both in vitro and in vivo. In our in vitro experiments, RPC’s effects on PARP1 overexpression strongly suggested that parthanatos would be prevented. We used these results, as well as other reports depicting nuclear AIF in the ischemic penumbra following MCAO, as the basis of our in vivo investigation [39, 40]. We specifically targeted the ischemic penumbra, because it is therapeutically viable [51]. RPC’s prophylaxis against parthanatos may prevent neurological damage in this region. Here we show that RPC prevents AIF nuclear translocation in the ischemic penumbra (Fig. 5). The translational quality of RPC is reflected in this data, suggesting that RPC’s mechanisms may operate within the heterogeneity of cell types residing in the brain, as opposed to exclusively within neurons. Additionally, RPC’s effects on PARP1 overexpression and parthanatos have the potential for cross-tolerance, as other neurodegenerative diseases have been shown to activate PARP1 [52]. Due to the sexual dimorphism of PARP1-mediated injury, RPC may block AIF nuclear translocation in female rats but fail to provide neuroprotection. A study showed that male PARP1−/− mice exhibited reduced infarct following MCAO compared to WT, but in female mice, the absence of PARP1 exacerbated ischemic injury. In ovariectomized WT female mice, exogenous restoration of 17β-Estradiol was able to protect against MCAO; however in ovariectomized PARP1−/− mice, 17β-estradiol exacerbated infarct [53]. Another study from this group showed that MCAO induced PAR production in the cerebral cortex of both sexes by 6 h of reperfusion, though males exhibited a larger increase over females. AIF nuclear translocation was also significantly increased in both sexes. Even with similar pathological manifestations, they again showed that female PARP1−/− mice had exacerbated injury [54]. A different study assessed the impact of male androgen signaling on PARP1-mediated injury following MCAO. The neuroprotection afforded by PARP1−/−, as well as PARP1 inhibition, was lost due to castration in male mice. This protection was restored with androgen replacement [55]. Thus, the sexual dimorphism of PARP1-dependent ischemic injury is likely sex hormone-based, and further studies are required to determine RPC’s effects on PARP1 in female rats.
This study demonstrates that RPC’s downregulation of PARP1 is independent of OGD. We observe that total PARP1 protein is significantly reduced 2 days following RPC, with no change in PARP1 mRNA. A strong candidate to mediate this downregulation is proteasomal degradation. Ubiquitin-dependent proteasomal degradation is a cellular mechanism that ensures the effective removal of waste and aggregated proteins. E3-ubiquitin ligases designate proteins for degradation through polyubiquitination [56]. A specific E3-ubiquitin ligase, known as Iduna, is PAR dependent and targets PARylated proteins, including PARP1 [57,58,59]. In a model of traumatic brain injury, Iduna overexpression was shown to inhibit parthanatos in HT22 cells. Overexpression improved NAD+ levels while blocking both PARP1 activation and AIF nuclear translocation [58]. Another E3 ubiquitin ligase, WW domain containing E3 ubiquitin protein ligase 2 (WWP2), has also been shown to target PARP1 for ubiquitination and degradation [60]. RPC’s influence on Iduna and WWP2 warrants investigation, specifically for any changes in expression or activity. Furthermore, inhibition of the proteasome would determine if this process is involved in PARP1’s downregulation [61]. Autophagy is another potential catabolic pathway to degrade PARP1. This process degrades damaged organelles, proteins, and other cellular debris through packaging in autophagosomes and fusion with lysosomes. Autophagy is controlled by AMP-activated protein kinase (AMPK), which is a metabolically sensitive kinase that regulates catabolic processes [62]. Our lab has shown that RSV treatment in astro-neuronal cultures increased AMPK phosphorylation and activity. This increase was dependent on the activity of protein kinase C epsilon (PKCε) [38]. In another study, RSV induced liver kinase B1 (LBK1)-dependent AMPK phosphorylation in the Neuro2A cell line [63]. These studies demonstrate a potential link between RSV and autophagy. Inhibition of autophagy may determine whether this mechanism is induced by RPC to downregulate PARP1. Specific isolation of the mechanism driving PARP1 downregulation would reduce off-target effects of RPC and provide more targeted prophylaxis.
Conclusion
This study reports a novel mechanism of ischemic tolerance induced by RPC. RPC significantly reduced OGD-induced cell death and prevented OGD-mediated PARP1 overexpression, likely reducing the propensity for PARP1 overactivation. Blocking increases in PARP1 translated to protection against parthanatos, depicted as the amelioration of NAD+/NADH loss and prevention of AIF nuclear translocation. Finally, RPC likely generates these protective effects through a long-term downregulation of nuclear PARP1 protein. Overall, this study describes a novel use of RPC to negatively regulate PARP1 and prevent PARP1-mediated neuronal injury.
Data Availability
The data that supports the current study is available upon reasonable request from the corresponding author.
References
Peters O, Back T, Lindauer U, et al. Increased formation of reactive oxygen species after permanent and reversible middle cerebral artery occlusion in the rat. J Cereb Blood Flow Metab. 1998;18:196–205. https://doi.org/10.1097/00004647-199802000-00011.
Shirley R, Ord EN, Work LM. Oxidative stress and the use of antioxidants in stroke. Antioxidants (Basel). 2014;3:472–501. https://doi.org/10.3390/antiox3030472.
Sun MS, Jin H, Sun X, et al. Free radical damage in ischemia-reperfusion injury: an obstacle in acute ischemic stroke after revascularization therapy. Oxid Med Cell Longev. 2018;2018:3804979. https://doi.org/10.1155/2018/3804979.
Koehler RC, Dawson VL, Dawson TM. Targeting parthanatos in ischemic stroke. Front Neurol. 2021;12: 662034. https://doi.org/10.3389/fneur.2021.662034.
Zhao H, Tang J, Chen H, et al. 14,15-EET reduced brain injury from cerebral ischemia and reperfusion via suppressing neuronal parthanatos. Int J Mol Sci. 2021;22(18):9660. https://doi.org/10.3390/ijms22189660.
Li X, Klaus JA, Zhang J, et al. Contributions of poly(ADP-ribose) polymerase-1 and -2 to nuclear translocation of apoptosis-inducing factor and injury from focal cerebral ischemia. J Neurochem. 2010;113:1012–22. https://doi.org/10.1111/j.1471-4159.2010.06667.x.
Wang Y, Kim NS, Haince JF, et al. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos). Sci Signal. 2011;4:ra20. https://doi.org/10.1126/scisignal.2000902.
Eliasson MJ, Sampei K, Mandir AS, et al. Poly(ADP-ribose) polymerase gene disruption renders mice resistant to cerebral ischemia. Nat Med. 1997;3:1089–95. https://doi.org/10.1038/nm1097-1089.
Tao Z, Gao P, Liu HW. Identification of the ADP-ribosylation sites in the PARP-1 automodification domain: analysis and implications. J Am Chem Soc. 2009;131:14258–60. https://doi.org/10.1021/ja906135d.
Satoh MS, Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356:356–8. https://doi.org/10.1038/356356a0.
Tanaka M, Miwa M, Hayashi K, et al. Separation of oligo(adenosine diphosphate ribose) fractions with various chain lengths and terminal structures. Biochemistry. 1977;16:1485–9. https://doi.org/10.1021/bi00626a037.
Gao Y, Wang Z, He W, et al. Mild hypothermia protects neurons against oxygen glucose deprivation via poly (ADP-ribose) signaling. J Matern Fetal Neonatal Med. 2019;32:1633–9. https://doi.org/10.1080/14767058.2017.1413548.
Andrabi SA, Kim NS, Yu SW, et al. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci U S A. 2006;103:18308–13. https://doi.org/10.1073/pnas.0606526103.
Wang Y, An R, Umanah GK, et al. A nuclease that mediates cell death induced by DNA damage and poly(ADP-ribose) polymerase-1. Science. 2016;354(6308):aad6872. https://doi.org/10.1126/science.aad6872.
Zhou Y, Liu L, Tao S, et al. Parthanatos and its associated components: promising therapeutic targets for cancer. Pharmacol Res. 2021;163: 105299. https://doi.org/10.1016/j.phrs.2020.105299.
Kim Y, Kim YS, Kim HY, et al. Early treatment with poly(ADP-ribose) polymerase-1 inhibitor (JPI-289) reduces infarct volume and improves long-term behavior in an animal model of ischemic stroke. Mol Neurobiol. 2018;55:7153–63. https://doi.org/10.1007/s12035-018-0910-6.
Nakajima H, Kakui N, Ohkuma K, et al. A newly synthesized poly(ADP-ribose) polymerase inhibitor, DR2313 [2-methyl-3,5,7,8-tetrahydrothiopyrano[4,3-d]-pyrimidine-4-one]: pharmacological profiles, neuroprotective effects, and therapeutic time window in cerebral ischemia in rats. J Pharmacol Exp Ther. 2005;312:472–81. https://doi.org/10.1124/jpet.104.075465.
Narayanan SV, Dave KR, Perez-Pinzon MA. Ischemic preconditioning and clinical scenarios. Curr Opin Neurol. 2013;26:1–7. https://doi.org/10.1097/WCO.0b013e32835bf200.
Garnier P, Ying W, Swanson RA. Ischemic preconditioning by caspase cleavage of poly(ADP-ribose) polymerase-1. J Neurosci. 2003;23:7967–73.
Morris KC, Lin HW, Thompson JW, et al. Pathways for ischemic cytoprotection: role of sirtuins in caloric restriction, resveratrol, and ischemic preconditioning. J Cereb Blood Flow Metab. 2011;31:1003–19. https://doi.org/10.1038/jcbfm.2010.229.
Raval AP, Dave KR, Perez-Pinzon MA. Resveratrol mimics ischemic preconditioning in the brain. J Cereb Blood Flow Metab. 2006;26:1141–7. https://doi.org/10.1038/sj.jcbfm.9600262.
Koronowski KB, Dave KR, Saul I, et al. Resveratrol preconditioning induces a novel extended window of ischemic tolerance in the mouse brain. Stroke. 2015;46:2293–8. https://doi.org/10.1161/STROKEAHA.115.009876.
Khoury N, Xu J, Stegelmann SD, et al. Resveratrol preconditioning induces genomic and metabolic adaptations within the long-term window of cerebral ischemic tolerance leading to bioenergetic efficiency. Mol Neurobiol. 2019;56:4549–65. https://doi.org/10.1007/s12035-018-1380-6.
Della-Morte D, Dave KR, DeFazio RA, et al. Resveratrol pretreatment protects rat brain from cerebral ischemic damage via a sirtuin 1-uncoupling protein 2 pathway. Neuroscience. 2009;159:993–1002. https://doi.org/10.1016/j.neuroscience.2009.01.017.
Narayanan SV, Dave KR, Perez-Pinzon MA. Ischemic preconditioning protects astrocytes against oxygen glucose deprivation via the nuclear erythroid 2-related factor 2 pathway. Transl Stroke Res. 2018;9:99–109. https://doi.org/10.1007/s12975-017-0574-y.
Narayanan SV, Dave KR, Saul I, et al. Resveratrol preconditioning protects against cerebral ischemic injury via nuclear erythroid 2-related factor 2. Stroke. 2015;46:1626–32. https://doi.org/10.1161/STROKEAHA.115.008921.
Xu J, Khoury N, Jackson CW, et al. Ischemic neuroprotectant PKCepsilon restores mitochondrial glutamate oxaloacetate transaminase in the neuronal NADH shuttle after ischemic injury. Transl Stroke Res. 2020;11:418–32. https://doi.org/10.1007/s12975-019-00729-4.
Lin HW, Saul I, Gresia VL, et al. Fatty acid methyl esters and Solutol HS 15 confer neuroprotection after focal and global cerebral ischemia. Transl Stroke Res. 2014;5:109–17. https://doi.org/10.1007/s12975-013-0276-z.
Cohan CH, Stradecki-Cohan HM, Morris-Blanco KC, et al. Protein kinase C epsilon delays latency until anoxic depolarization through arc expression and GluR2 internalization. J Cereb Blood Flow Metab. 2017;37:3774–88. https://doi.org/10.1177/0271678X17712178.
Koronowski KB, Khoury N, Saul I, et al. Neuronal SIRT1 (silent information regulator 2 homologue 1) regulates glycolysis and mediates resveratrol-induced ischemic tolerance. Stroke. 2017;48:3117–25. https://doi.org/10.1161/STROKEAHA.117.018562.
Nagayama T, Simon RP, Chen D, et al. Activation of poly(ADP-ribose) polymerase in the rat hippocampus may contribute to cellular recovery following sublethal transient global ischemia. J Neurochem. 2000;74:1636–45. https://doi.org/10.1046/j.1471-4159.2000.0741636.x.
Narasimhan P, Fujimura M, Noshita N, et al. Role of superoxide in poly(ADP-ribose) polymerase upregulation after transient cerebral ischemia. Brain Res Mol Brain Res. 2003;113:28–36. https://doi.org/10.1016/s0169-328x(03)00062-7.
Rajamohan SB, Pillai VB, Gupta M, et al. SIRT1 promotes cell survival under stress by deacetylation-dependent deactivation of poly(ADP-ribose) polymerase 1. Mol Cell Biol. 2009;29:4116–29. https://doi.org/10.1128/MCB.00121-09.
Chen J, Jin K, Chen M, et al. Early detection of DNA strand breaks in the brain after transient focal ischemia: implications for the role of DNA damage in apoptosis and neuronal cell death. J Neurochem. 1997;69:232–45. https://doi.org/10.1046/j.1471-4159.1997.69010232.x.
Liu L, Li J, Ke Y, et al. The key players of parthanatos: opportunities for targeting multiple levels in the therapy of parthanatos-based pathogenesis. Cell Mol Life Sci. 2022;79:60. https://doi.org/10.1007/s00018-021-04109-w.
Wang S, Yang X, Lin Y, et al. Cellular NAD depletion and decline of SIRT1 activity play critical roles in PARP-1-mediated acute epileptic neuronal death in vitro. Brain Res. 2013;1535:14–23. https://doi.org/10.1016/j.brainres.2013.08.038.
Wang S, Xing Z, Vosler PS, et al. Cellular NAD replenishment confers marked neuroprotection against ischemic cell death: role of enhanced DNA repair. Stroke. 2008;39:2587–95. https://doi.org/10.1161/STROKEAHA.107.509158.
Morris-Blanco KC, Cohan CH, Neumann JT, et al. Protein kinase C epsilon regulates mitochondrial pools of Nampt and NAD following resveratrol and ischemic preconditioning in the rat cortex. J Cereb Blood Flow Metab. 2014;34:1024–32. https://doi.org/10.1038/jcbfm.2014.51.
Ferrer I, Friguls B, Dalfo E, et al. Caspase-dependent and caspase-independent signalling of apoptosis in the penumbra following middle cerebral artery occlusion in the adult rat. Neuropathol Appl Neurobiol. 2003;29:472–81. https://doi.org/10.1046/j.1365-2990.2003.00485.x.
Yao H, Takasawa R, Fukuda K, et al. DNA fragmentation in ischemic core and penumbra in focal cerebral ischemia in rats. Brain Res Mol Brain Res. 2001;91:112–8. https://doi.org/10.1016/s0169-328x(01)00135-8.
Caprio FZ, Sorond FA. Cerebrovascular disease: primary and secondary stroke prevention. Med Clin North Am. 2019;103:295–308. https://doi.org/10.1016/j.mcna.2018.10.001.
Liu J, Gu Y, Guo M, et al. Neuroprotective effects and mechanisms of ischemic/hypoxic preconditioning on neurological diseases. CNS Neurosci Ther. 2021;27:869–82. https://doi.org/10.1111/cns.13642.
Strosznajder JB, Jesko H, Strosznajder RP. Effect of amyloid beta peptide on poly(ADP-ribose) polymerase activity in adult and aged rat hippocampus. Acta Biochim Pol. 2000;47:847–54.
Endres M, Wang ZQ, Namura S, et al. Ischemic brain injury is mediated by the activation of poly(ADP-ribose)polymerase. J Cereb Blood Flow Metab. 1997;17:1143–51. https://doi.org/10.1097/00004647-199711000-00002.
Abdelkarim GE, Gertz K, Harms C, et al. Protective effects of PJ34, a novel, potent inhibitor of poly(ADP-ribose) polymerase (PARP) in in vitro and in vivo models of stroke. Int J Mol Med. 2001;7:255–60.
Zhang J, Dawson VL, Dawson TM, et al. Nitric oxide activation of poly(ADP-ribose) synthetase in neurotoxicity. Science. 1994;263:687–9. https://doi.org/10.1126/science.8080500.
Moroni F. Poly(ADP-ribose)polymerase 1 (PARP-1) and postischemic brain damage. Curr Opin Pharmacol. 2008;8:96–103. https://doi.org/10.1016/j.coph.2007.10.005.
Khoury N, Koronowski KB, Young JI, et al. The NAD(+)-dependent family of sirtuins in cerebral ischemia and preconditioning. Antioxid Redox Signal. 2018;28:691–710. https://doi.org/10.1089/ars.2017.7258.
Alano CC, Garnier P, Ying W, et al. NAD+ depletion is necessary and sufficient for poly(ADP-ribose) polymerase-1-mediated neuronal death. J Neurosci. 2010;30:2967–78. https://doi.org/10.1523/JNEUROSCI.5552-09.2010.
Kim Y, Kim YS, Noh MY, et al. Neuroprotective effects of a novel poly (ADP-ribose) polymerase-1 inhibitor, JPI-289, in hypoxic rat cortical neurons. Clin Exp Pharmacol Physiol. 2017;44:671–9. https://doi.org/10.1111/1440-1681.12757.
Hakim AM. Ischemic penumbra: the therapeutic window. Neurology. 1998;51:S44–6. https://doi.org/10.1212/wnl.51.3_suppl_3.s44.
Mao K, Zhang G. The role of PARP1 in neurodegenerative diseases and aging. FEBS J. 2022;289:2013–24. https://doi.org/10.1111/febs.15716.
McCullough LD, Zeng Z, Blizzard KK, et al. Ischemic nitric oxide and poly (ADP-ribose) polymerase-1 in cerebral ischemia: male toxicity, female protection. J Cereb Blood Flow Metab. 2005;25:502–12. https://doi.org/10.1038/sj.jcbfm.9600059.
Yuan M, Siegel C, Zeng Z, et al. Sex differences in the response to activation of the poly (ADP-ribose) polymerase pathway after experimental stroke. Exp Neurol. 2009;217:210–8. https://doi.org/10.1016/j.expneurol.2009.02.012.
Vagnerova K, Liu K, Ardeshiri A, et al. Poly (ADP-ribose) polymerase-1 initiated neuronal cell death pathway–do androgens matter? Neuroscience. 2010;166:476–81. https://doi.org/10.1016/j.neuroscience.2009.12.041.
Ikeda F. Ubiquitin conjugating enzymes in the regulation of the autophagy-dependent degradation pathway. Matrix Biol. 2021;100–101:23–9. https://doi.org/10.1016/j.matbio.2020.11.004.
Kang HC, Lee YI, Shin JH, et al. Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc Natl Acad Sci U S A. 2011;108:14103–8. https://doi.org/10.1073/pnas.1108799108.
Xu H, Li X, Wu X, et al. Iduna protects HT22cells by inhibiting parthanatos: The role of the p53-MDM2 pathway. Exp Cell Res. 2019;384:111547. https://doi.org/10.1016/j.yexcr.2019.111547.
Andrabi SA, Kang HC, Haince JF, et al. Iduna protects the brain from glutamate excitotoxicity and stroke by interfering with poly(ADP-ribose) polymer-induced cell death. Nat Med. 2011;17:692–9. https://doi.org/10.1038/nm.2387.
Zhang N, Zhang Y, Qian H, et al. Selective targeting of ubiquitination and degradation of PARP1 by E3 ubiquitin ligase WWP2 regulates isoproterenol-induced cardiac remodeling. Cell Death Differ. 2020;27:2605–19. https://doi.org/10.1038/s41418-020-0523-2.
Fricker LD. Proteasome inhibitor drugs. Annu Rev Pharmacol Toxicol. 2020;60:457–76. https://doi.org/10.1146/annurev-pharmtox-010919-023603.
Tamargo-Gomez I, Marino G. AMPK: regulation of metabolic dynamics in the context of autophagy. Int J Mol Sci. 2018;19(12):3812. https://doi.org/10.3390/ijms19123812.
Dasgupta B, Milbrandt J. Resveratrol stimulates AMP kinase activity in neurons. Proc Natl Acad Sci U S A. 2007;104:7217–22. https://doi.org/10.1073/pnas.0610068104.
Funding
This work was supported by the National Institutes of Health grants (3R01NS034773; R01NS45676, R01NS054147) and an American Heart Association predoctoral fellowship (20PRE35120403).
Author information
Authors and Affiliations
Contributions
All authors contributed to the conception and experimental design of the study. CWJ and IS conducted experiments with IE and EF as contributors. CWJ, IE, and EF conducted data processing and analysis. CWJ, KRD, and MAPP drafted the manuscript, and all other authors reviewed the manuscript. This manuscript is approved by all authors for publication.
Corresponding author
Ethics declarations
Ethics Approval
Animal use in this study was approved by the University of Miami Institutional Animal Care and Use Committee. To ensure animal welfare, experiments using animals abided by the ARRIVE Guidelines, the 3R’s concept, and the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals.
We declare that this work has not been published elsewhere, nor is it being considered for publication by another publisher.
We declare that all authors have reviewed and approved this manuscript for publication.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jackson, C.W., Xu, J., Escobar, I. et al. Resveratrol Preconditioning Downregulates PARP1 Protein to Alleviate PARP1-Mediated Cell Death Following Cerebral Ischemia. Transl. Stroke Res. 15, 165–178 (2024). https://doi.org/10.1007/s12975-022-01119-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12975-022-01119-z