Abstract
Background
Gastric cancer (GC) is a highly chemoresistant malignancy with a poor prognosis. Paclitaxel’s low response rate as second-line chemotherapy for advanced GC has prompted intensive research into its molecular basis and prospective targeted therapies to enhance its therapeutic efficacy. The objective of this study was to investigate the synergistic effects of NRF2 silencing in combination with paclitaxel treatment on GC cell viability, apoptosis, proliferation, autophagy, and migration.
Methods
\After the siRNA-mediated silencing of NRF2 in AGS cells, the transfection efficacy was evaluated by qRT-PCR. The MTT assay was then applied to assess cell viability, followed by flow cytometry analysis for apoptosis, proliferation, and autophagy in AGS cells treated with NRF2 siRNA, paclitaxel, or their combination. Thereafter, the migration of cells was measured using a wound-healing assay. Ultimately, the relative gene expression levels of apoptotic (Bax, Caspase-3, and Caspase-9), anti-apoptotic (Bcl-2), metastatic (MMP-2), and cell cycle (P53) genes were measured by qRT-PCR in all experiment groups to further assess the molecular basis for the combination therapy.
Results
NRF2 siRNA transfection significantly enhanced paclitaxel-induced apoptosis and sensitized AGS cells to paclitaxel via modulating the expression of apoptosis-related genes including Bcl-2, Bax, Caspase-3, and Caspase-9. Besides, NRF2 siRNA and paclitaxel synergistically induced cell cycle arrest at the G2 phase, promoted autophagy activation, and inhibited AGS cell migration via MMP-2 downregulation. Additionally, P53, a key regulator of cell growth, was significantly upregulated in the treated groups compared to the control group.
Conclusions
Our findings suggest that paclitaxel combined with siRNA-mediated silencing of NRF2 might represent a promising therapeutic strategy for GC, however further translational and clinical research are warranted.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Gastric cancer (GC) is a major global concern, ranking as the fifth most common cancer diagnosed worldwide [1]. Despite tremendous breakthroughs in surgical techniques and chemoradiotherapy, silent progression and poor screening practices have made early diagnosis of GC elusive. Consequently, the vast majority of patients (> 70%) still receive an advanced cancer diagnosis with regional or distant metastases, making GC the third most lethal malignancy [2]. Advanced GC is clinically approached by sequential lines of chemotherapy, commencing with fluoropyrimidines and platinum compounds in the first line, which fails in more than 95% of patients and is then followed by paclitaxel combined with or without ramucirumab in the second line [3]. However, the response rate to paclitaxel in advanced GC patients is roughly 16–22%, underscoring the essential need for further research into the molecular basis of chemoresistance and prospective targeted therapy-based translational strategies for chemosensitization [4]. Although, nowadays the treatment options for gastric cancer have greatly improved over the past few years, with new immunotherapies and targeted therapies emerging for patients at various stages of the disease [5]. In tumor biology, molecular targeted therapy has garnered significant attention for its potential to enhance the specificity of anti-cancer treatments and markedly reduce non-selective resistance and toxicity [6].
Paclitaxel, a member of taxanes, is a microtubule-stabilizing drug that hinders microtubule depolymerization and dynamics, hence suppressing metaphase-anaphase transitions and consequently disrupting cell mitosis while triggering apoptosis and autophagy [7]. Besides, recent findings on the therapeutic mechanisms of paclitaxel in cancer are suggestive of its potential to drive early reactive oxygen species (ROS) and hydrogen peroxide (H2O2) generation, implicating in paclitaxel-induced cancer cell death [8]. Therefore, interventions in line to inhibit cancer cells' cytoprotective activities against oxidative stress may enhance paclitaxel's cytotoxicity and hence sensitize GC cells to paclitaxel.
Nuclear factor E2-related factor 2 (NRF2) is a transcription factor essential for cytoprotective activities against oxidation, electrophilic stress, and xenobiotic processes [9]. Studies have demonstrated that NRF2 is aberrantly expressed in tumor cells and serves a pro-oncogenic role by enhancing tumor cell viability, proliferation, invasion, and chemoresistance [10,11,12]. NRF2 is a complex protein that serves as cancer marker due to its high expression in cancer cells and low in normal tissues. It is linked to poor outcomes and drug resistance in cancer. While it protects normal cells, it also helps cancer cells survive therapy, making it a potential target for new cancer treatments [13]. Therefore, it is conceivable that silencing NRF2 expression in tumor cells might be an effective anti-cancer therapeutic strategy [14]. RNA interference is a common strategy that has been used in various studies to achieve this objective. The selective and effective regulation of homology-dependent post-transcriptional gene silencing by small interfering RNA (siRNA) have been shown to inhibit tumor cell proliferation [15].
Given the above reflection, we speculated that suppressing NRF2's cytoprotective effects by siRNA-mediated silencing of its gene may act synergistically with paclitaxel. Therefore, we aimed to investigate the effects of monotherapy and additive effects of combination therapy with NRF2 siRNA and paclitaxel on GC cell viability, apoptosis, proliferation, autophagy, and migration.
2 Materials and methods
2.1 Cell culture
The GC cell line, AGS, was purchased from the cell bank of Pasteur Institute (Tehran, Iran). The supplier confirmed the authenticity of the cell line through short tandem repeat (STR) profiling and ensured it was free of mycoplasma contamination. Cells were cultured in Roswell Park Memorial Institute medium (RPMI)-1640 medium enriched with 10% fetal bovine serum (FBS; GIBCO, Carlsbad, CA) and were incubated at 37◦ C, with 95% humidity, and a 5% CO2 atmosphere.
2.2 Small interfering RNA transfection
NRF2 siRNA was obtained as a lyophilized cocktail containing a duplex (Table 1) of siRNA from Metabion International AG (Germany). Cells were cultured at a concentration of 3 × 105 per well in six-well plates containing RPMI-1640 medium with 10% FBS. They were then electroporated with 100 pmol of siRNA concentration using a Gene Pulser Xcell (Bio-Rad, USA). After 48 h of transfection, cells were collected for further experiments.
2.3 Proliferation assay
The MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide) assay was applied to assess NRF2 siRNA and paclitaxel-related cytotoxicity in AGS cells. Briefly, cells were seeded at a density of 15 × 103 cells per well in 96-well culture plates. After 24 h, cells were treated with different concentrations of paclitaxel, and MTT dye reduction was measured to determine the half-maximal inhibitory concentration (IC50) of the drug.
To investigate the viability of AGS cells after NRF2 siRNA transfection, seeded cells were categorized into four experimental groups: control (untransfected), NRF2 siRNA transfected, negative control (scrambled siRNA), and positive control (cells treated with DMSO). After overnight incubation, the culture medium was withdrawn and 50 μl of MTT (2 mg/ml) was added to each well. After 4 h of incubation, 150 µL of dimethyl sulfoxide (DMSO) was added for 20 min to dissolve the formazan crystals. Ultimately, the optical density (OD) of each well was measured using an enzyme-linked immunosorbent assay (ELISA) reader (Sunrise™, Tecan, Switzerland).
2.4 Flow cytometry analysis of apoptosis
An annexin V/propidium iodide (PI) assay was applied to assess apoptosis. In a six-well plate, cells were seeded at a density of 3 × 105 cells per well and incubated for 24 h. The experiments were subdivided into four groups: control, NRF2 siRNA, paclitaxel, and combined NRF2 siRNA/paclitaxel. After 24 h of NRF2 siRNA transfection, the cells were treated with paclitaxel and incubated for another 24 h. The cells were then stained with annexin V and PI according to the manufacturer's instructions (EXBIO, Vestec, Czech Republic). For the annexin V staining procedure, 300 μl of binding buffer, 3 μl of annexin V, and 1 μl PI were added to each group. After that, they were incubated at room temperature in the darkness for 15 min. Flow cytometry (MiltenyBiotecTM FACS Quant 10; MiltenyBiotec, Germany) was used to examine the stained cells and the apoptosis rate was determined using FlowJo software (Tree Star, San Carlos, CA).
2.5 Flow cytometry analysis of cell cycle arrest
Cells were seeded at a density of 3 × 105 cells per well in a six-well plate and incubated for 24 h. Then, four groups were described: control, NRF2 siRNA, paclitaxel, and combined NRF2 siRNA/paclitaxel. The cells were prepared for the experiment after being transfected with NRF2 siRNA and treated with paclitaxel. Thereafter, the cells were rinsed in PBS and fixed in 70% ethanol overnight at -20 °C. Cells were then resuspended in PBS containing 5 μg/ml RNase A, incubated at 37◦ C for 30 min, and stained with DAPI solution (1 μg/ml DAPI, 1 μg/ml Triton X100 in PBS) for analysis. Finally, after 10 min of incubation on ice in the dark, a flow cytometry instrument (Milteny Biotec MACSQuant 10) was used to evaluate the cell cycle. The data were processed by the FlowJo fluorescence activated cell sorting (FACS) analysis software.
2.6 Autophagy assay
MDC (monodansyl cadaverine) staining was used to examine autophagy activation in different treatment groups. This approach is a simple and rapid method for assessing autophagy in vitro. MDC, a blue fluorescent dye, accumulates in autophagic vesicles exclusively by ion trapping and specific interactions with vesicle membrane lipids [16]. The AGS cells were then transfected with NRF2 siRNA and seeded into a 6-well plate (3 × 105 cells/well) for 24 h before being treated with paclitaxel and incubated for another 24 h. Then, the cells were washed with PBS and stained with the MDC (50 μM). After 10 min of incubation, the cells were washed with PBS again and detached with trypsin. Consequently, the autophagy rate was measured by a flow cytometry device, and the data was analyzed with FlowJo software.
2.7 Scratch-wound migration assay
The migration of AGS cells was determined using a wound-healing assay (scratch test). In a 24-well plate, cells (3 × 105 cells per well) were seeded for 24 h (the transfected groups were already transfected with NRF2 siRNA). After that, four groups were considered: control, NRF2 siRNA, paclitaxel, and combined NRF2 siRNA/paclitaxel. Then, a scratch was made in the center of each well using (10–100) µL pipette tips across the cell monolayer to create an open gap. Images were captured at 0, 24, and 48 h by an inverted microscope to monitor the migration of the cells.
2.8 Evaluation of gene expression by quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated by Trizol reagent (RiboEx Kit, GeneAll, South Korea) according to the manufacturer’s instructions. The extracted RNA samples were then examined for quantity and purity by nanodrop (Thermo Fisher Scientific, Lenexa, KS) device. Following that, complementary DNA (cDNA) synthesis was carried out using a specific kit (Biofact, South Korea) and a thermal cycler system (Bio-Rad, Hercules, CA). The primerblast software from the NCBI website (http://www.nchi.nlm.nih.gov) was used to blast all pair primer sequences prior to the experiment. The SYBR green master mix and specific primers for NRF2, P53, B-cell lymphoma 2 (Bcl-2), Bcl-2-associated X protein (Bax), matrix metalloproteinases-2 (MMP-2), Caspase-3, and Caspase-9 (Table 2) were used to evaluate mRNA expression using the STEP ONE PLUS qRT-PCR system (Applied Biosystems, Foster City, USA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was considered an internal control, and the gene expression ratio was calculated by the 2− ΔΔCt method.
2.9 Statistical analysis
The test results were analyzed with a one-way analysis of variance (ANOVA) and a student’s t-test. GraphPad Prism version 7.0 (GraphPad Software, La Jolla, CA, USA) was used to create the relevant diagrams. The results were presented in this study as mean standard deviation (SD), and a p-value of less than 0.05 was considered significant.
3 Results
3.1 NRF2-siRNA is effectively transfected in AGS cells
By electroporating FITC-conjugated siRNA control, the capacity of cellular transfection of AGS cells using flow cytometry was evaluated, and cells without electroporation were examined as control groups. The control group cells rarely showed any fluorescence, whereas the AGS electroporated cells with FITC- attached siRNA control clearly showed fluorescence (Fig. 1). Using flow cytometry, the initial NRF2- siRNA transfection efficiency was determined to be 66% efficient. Compared to the control group, the electroporated cells displayed the highest fluorescence intensity of AGS cells.
3.2 NRF2 siRNA transfection significantly downregulated NRF2 mRNA expression in AGS cells
To investigate the effect of NRF2 suppression in GC, we transfected the AGS cells with NRF2 siRNA. Non-transfected cells and negative control cells transfected with non-target siRNA (scrambled siRNA) were also employed as controls. The qRT-PCR analysis demonstrated that the level of NRF2 mRNA expression in the transfected group was significantly lower compared to the control group (Fig. 2).
3.3 NRF2 suppression reduced viability of AGS cells
MTT assay was used to evaluate the cytotoxic effect of NRF2 inhibition on the viability of AGS cells transfected with NRF2 siRNA. Transfection with NRF2 siRNA was revealed to significantly decrease cell viability and proliferation after 48 h of incubation as compared to the control group. Additionally, it was found that the viability of the cell was not adversely affected by transfection reagents when scrambled-siRNA was used. While using DMSO as a positive control led to a sharp and significant decrease in cell viability. All results are expressed as mean SD and are compared to controls (non-transfected cells) (Fig. 3).
Effect of NRF2 suppression on the viability of AGS cells according to MTT assay. After 48 h, transfection of AGS cells with NRF2 siRNA significantly decreased cell proliferation and viability compared to the control group. In addition, scrambled siRNA had no cytotoxic effect on the cell’s viability. (n = 3;*** p < 0.001 **** p < 0.0001)
3.4 Paclitaxel decreased viability of AGS cells
To investigate the effects of paclitaxel on AGS cell proliferation, we employed the MTT assay and varied concentrations of paclitaxel to treat the cells. After 24 h of treatment, our results showed a constant decrease in the AGS cell survival. As shown in (Fig. 4), the IC50 of paclitaxel that reduced cell viability to 50% compared to the control group was determined to be 4.328 μM, and it was employed in the subsequent experiment.
3.5 NRF2 siRNA/ paclitaxel combination induced apoptosis in AGS cells
The annexin V/PI assay was used to investigate the effects of a single NRF2 siRNA and a combination of NRF2 siRNA/paclitaxel on AGS cell apoptosis. We found that using NRF2 siRNA and paclitaxel combination significantly enhances apoptosis compared to using either agent alone. NRF2 siRNA and paclitaxel-induced early and late apoptosis in AGS cells at rates of 29.30% and 40.7%, respectively. As NRF2 siRNA was combined with paclitaxel, the rate of apoptosis was significantly increased (Fig. 5). As a result, we observed a significant increase in apoptosis when NRF2 siRNA and paclitaxel were employed simultaneously. These findings demonstrated that suppressing NRF2 could render AGS cells more sensitive to paclitaxel.
3.6 NRF2 siRNA/ paclitaxel combination arrested AGS cells in the G2 phase of the cell cycle
To assess the effect of NRF2 siRNA and paclitaxel on cell proliferation, we evaluated the cell cycle distribution of AGS cells after NRF2 siRNA transfection and paclitaxel treatment. According to flow cytometry analysis, NRF2 siRNA raised the number of cells in the G1 phase cells from 61.4% to 73.6%, and paclitaxel treatment raised the number of cells in the G2 phase from 20.8 to 75.9% as compared to controls. Interestingly, as NRF2 siRNA was combined with paclitaxel, the number of cells in the G2 phase increased from 20.8% to 88.4%. Thus, the NRF2 siRNA/paclitaxel combination was demonstrated to have a more significant effect on enhancing the proportion of AGS cells in the G2 phase than either agent alone (Fig. 6).
3.7 NRF2 siRNA/ paclitaxel combination induced autophagy in AGS cells
To evaluate the effect of NRF2 siRNA and paclitaxel treatment on autophagy in AGS cells, flow cytometry was used to detect the MDC-labeled cells. Flow cytometry findings demonstrated that NRF2 siRNA transfection raised the percentage of MDC-positive cells from 0.26% in the control group to 9.85%. Also, paclitaxel treatment increased the percentage of positive autophagy cells (MDC POS) by 0.26–10.1% as compared to controls. Furthermore, these findings showed that the combination treatment by NRF2 siRNA and paclitaxel increased the percentage of MDC POS AGS cells to 18.6%. Thus, we revealed that the combination of NRF2 siRNA and paclitaxel significantly promoted autophagy in AGS cells (Fig. 7).
3.8 NRF2 siRNA/ paclitaxel combination inhibited migration of AGS cells
Given that cancer cell invasion and migration are major steps in metastasis, we performed the wound scratch method to determine whether siRNA-mediated NRF2 suppression affected AGS cell migration. The results showed that the siRNA-mediated NRF2 silencing or paclitaxel treatments alone can significantly reduce AGS cell migration (Fig. 8). Furthermore, as compared to control cells, the NRF2 siRNA/paclitaxel combination significantly inhibited AGS cell migration 48 h after siRNA transfection. These findings suggest that NRF2 may promote the migration potential of cancer cells, and the use of NRF2 siRNA in combination with paclitaxel can result in a significant decrease in the AGS cells' ability to migrate.
3.9 NRF2 siRNA/ paclitaxel combination altered the expression level of apoptotic, anti-apoptotic, metastatic, and cell cycle genes
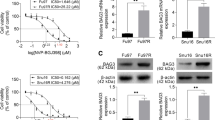
Following the promising results of NRF2 siRNA transfection and combination therapy in reducing tumor cell viability, inducing apoptosis, arresting the cell cycle, and minimizing migration, we investigated the molecular mechanisms underlying these effects. As shown in Fig. 9A, the expression level of apoptotic genes including Bax, Caspase-3, and Caspase-9 were significantly increased in the siRNA and combination groups. In contrast, Bcl-2 (anti-apoptotic) and MMP-2 (metastatic) gene expression were down-regulated in NRF2 siRNA transfection and paclitaxel treatment groups (Fig. 9B). The NRF2 siRNA and paclitaxel treatment also altered the expression of the cell cycle gene P53. All of the experimental groups had significantly higher levels of relative P53 expression (Fig. 9C). However, the combination group exhibited the most pronounced effects on P53 gene expression, which was also evident in other gene alterations.
The NRF2-siRNA and combined therapy of paclitaxel and NRF2 siRNA transfection significantly increased mRNA expression of apoptotic genes (Bax, Caspase-3, and Caspase-9) in GC cells. A Relative expression of Bcl-2 (anti-apoptotic) and MMP-2 (metastatic) genes was decreased in paclitaxel treated, NRF2 siRNA transfected, and combination-treated cells. B The mRNA expression of P53 expression as a cell cycle-related gene was enhanced in all experimental groups. C (n = 3); *p < 0.05, **p < 0.01, ****p < 0.000
4 Discussion
GC is the most common malignancy of the digestive system, with high morbidity and mortality rates worldwide [17]. Despite developments in treatment over the last few decades, patients with GC still have a poor prognosis [18, 19]. Although cytotoxic chemotherapy remains the cornerstone of treatment for metastatic GC, recent developments in the molecular understanding of the disease have reawakened hope that targeted therapies can be used to improve survival and reduce toxicity [20].
NRF2 is a particular transcription factor that strongly regulates antioxidant gene expression. Evidence suggests that malignant tumors, including GC, have higher levels of NRF2 expression and activation, which is linked to enhanced antioxidant capacity, chemoresistance, and a poor clinical prognosis [21]. NRF2 has been shown to promote the growth and migration of malignant cells. According to studies, knockdown of NRF2 by specific siRNA reduces the progression of colon cancer [22] and migration of human glioma tumors [23]. Also, one of the main reasons and ideas behind initiating this research is that we previously conducted a similar study on a pancreatic cancer cell line (Miapaca-2). This study demonstrates that the suppression of NRF2 via siRNA, in combination with paclitaxel treatment, significantly enhances apoptosis and reduces the migration of pancreatic cancer cells [24]. To investigate the responsiveness of this combination, on cells from another type of cancer and to explore the potential involvement of new cellular pathways, we initiated this study on gastric cancer cells.
In several malignancies, NRF2 has been found to have a significant impact on cell survival. In this study, we investigated the cytotoxic effect of NRF2 gene suppression on AGS cell survival by MTT assay. The results showed that transfection of cells with NRF2 siRNA leads to a considerable decrease in cell survival compared to the control group. In this way, the results of a study by Lee et al. on SW480 cells from colon cancer showed that inhibition of the NRF2 gene by specific siRNA reduces cell survival and progression [22]. Furthermore, Zhang et al. discovered that when the NRF2 gene was knocked down by short hairpin RNA (shRNA) in hepatocellular carcinoma cells the viability of cells was significantly reduced [25].
Consistent with MTT findings, annexin-V/PI staining results demonstrated that NRF2 siRNA stimulates apoptosis. However, the increased rate of apoptosis in the combination use of NRF2 siRNA and paclitaxel was more significant. In line with these findings, Bao et al. found that siRNA-mediated NRF2 knockdown promoted cisplatin-induced apoptosis in ovarian carcinoma cells [26]. Likewise, another study by Ma et al. discovered that cisplatin treatment and NRF2 gene silencing by shRNA dramatically enhanced the apoptosis rate in cervical cancer as compared to control groups [27].
To investigate the underlying mechanism, we assessed the expression levels of genes implicated in the apoptotic pathway. The Bcl-2 family comprises the proteins Bcl-2, a pro-apoptotic regulator, Bax, and Caspase-3 and Caspase-9, endogenous apoptotic pathway internal caspases that can regulate cell death via various mechanisms [28]. The relative mRNA expression of the genes under investigation was assessed by qRT-PCR in control cells, NRF2 siRNA transfected cells, and the combination group. qRT-PCR findings showed that paclitaxel and NRF2 siRNA both upregulated the Bax gene while downregulating the Bcl-2 gene. These alterations in gene expression were more significant when siRNA and paclitaxel were used simultaneously. In Line with our findings, a study by Lee et al. on the SW480 colon cancer cell line found that Bcl-2 expression declines after siRNA-mediated NRF2 knockdown [22]. The study by Surikant et al. also showed that NRF2 in lung cancer reduces apoptosis in cancer cells by increasing the expression of Bcl-2 and subsequently enhancing the Bax gene expression [29]. The results of qRT-PCR in the current investigation showed that NRF2 siRNA and paclitaxel alone increased the expression of Caspase-3 and Caspase-9 genes. However, this change was more prominent when NRF2 siRNA and paclitaxel were used together. In agreement with our findings, a study by Pan et al. showed that NRF2 suppression by siRNA in the U251 glioblastoma cell line resulted in enhanced apoptosis and upregulated expression and activity of Caspase-3 and -9 [30]. Therefore, it is plausible that NRF2 could inhibit GC cell apoptosis by altering major regulators of apoptosis pathways.
In addition, we assessed cell cycle state in treatment groups to determine the antigrowth effect of NRF2 siRNA and paclitaxel on GC. According to the findings, the number of NRF2-silenced cells was increased during the G1 phase. We also showed that paclitaxel treatment, both alone and in combination with NRF2 siRNA, caused cell cycle arrest in the G2 phase, implying that NRF2 siRNA and paclitaxel could decrease AGS cell proliferation by inducing cell cycle arrest in these phases. In this regard, Homma et al. demonstrated that the knockdown of NRF2 by siRNA arrested the cell cycle at the G1 phase in a human lung cancer cell line [31]. Likewise, Choi et al. investigated the effect of taxol treatment on cell cycle progression in human breast cancer and found that the percentage of G2/M cells was significantly increased compared to untreated controls [32].
Moreover, the antigrowth effects of NRF2 siRNA and paclitaxel were assessed by measuring the levels of P53 expression in treatment groups. P53 is a transcription factor that is significantly induced by a variety of stress signals, with cell cycle arrest and apoptosis being the most prominent biological effects [33]. According to qRT-PCR analysis, P53 expression was dramatically upregulated in cells transfected with NRF2 siRNA and treated with paclitaxel alone; however, the highest expression of P53 was found in combination therapy. In this way, You et al. performed a study on the link between NRF2 and murine double minute 2 (Mdm2). According to their findings, blocking NRF2 led to the downregulation of Mdm2 in ovarian cancer cells, which then positively regulated P53 signaling and promoted cell death [34].
Autophagy is an evolutionarily conserved catabolic mechanism that is essential in cancer cell death and survival, as well as cell protection against carcinogenesis [35]. In the current study, we evaluated the impact of NRF2 siRNA in combination with paclitaxel on the induction of autophagy in AGS cells. Our results showed that paclitaxel and NRF2 siRNA transfection enhanced the induction of autophagy compared to the control, suggesting that NRF2 regulates autophagy in GC cells. In this regard, Zhou et al. demonstrated that the silencing of NRF2 enhances baseline autophagy levels in the U251 glioma cell line. Additionally, levels of autophagy were markedly elevated after temozolomide (TMZ) treatment [36].
One of the most intriguing findings of this research was that suppressing NRF2 could inhibit GC cell migration. The wound healing assay (scratch) results revealed that using either NRF2 siRNA or paclitaxel alone inhibited AGS cell migration. However, when NRF2 siRNA and paclitaxel were combined, migration was more dramatically decreased as compared to the control group, suggesting that NRF2 siRNA and paclitaxel have synergistic anti-metastatic effects on GC cells. In a study by Shen et al., esophageal squamous cell carcinoma cells had enhanced levels of NRF2 expression, and there was a significant protein-level association between NRF2 and hypoxia-inducible factor (HIF)-1α. In addition, the results of the wound healing experiment showed that NRF2 suppression via shRNA reduced HIF-1α expression, hence limiting cell migration [37]. Similar findings were reported in research on human glioma cells, which showed that siRNA-mediated NRF2 gene suppression decreased the expression of MMP-9, as well as cell migration [23]. Furthermore, Payandeh et al. studied colorectal cancer cells and discovered that siRNA-mediated suppression of NRF2 in combination with Oxaliplatin might dramatically limit cell migration [38].
Consequently, we measured MMP-2 expression levels in treatment groups as a metastasis promoter to further examine the mechanism by which NRF2 siRNA may serve as an anti-metastasis agent in GC. According to our findings, MMP-2 was downregulated in cells treated with NRF2 siRNA and paclitaxel, either alone or in combination. Overexpression of MMP-2 has been linked to the depth of invasion and lymph node metastasis of GC [39]. In agreement with our findings, Shen et al. found that silencing of NRF2 gene expression by siRNA could significantly downregulate the MMP-2 gene in esophageal squamous cell carcinoma, resulting in decreased cell migration and invasion [37].
There are a few limitations to the current investigation. First of all, the AGS cell line was the sole gastric cancer cell line on which the experiments were run. Secondly, we were unable to perform protein-based assays to investigate the protein expression of the studied genes through the use of protein-based assays. Thirdly, in vivo tests were not possible. Consequently, it is important to evaluate the results while taking these constraints into account.
5 Conclusions
Taken together, our findings showed that suppressing NRF2 expression might trigger apoptosis and improve the sensitivity of AGS cells to paclitaxel treatment by modulating the expression of apoptosis-related genes such as Caspase-3, Caspase-9, Bax, and Bcl-2. Additionally, it was shown that the combination of NRF2 siRNA with paclitaxel induces cell cycle arrest at the G2 phase. Similarly, NRF2 siRNA and paclitaxel combination decreased AGS cell migration by downregulating MMP-2. As well, P53, a key regulator of cell cycle and growth, was expressed at a greater level in the treated groups compared to the control group. Therefore, NRF2 knockdown might be employed to render cancer cells more susceptible to paclitaxel in the treatment of GC; however, further studies, especially in vivo investigations, are necessary to validate the efficacy of this combination therapy as a potential therapeutic strategy for GC.
Data availability
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
References
Rawla P, Barsouk A. Epidemiology of gastric cancer: global trends, risk factors and prevention. Gastroenterol Rev. 2019;14(1):26.
Digklia A, Wagner ADJ. Advanced gastric cancer: current treatment landscape and future perspectives. World J Gastroenterol. 2016;22(8):2403.
Smyth EC, et al. Gastric cancer. Lancet. 2020;396(10251):635–48.
Bang YJ, et al. Docetaxel 75 mg/m(2) is active and well tolerated in patients with metastatic or recurrent gastric cancer: a phase II trial. Jpn J Clin Oncol. 2002;32(7):248–54.
Guan W-L, et al. Gastric cancer treatment: recent progress and future perspectives. J Hematol Oncol. 2023;16(1):57.
Xu W, et al. Molecular targeted therapy for the treatment of gastric cancer. J Exp Clin Cancer Res. 2016;35:1–11.
Zhu L, Chen L. Progress in research on paclitaxel and tumor immunotherapy. Cell Mol Biol Lett. 2019;24(1):1–11.
Alexandre J, et al. Novel action of paclitaxel against cancer cells: bystander effect mediated by reactive oxygen species. Cancer Res. 2007;67(8):3512–7.
Cullinan SB, et al. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24(19):8477–86.
Stacy DR, et al. Increased expression of nuclear factor E2 p45-related factor 2 (NRF2) in head and neck squamous cell carcinomas. Head Neck. 2006;28(9):813–8.
Jaramillo MC, Zhang DD. The emerging role of the Nrf2–Keap1 signaling pathway in cancer. Genes Dev. 2013;27(20):2179–91.
De Nicola GM, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475(7354):106–9.
Lin L, et al. Nrf2 signaling pathway: current status and potential therapeutic targetable role in human cancers. Front Oncol. 2023;13:1184079.
Wu S, Lu H, Bai YJ. Nrf2 in cancers: a double-edged sword. Cancer Med. 2019;8(5):2252–67.
Yin JQ, et al. siRNA agents inhibit oncogene expression and attenuate human tumor cell growth. J Exp Ther. 2003;3(4):194–204.
Murugan S, Amaravadi RK. Methods for studying autophagy within the tumor microenvironment. Cham: Springer; 2016. p. 145–66.
Johnston FM, Beckman MJ. Updates on management of gastric cancer. Curr Oncol Rep. 2019;21(8):1–9.
D’Ugo D, et al. Global updates in the treatment of gastric cancer: a systematic review Part 2: perioperative management, multimodal therapies, new technologies, standardization of the surgical treatment and educational aspects. Updates Surg. 2020;72(2):355–78.
Tan Z. Recent advances in the surgical treatment of advanced gastric cancer: a review. Med Sci Monit. 2019;25:3537.
Patel TH, Cecchini M. Targeted therapies in advanced gastric cancer. Curr Treat Option Oncol. 2020;21(9):1–14.
Farkhondeh T, et al. Roles of Nrf2 in gastric cancer: targeting for therapeutic strategies. Molecules. 2021;26(11):3157.
Lee YJ, et al. Overexpression of Nrf2 promotes colon cancer progression via ERK and AKT signaling pathways. Annal Surg Treat Res. 2020;98(4):159.
Pan H, et al. The role of Nrf2 in migration and invasion of human glioma cell U251. World Neurosurg. 2013;80(3–4):363–70.
Riazi-Tabrizi N, et al. NRF2 Suppression Enhances the Susceptibility of Pancreatic Cancer Cells, Miapaca-2 to Paclitaxel. Mol Biotechnol. 2023;23:1–14.
Zhang M, et al. Nrf2 is a potential prognostic marker and promotes proliferation and invasion in human hepatocellular carcinoma. BMC Cancer. 2015;15(1):1–12.
Bao L-J, et al. Nrf2 induces cisplatin resistance through activation of autophagy in ovarian carcinoma. Int J Clin Exp Pathol. 2014;7(4):1502.
Ma X, et al. Nrf2 knockdown by shRNA inhibits tumor growth and increases efficacy of chemotherapy in cervical cancer. Cancer Chemother Pharmacol. 2012;69(2):485–94.
Tsujimoto YJ. Role of Bcl-2 family proteins in apoptosis: apoptosomes or mitochondria? Genes Cells. 1998;3(11):697–707.
Niture SK, Jaiswal AK. Nrf2 protein up-regulates antiapoptotic protein Bcl-2 and prevents cellular apoptosis. J Biol Chem. 2012;287(13):9873–86.
Pan H, et al. The involvement of Nrf2–ARE pathway in regulation of apoptosis in human glioblastoma cell U251. Neurol Res. 2013;35(1):71–8.
Homma S, et al. Nrf2 enhances cell proliferation and resistance to anticancer drugs in human lung cancer. Clin Cancer Res. 2009;15(10):3423–32.
Choi YH, Yoo YHJ. Taxol-induced growth arrest and apoptosis is associated with the upregulation of the Cdk inhibitor, p21WAF1/CIP1, in human breast cancer cells. Onclo Rep. 2012;28(6):2163–9.
Joruiz SM, Bourdon J-C. p53 isoforms: key regulators of the cell fate decision. Cold Spring Harb Perspect Med. 2016;6(8):a026039.
You A, et al. Transcription factor Nrf2 maintains the basal expression of Mdm2: an implication of the regulation of p53 signaling by Nrf2. Archiv Biochem Biophys. 2011;507(2):356–64.
Sui X, et al. Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis. 2013;4(10):e838–e838.
Zhou Y, et al. Knockdown of Nrf2 enhances autophagy induced by temozolomide in U251 human glioma cell line. Oncol Rep. 2013;29(1):394–400.
Shen H, et al. Blockage of Nrf2 suppresses the migration and invasion of esophageal squamous cell carcinoma cells in hypoxic microenvironment. Dis Esophagus. 2014;27(7):685–92.
Payandeh Z, et al. The Impact of Nrf2 Silencing on Nrf2-PD-L1 Axis to Overcome Oxaliplatin Resistance and Migration in Colon Cancer Cells. Avicenna J Med Biotechnol. 2021;13(3):116.
Zheng H, et al. Expressions of MMP-2, MMP-9 and VEGF are closely linked to growth, invasion, metastasis and angiogenesis of gastric carcinoma. Avicenna J Med Biotechnol. 2006;26(5A):3579–83.
Acknowledgements
All the authors would like to thank the Immunology Research Center of Tabriz University of Medical Sciences for their kind support during this study.
Funding
No funding was received.
Author information
Authors and Affiliations
Contributions
Shima Hasani. The first author of the manuscript performed the experiment, contributed to the cellular and molecular assays, and wrote the initial version of the manuscript. Mohammad Khalaj-Kondori. The corresponding author of the manuscript supervised the project and revised the main text of the manuscript. Sahar Safaei. and Mohammad Amini. Contributed to the cellular and molecular assays, analyzed the data, and formal analysis. Negin Riazi Tabrizi. and Mohadeseh Maghsoudi. Helped with the data categorization and interpreted the results. Behzad Baradaran. The corresponding author of the manuscript supervised the project and revised the main text of the manuscript. The authors declare that all data were generated in-house and that no paper mill was used.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The research protocol was approved by the Ethics Committie of Tabriz University of Medical sciences, Iran (IR.TBZMED.VCR.REC.1400.329).
Competing interests
No potential competing interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hasani, S., Khalaj-Kondori, M., Safaei, S. et al. Co-targeting NRF2 potentially enhances the in vitro anticancer effects of paclitaxel in gastric cancer cells. Discov Onc 15, 424 (2024). https://doi.org/10.1007/s12672-024-01312-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12672-024-01312-6