Abstract
This is a first-time report of structural characterization and assessment of the antibacterial activity of silver nanoparticles synthesized using the fungus Aspergillus hortai isolated from domestic wastewater. Silver nanoparticles were synthesized using cell-free filtrate of Aspergillus hortai cultured in potato dextrose broth at 60 °C in the presence of 2-mM silver nitrate for 24 h. The appearance of reddish brown color and a surface plasmon resonance peak at 420 nm confirmed the formation of the silver nanoparticles. Dynamic light scattering analysis reported an average particle size of 57 nm having a polydispersity index of 0.21 and zeta potential of − 19 mV. Transmission electron micrographs confirmed almost spherical shape of the nanoparticles and the selected area electron diffraction pattern corresponded to the crystal lattice structure of silver. The antibacterial potential of the silver nanoparticles was evaluated using a growth curve assay, well diffusion method, and DREAM assay on Escherichia coli (ATCC 35218) and Staphylococcus aureus (ATCC 25923). The assessment of the antibacterial potential of the silver nanoparticles demonstrated activity comparable to that of streptomycin, which was used as a positive control in all the experiments.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Excessive and indiscriminate use of antibiotics has led to a surge in multi-drug resistant bacterial strains termed as superbugs [1, 2]. This has triggered the search for new drugs or agents that are effective against these superbugs. Silver is a known antimicrobial agent that has been used in different forms to effectively treat infections caused by a range of microorganisms. Silver nanoparticles are more effective than silver nitrate or free silver because of the capping agents that confer stability and make them less toxic and more specific in their activity [3, 4]. In recent times, the antimicrobial capability of silver nanoparticles has been harnessed in cleaning agents, water treatment systems, and as coatings in medical devices [5].
Conventional physical and chemical methods of synthesis of silver nanomaterials involve hazardous reagents and stabilizing agents, expensive equipment, and laborious protocols. Alternative green synthesis routes utilize biological systems such as bacteria, fungi, and plants wherein biomolecules like proteins, polysaccharides, and lipids serve as reducing and capping agents [6, 7]. Among them, fungi which exude large quantities of extracellular biomolecules such as proteins, enzymes, lipids, and polysaccharides [9,10,11] are more advantageous due to ease of handling, higher capacity for metal accumulation, and economic viability in scale-up operations.
Microorganisms involved in the biological treatment of wastewater include such organisms which are capable of thriving under stress from pollutants, physical pressure, and other microorganisms in its surroundings. Fungi found in these conditions are capable of co-existing in a microbial community and work in tandem with bacteria for the breakdown of organic pollutants. Due to their large surface area and ability to provide a substratum for other microorganisms to bind and trap molecules leading to the formation of flocs, fungi play a significant role in wastewater treatment plants. Filamentous fungi that are able to adapt to and survive in such hostile environments containing a variety of organic and inorganic pollutants are of great significance in bioremediation processes.
Recently, a cost-effective and rapid dye reduction-based electron-transfer activity monitoring (DREAM) assay was employed to investigate the antibacterial activity of plant extracts [12]. Although this assay has been used previously to evaluate the electron transfer activity of bacteria in the presence of nanoparticles [13], it has not been employed hitherto for assessing the antibacterial activity of silver nanoparticles. This technique can overcome the problem of ineffective diffusion of silver nanoparticles across the agar medium in diffusion assays.
This study presents a first-time report of mycosynthesis of silver nanoparticles using extracts of the fungus Aspergillus hortai isolated from a domestic wastewater treatment plant, structural characterization of the nanoparticles, and evaluation of their antimicrobial potential.
2 Materials and Methods
2.1 Mycosynthesis of Silver Nanoparticles
Isolate MWWRS1c (A. hortai) was isolated from activated sludge of a domestic wastewater treatment plant located in the Prasanthi Nilayam township and was cultured in flasks containing potato dextrose broth maintained at 25 °C at 130 rpm in a temperature-controlled shaker for 7 days. Fungal biomass was then harvested and rinsed twice with sterile deionized water to remove media components. Ten grams of biomass was then suspended in 100 ml of sterile deionized water for 3 days at 25 °C at 130 rpm. The cell-free filtrate (CFF) was obtained after discarding the biomass and filtering the medium with Whatman filter paper No.1. Fifty milliliters of the CFF was added to an equal volume of different concentrations of silver nitrate solution (1 mM, 2 mM, and 5 mM) and incubated at different temperatures (30 °C, 40 °C, 50 °C, and 60 °C) in the dark for 24 h [14].
2.2 Characterization of Mycosynthesized Silver Nanoparticles
Synthesis of silver nanoparticles (AgNPs) was confirmed by a typical change in color of the reaction mixture to brown. The surface plasmon resonance peak was recorded by a wavelength scan in the range of 200 to 800 nm using a UV–visible spectrophotometer (Spectramax M5, Molecular Devices). Size distribution and zeta potential of AgNPs were analyzed by dynamic light scattering (DLS) using a particle size analyzer (Litesizer 500, Anton Paar) at room temperature. The morphology and crystalline structure of AgNPs were characterized by transmission electron microscopy (TEM) (JEOL HRTEM JEM-2100Plus) at an accelerating voltage of 200 kV. For determining the capping agents on the surface of AgNPs, the reaction mixture was centrifuged at 13,000 rpm for 10 min. The pellet was washed thrice with double distilled H2O to remove remnants of the CFF and dried overnight at 60 °C. The resultant AgNP powder was mixed with potassium bromide (KBr) in a 1:80 ratio to make KBr pellets that were used to perform FTIR scan (Spectrum Two, Perkin Elmer) at room temperature in transmittance mode between 4000 and 400 cm−1 at a resolution of 1 cm−1.
2.3 Antibacterial Activity of AgNPs
2.3.1 Bacterial Growth Curve Assay
Escherichia coli (ATCC 35218) (Gram negative) and Staphylococcus aureus (ATCC 25923) (Gram positive) were used for this study. Each of these strains was inoculated in 50 ml of sterile Luria Bertani (LB) broth in 250-ml conical flasks and incubated overnight at 37 °C at 180 rpm. 0.5 ml of the overnight culture was added to 50 ml of sterile LB broth to initiate the growth curve experiment. The AgNP sample was added during the 2nd hour of the growth phase of the bacteria after inoculation. One milligram Streptomycin ml−1 was used as a positive control. The bacterial growth curve was plotted based on the OD600 values for 8 h at 1-h intervals using a UV–visible spectrophotometer [15]. All the experiments were performed in triplicates.
2.3.2 Well Diffusion Method
Overnight cultures of E. coli and S. aureus in LB broth after obtaining 0.5 McFarland standard were inoculated on LB agar plates using sterile cotton swabs. After allowing the plates to stand for 10 min, 6 mm wells were bored into the medium. Fifty microliter of the samples were added to the wells and the plates were incubated overnight at 37 °C. One milligram Streptomycin ml−1 was used as a positive control [16]. Experiments were performed in triplicates.
2.3.3 DREAM Assay
DREAM assay was employed with slight modifications. Samples were added to different conical flasks containing freshly inoculated bacterial cultures at 37 °C. Methylene blue reduction kinetics were recorded as A660 for 3 min at 1-h intervals along the growth curve experiment for 8 h using a UV–visible spectrophotometer. One milligram Streptomycin ml−1 was used as a positive control. All the experiments were done in triplicates. The methylene blue reduction trajectory at A660 was plotted as a function of time to obtain the microbial activity profile.
3 Results and Discussion
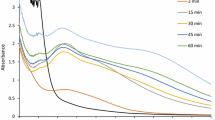
A. hortai, first described by Langeron (1922), was initially considered as a synonym of A. terreus and was later found to be distinct due to its distinct extrolite profile [18]. Figure 1a and b present the cultural characteristics of the isolate MWWRS1c and the phylogenetic analysis based on ITS sequencing (Indiseq Genomics, Chennai) respectively which confirm its identity as A. hortai. The sequence was deposited in GenBank with accession number MZ678771. This isolate showed the best response in terms of biosynthesis of silver AgNPs compared to all the other fungal specimens isolated from the wastewater samples. Formation of AgNPs upon addition of CFF to silver nitrate solution resulted in the reduction of Ag+ ions and was visualized as a change in color from pale yellow to dark reddish brown (Fig. 2 inset) which is typical for AgNPs. The UV–visible spectrum (Fig. 2) showed a surface plasmon resonance peak at 420 nm which is within the 380 to 450 nm range described for AgNPs [19].
Results of the studies on the optimization of reaction temperature for AgNP biosynthesis using A. hortai (Fig. 3a) showed that AgNP formation was better at higher temperatures. The rate of formation, size, and stability of the particles depends on the reaction temperature which is responsible for creating a conducive environment for the nucleation of AgNPs [20] and would vary depending on the fungal species involved [21, 22]. Since the further increase in temperature can result in the denaturation of enzymes responsible for the reduction of Ag+ ions [23, 24], the temperature was maintained at 60 °C for the reaction time of 24 h. Similarly, higher AgNO3 concentrations (1 mM, 2 mM, and 5 mM) resulted in a corresponding increase in the peak intensity as shown in Fig. 3b. But it was observed that the peak broadened at higher concentrations of silver nitrate over a period of time probably due to aggregation of the AgNPs. It is also known that excess metal ions can lead to the formation of larger nanoparticles with irregular morphology due to the contention between Ag+ ions and functional groups from the CFF [20, 22, 24]. As a result, a 2-mM concentration of AgNO3 was used for the synthesis of AgNPs in this study.
Dynamic light scattering analysis (Fig. 4a) showed that the average size distribution of the AgNPs was 4.08 nm (3.35%), 57.65 nm (72.67%), and 458.4 nm (23.98%) with a polydispersity index of 0.21. It was noticed that with an increase in temperature, the size and polydispersity index of the AgNPs reduced. The zeta potential value of AgNPs was − 19.1 mV (Fig. 4b) which indicates the stability of the AgNPs due to repulsion between the nanoparticles [21]. The negative charge can be attributed to the proteins present on the surface of nanoparticles as capping agents during synthesis, because of which, the electrostatic repulsive force between the nanoparticles results in the prevention of aggregation and agglomeration in lower silver nitrate concentrations [22, 25]. TEM and HR-TEM micrographs confirmed the polydispersity of the near-spherical AgNPs (Fig. 5a) and their crystalline structure (Fig. 5b). The selected area electron diffraction (SAED) pattern (Fig. 5c) suggests a correspondence of the AgNPs to the crystal lattice structure of metallic silver.
FTIR spectrum of the mycosynthesized AgNPs (Fig. 6) showed a broad peak at 3283 cm−1 corresponding to alcohol or phenol. The peaks at 3283 cm−1 and 2919 cm−1 correspond to the stretching vibrations of primary and secondary amines respectively, and their corresponding bending vibrations are observed at 1643 cm−1 and 1555 cm−1 respectively [26]. These bands confirm the role of proteins in the formation of capping agents on the surface of the nanoparticles thereby conferring stability [19, 27]. Biomolecules exuded from fungi are known to have an affinity towards metal surfaces. The biomolecules responsible for the reduction of the silver ions are activated as soon as silver nitrate is added to the CFF. These protein and amino acid residues in the CFF simultaneously bind to the metal surface during the nanoparticle synthesis as capping agents and confer stability [22, 28].
Growth curve plots of E. coli and S. aureus in the presence of the mycosynthesized AgNPs showed that bacterial growth was inhibited upon the addition of the AgNPs to the bacterial culture 2 h after initiation of the culture (Fig. 7). CFF, which was used as a negative control, had no inhibitory effect on the growth of bacterial strains while streptomycin, added to the culture along bacterial inoculation as a positive control, inhibited the bacterial growth. This method indicates the detrimental effect of silver nanoparticles on the bacterial growth. Silver which has a high affinity to sulfur and phosphorous binds to the cell wall and disrupts its integrity. AgNPs are also known to inhibit protein translation by binding to translation factors [1, 2]. These results point to the antibacterial potential of the AgNPs synthesized using A. hortai.
The inhibitory activity of the mycosynthesized AgNPs on bacterial proliferation was tested using the agar well diffusion assay. Upon observation of the plates 24 h after inoculation, the zone of inhibition was found to be the largest for the highest concentration of AgNPs (Table 1). It was interesting to note that the antibacterial activity was more pronounced in S. aureus compared to E. coli. Gram-positive bacteria are generally expected to be less susceptible to inhibitory activity due to the presence of a thick peptidoglycan layer. This could be the reason for previously reported AgNPs having no effect on Gram-positive bacteria [29, 30]. It must also be noted that the size of the nanoparticles has an impact on antibacterial activity [3] due to variations in their dispersion across the agar medium [15]. Bactericidal activity can also be attributed to the proteins on the surface of the mycosynthesized AgNPs [4, 5]. It would be interesting to carry out further studies to identify the reasons for the more pronounced antibacterial activity in the Gram-positive S. aureus.
DREAM assay which relies on the transfer of electrons, resulting from the microbial breakdown of the organic substrates, to methylene blue helps in understanding the viability of bacteria rather than merely the bacterial cell load [12, 17]. It can be evidenced from the methylene blue reduction profiles in Fig. 8 that the addition of AgNPs to the bacterial cultures disrupted their cellular function leading to loss of viability and eventually death. The reduction of methylene blue was almost negligible when the bacterial cultures were grown with the AgNPs, similar to that observed in the presence of the positive control streptomycin. The addition of only CFF to the cultures did not significantly impact bacterial viability. Preliminary studies carried out to ascertain the ability of the CFF or AgNPs by itself to reduce methylene blue ruled out the possibility.
Although the precise mechanism of antibacterial activity of AgNPs is not completely understood, a mixed approach involving different assays will help in gaining perspective about the different modes of action that are possible. AgNPs could inhibit bacterial growth and activity by a combination of the following: (i) adhering to the cell membrane, altering membrane structure and permeability resulting in impaired membrane transport and hampered ATP production [31]; (ii) penetrating the cell, interacting with cell organelles and disrupting their function [32, 33]; (iii) disrupting signalling pathways by interacting with associated proteins and rendering the cell incapable of performing basic physiological functions [34, 35]; (iv) oxidizing proteins, lipids and DNA bases resulting in cellular toxicity [36, 37]; and (v) releasing reactive oxygen species (ROS) and free radicals leading to oxidative stress [38].
4 Conclusions
The rising trend of bacteria acquiring resistance to antibiotics has spurred the search for alternative approaches to counter microbial contamination and infections. The antibacterial properties of silver have been successfully exploited in the use of AgNPs synthesized from biological sources as alternatives to antibiotics. This first-time report of AgNPs synthesized using A. hortai isolated from domestic wastewater showcases their antibacterial potential. The AgNPs were structurally characterized using UV–vis and FTIR spectroscopy, DLS analysis, and TEM. The antibacterial potential of these mycosynthesized AgNPs, ascertained using growth curve analysis, agar plate well diffusion, and the DREAM assay, was found to be comparable to that of streptomycin. Future work would involve further physico-chemical characterization and testing the capacity of these AgNPs as broad-spectrum antibacterial agents.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Prasher, P., Singh, M., & Mudila, H. (2018). Silver nanoparticles as antimicrobial therapeutics: current perspectives and future challenges. 3 Biotech, 8(10), 1–23. https://doi.org/10.1007/s13205-018-1436-3
Wahab, S., Khan, T., Adil, M., & Khan, A. (2021). Mechanistic aspects of plant-based silver nanoparticles against multi-drug resistant bacteria. Heliyon, 7(7), e07448. https://doi.org/10.1016/j.heliyon.2021.e07448
Abdulsahib, S. S. (2021). Synthesis, characterization and biomedical applications of silver nanoparticles. Biomedicine, 41(2), 458–464. https://doi.org/10.51248/.v41i2.1058
Zhao, X., Zhou, L., Riaz Rajoka, M. S., Yan, L., Jiang, C., Shao, D., … Jin, M. (2018). Fungal silver nanoparticles: synthesis, application and challenges. Critical Reviews in Biotechnology, 38(6), 817–835.https://doi.org/10.1080/07388551.2017.1414141
Prabhu, S., & Poulose, E. K. (2012). Silver nanoparticles: Mechanism of antimicrobial. International Nano Letters, 2, 32–41
Osorio-Echavarría, J., Osorio-Echavarría, J., Ossa-Orozco, C. P., & Gómez-Vanegas, N. A. (2021). Synthesis of silver nanoparticles using white-rot fungus Anamorphous Bjerkandera sp. R1: Influence of silver nitrate concentration and fungus growth time. Scientific Reports, 11(1), 1–14. https://doi.org/10.1038/s41598-021-82514-8
Saxena, J., Sharma, M. M., Gupta, S., Singh, A., Industrial, M., Jaipur, A., … Express, J. (2014). Emerging role of fungi in nanoparticle synthesis and. World Journal of Pharmacy and Pharmaceutical Sciences, 3(9), 1586–1613
Li, P. J., Pan, J. J., Tao, L. J., Li, X., Su, D. L., Shan, Y., & Li, H. Y. (2021). Green synthesis of silver nanoparticles by extracellular extracts from Aspergillus japonicus PJ01. Molecules, 26(15). https://doi.org/10.3390/molecules26154479
Abbasi, E., Milani, M., Fekri Aval, S., Kouhi, M., Akbarzadeh, A., Tayefi Nasrabadi, H., … Samiei, M. (2014). Silver nanoparticles: Synthesis methods, bio-applications and properties. Critical Reviews in Microbiology, 42(2), 1–8.https://doi.org/10.3109/1040841X.2014.912200
Sastry, M., Ahmad, A., Islam Khan, M., & Kumar, R. (2003). Biosynthesis of metal nanoparticles using fungi and actinomycete. Current Science, 85(2), 162–170
Lotfy, W. A., Alkersh, B. M., Sabry, S. A., & Ghozlan, H. A. (2021). Biosynthesis of silver nanoparticles by Aspergillus terreus: Characterization, optimization, and biological activities. Frontiers in Bioengineering and Biotechnology, 9(April). https://doi.org/10.3389/fbioe.2021.633468
Aiyer, K. S., Rai, R., & Vijayakumar, B. S. (2019). Assessing Activity of antimicrobial agents and screening antibiotic-resistant bacteria through DREAM assay. Applied Biochemistry and Biotechnology, 188(4), 1158–1167. https://doi.org/10.1007/s12010-019-02981-8
Vishwanathan, A. S., Aiyer, K. S., Chunduri, L. A. A., Venkataramaniah, K., Siva Sankara Sai, S., & Rao, G. (2016). Carbon quantum dots shuttle electrons to the anode of a microbial fuel cell. 3 Biotech, 6(2), 228. https://doi.org/10.1007/s13205-016-0552-1
Gaikwad, S. C., Birla, S. S., Ingle, A. P., Gade, A. K., Marcato, P. D., Rai, M., & Duran, N. (2013). Screening of different Fusarium species to select potential species for the synthesis of silver nanoparticles. Journal of the Brazilian Chemical Society, 24(12), 1974–1982
Wu, Y., Yang, Y., Zhang, Z., Wang, Z., Zhao, Y., & Sun, L. (2018). A facile method to prepare size-tunable silver nanoparticles and its antibacterial mechanism. Advanced Powder Technology, 29(2), 407–415. https://doi.org/10.1016/j.apt.2017.11.028
Balakumaran, M. D., Ramachandran, R., Balashanmugam, P., Mukeshkumar, D. J., & Kalaichelvan, P. T. (2016). Mycosynthesis of silver and gold nanoparticles: Optimization, characterization and antimicrobial activity against human pathogens. Microbiological Research, 182, 8–20. https://doi.org/10.1016/j.micres.2015.09.009
Vishwanathan, A. S., Devkota, R., Siva Sankara Sai, S., & Rao, G. (2015). DREAM assay for studying microbial electron transfer. Applied Biochemistry and Biotechnology, 177(8), 1767–1775.https://doi.org/10.1007/s12010-015-1852-3
Samson, R. A., Peterson, S. W., Frisvad, J. C., & Varga, J. (2011). New species in Aspergillus section Terrei. Studies in Mycology, 69, 39–55. https://doi.org/10.3114/sim.2011.69.04
Jain, N., Bhargava, A., Majumdar, S., Tarafdar, J. C., & Panwar, J. (2011). Extracellular biosynthesis and characterization of silver nanoparticles using Aspergillus flavus NJP08: A mechanism perspective. Nanoscale, 3(2), 635–641. https://doi.org/10.1039/c0nr00656d
Liu, H., Zhang, H., Wang, J., & Wei, J. (2020). Effect of temperature on the size of biosynthesized silver nanoparticle: Deep insight into microscopic kinetics analysis. Arabian Journal of Chemistry, 13(1), 1011–1019. https://doi.org/10.1016/j.arabjc.2017.09.004
Elamawi, R. M., Al-Harbi, R. E., & Hendi, A. A. (2018). Biosynthesis and characterization of silver nanoparticles using Trichoderma longibrachiatum and their effect on phytopathogenic fungi. Egyptian Journal of Biological Pest Control, 28(1), 1–11. https://doi.org/10.1186/s41938-018-0028-1
Guilger-Casagrande, M., & de Lima, R. (2019). Synthesis of silver nanoparticles mediated by fungi: A review. Frontiers in Bioengineering and Biotechnology, 7(October), 1–16. https://doi.org/10.3389/fbioe.2019.00287
Birla, S. S., Gaikwad, S. C., Gade, A. K., & Rai, M. K. (2013). Rapid synthesis of silver nanoparticles from Fusarium oxysporum by optimizing physicocultural conditions. The Scientific World Journal, 2013,. https://doi.org/10.1155/2013/796018
AbdelRahim, K., Mahmoud, S. Y., Ali, A. M., Almaary, K. S., Mustafa, A. E. Z. M. A., & Husseiny, S. M. (2017). Extracellular biosynthesis of silver nanoparticles using Rhizopus stolonifer. Saudi Journal of Biological Sciences, 24(1), 208–216. https://doi.org/10.1016/j.sjbs.2016.02.025
Phanjom, P., & Ahmed, G. (2015). Biosynthesis of silver nanoparticles by Aspergillus oryzae (MTCC No. 1846) and its characterizations. Nanoscience and Nanotechnology, 5(1), 14–21. https://doi.org/10.5923/j.nn.20150501.03
Vigneshwaran, N., Ashtaputre, N. M., Varadarajan, P. V., Nachane, R. P., Paralikar, K. M., & Balasubramanya, R. H. (2007). Biological synthesis of silver nanoparticles using the fungus Aspergillus flavus. Materials Letters, 61(6), 1413–1418. https://doi.org/10.1016/j.matlet.2006.07.042
Nandiyanto, A. B. D., Oktiani, R., & Ragadhita, R. (2019). How to read and interpret ftir spectroscope of organic material. Indonesian Journal of Science and Technology, 4(1), 97–118. https://doi.org/10.17509/ijost.v4i1.15806
Abdel-Kareem, M., Zohri, A.-N., & Rasmey, A.-H. (2021). Biosynthesis of silver nanoparticles by Aspergillus sakultaensis and its antibacterial activity against human pathogens. Egyptian Journal of Microbiology, 0(0), 0–0. https://doi.org/10.21608/ejm.2021.46387.1177
Rahimi, G., Alizadeh, F., & Khodavandi, A. (2016). Mycosynthesis of silver nanoparticles from Candida albicans and its antibacterial activity against Escherichia coli and Staphylococcus aureus. Tropical Journal of Pharmaceutical Research, 15(2), 371–375. https://doi.org/10.4314/tjpr.v15i2.21
Mistry, H., Thakor, R., Patil, C., Trivedi, J., & Bariya, H. (2021). Biogenically proficient synthesis and characterization of silver nanoparticles employing marine procured fungi Aspergillus brunneoviolaceus along with their antibacterial and antioxidative potency. Biotechnology Letters, 43(1), 307–316. https://doi.org/10.1007/s10529-020-03008-7
Kim, S. H., Lee, H. S., Ryu, D. S., Choi, S. J., & Lee, D. S. (2011). Antibacterial activity of silver-nanoparticles against Staphylococcus aureus and Escherichia coli. Korean Journal of Microbiology and Biotechnology, 39(1), 77–85
Morones, J. R., Elechiguerra, J. L., Camacho, A., Holt, K., Kouri, J. B., Ramírez, J. T., & Yacaman, M. J. (2005). The bactericidal effect of silver nanoparticles. Nanotechnology, 16(10), 2346–2353. https://doi.org/10.1088/0957-4484/16/10/059
Radzig, M. A., Nadtochenko, V. A., Koksharova, O. A., Kiwi, J., Lipasova, V. A., & Khmel, I. A. (2013). Antibacterial effects of silver nanoparticles on gram-negative bacteria: Influence on the growth and biofilms formation, mechanisms of action. Colloids and Surfaces B: Biointerfaces, 102, 300–306. https://doi.org/10.1016/j.colsurfb.2012.07.039
Shrivastava, S., Bera, T., Roy, A., Singh, G., Ramachandrarao, P., & Dash, D. (2007). Characterization of enhanced antibacterial effects of novel silver nanoparticles. Nanotechnology, 18(22). https://doi.org/10.1088/0957-4484/18/22/225103
Waseem, M., & Nisar, M. A. (2016). Fungal-derived nanoparticles as novel antimicrobial and anticancer agents. Functionalized Nanomaterials. https://doi.org/10.5772/66922
Dakal, T. C., Kumar, A., Majumdar, R. S., & Yadav, V. (2016). Mechanistic basis of antimicrobial actions of silver nanoparticles. Frontiers in Microbiology, 7(NOV), 1–17. https://doi.org/10.3389/fmicb.2016.01831
Anees Ahmad, S., Sachi Das, S., Khatoon, A., Tahir Ansari, M., Afzal, M., SaquibHasnain, M., & Kumar Nayak, A. (2020). Bactericidal activity of silver nanoparticles: A mechanistic review. Materials Science for Energy Technologies, 3, 756–769. https://doi.org/10.1016/j.mset.2020.09.002
Xie, H., Mason, M. M., & Wise, J. P. (2011). Genotoxicity of metal nanoparticles. Reviews on Environmental Health, 26(4), 251–268. https://doi.org/10.1515/REVEH.2011.033
Acknowledgements
The authors dedicate this work to Bhagawan Sri Sathya Sai Baba, the Founder Chancellor of Sri Sathya Sai Institute of Higher Learning. Research facilities provided by the Central Research Instruments Facility (CRIF) and Central Research Laboratory (CRL), SSSIHL are gratefully acknowledged. The authors express their thanks to Dr. B. E. Pradeep, Department of Biosciences, SSSIHL, for providing the bacterial strains for the study.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Research Involving Humans and Animals.
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rai, R., Vishwanathan, A.S. & Vijayakumar, B.S. Antibacterial Potential of Silver Nanoparticles Synthesized Using Aspergillus hortai. BioNanoSci. 13, 203–211 (2023). https://doi.org/10.1007/s12668-022-01043-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12668-022-01043-4