Abstract
A malignant tumor in brain is detected using images from Magnetic Resonance scanners. Malignancy detection in brain and separation of its tissues from normal brain cells allows to correctly localizing abnormal tissues in brain’s Magnetic Resonance Imaging (MRI). In this article, a new method is proposed for the segmentation and classification of brain tumor based on improved saliency segmentation and best features selection approach. The presented method works in four pipe line procedures such as tumor preprocessing, tumor segmentation, feature extraction and classification. In the first step, preprocessing is performed to extract the region of interest (ROI) using manual skull stripping and noise effects are removed by Gaussian filter. Then tumor is segmented in the second step by improved thresholding method which is implemented by binomial mean, variance and standard deviation. In the third step, geometric and four texture features are extracted. The extracted features are fused by a serial based method and best features are selected using Genetic Algorithm (GA). Finally, support vector machine (SVM) of linear kernel function is utilized for the classification of selected features. The proposed method is tested on two data sets including Harvard and Private. The Private data set is collected from Nishtar Hospital Multan, Pakistan. The proposed method achieved average classification accuracy of above 90% for both data sets which shows its authenticity.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
In the domain of computer vision, detection and classification of medical infections gained much attention due to their emerging applications as medical imaging (Havaei et al. 2017; Irum et al. 2015; Masood et al. 2013). In this modern world with developments in the domain of information technology, several image processing and machine learning methods are introduced for accurate detection and classification of disease symptoms. These methods detect and classify disease regions accurately and then classify them into the respective category. This process can help doctors in fast and accurate diagnosis and categorization of tumor or lesion region. Brain tumor tissues affect normal growth of brain. Several factors exist for tumor threat such as location, tumor shape, size, and tumor texture. The tumor tissues consume space in brain and affect brain cells. Normally brain tumor is known as malignant. Human body parts suffer due to attack on brain tissues (Calabrese et al. 2007; Kadam et al. 2012) which can cause many problems for human health. Therefore, early detection of tumor is necessary to save humans from severe health loss and decrease the death rate. Moreover, correct identification of brain tumor is also important because wrong detection of tumor can cause causalities (Ajaj and Syed 2015; Raza et al. 2012).
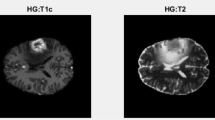
A brain tumor consists of two types including malignant or benign. The malignant tumor cultivates rapidly producing weight inside the brain cells. If this problem is not detected accurately and timely, it can cause death (Mosquera et al. 2013). Primary and secondary are two major types of tumor. Tumor which starts inside the brain is called as Primary brain tumor. This kind of tumor is most malignant one. Secondary brain tumor is found in other parts of body and then spread to the brain or spinal tissues (Ohgaki and Kleihues 2013). MRI of brain is an easy and protected test which utilizes radio waves and magnetic field to get the comprehensive image of a human brain (Chua et al. 2015). Normally four MRI modalities including T1, T1c, T2 and T2f weighted are used to diagnose tumor part in the image (Bauer et al. 2011). These MR images have some noise due to thermal effects. Therefore, before segmentation of brain tumor, the removal of noise is important.
In this article, MR images are used as shown in Fig. 1. In figure, T1 weighted image is obtained by using Time to Echo (TE) and Repetition time (TR), T2 image is shaped by using long TE and TR as compared to T1 weighted imaged and FLAIR image is composed in the same way as T1 or T2 except that its TE and TR are longer as compared to both.
Brain MRI Types (Amin et al. 2018)
1.1 Motivation and problem statement
Image processing and Computer vision (CV) are interdisciplinary areas that deal with images and videos. In these areas, images are processed by external sources and generate required results. The generated results are in the form of images or numeric values. These areas including machine learning gained much attention in the field of medical imaging. Brain tumor, skin cancer and stomach infections are major areas of medical imaging that deal with computer based methods using CV and machine learning approaches. In this article, brain tumor is dealt from MR images. The doctors utilize MR images for brain tumor segmentation and classification. For this purpose, computer based systems are introduced that comprise of four primary steps including tumor contrast stretching, tumor segmentation, feature extraction and classification. But still several problems exist in these systems that affect the system accuracy. The major problems include tumor shape, size, location, irregularity, and tumor diameter. To resolves these issues, in this article, an improved thresholding method is proposed for tumor segmentation and further multi model feature optimization based classification is performed.
1.2 Contribution
The proposed method comprises of four primary steps. In the first step, preprocessing is performed to remove noise factor in images. Then in the second step, tumor region is segmented by improved thresholding method. In the third step, nine geometric and four Harlick features are extracted which are fused by a serial method. In the very next step, GA is implemented for best features selection and classified by SVM. The major contributions are given below:
-
1.
An improved segmentation method is implemented based on three stages. In the first stage, binomial mean is calculated from the preprocessed image, then in the second step their variance is computed and afterwards these values are substituted in a threshold function. The threshold function gives a binary image which is later optimized by morphological operations.
-
2.
Information of nine geometric and four texture features is fused and a GA is implemented for the optimization of fused features. GA selects best features based on their fitness function.
-
3.
A Private data set for the evaluation of proposed method is designed and also validated for each single, fused and selected feature vector to show the system authenticity.
1.3 Paper organization
The organization of this manuscript is as follows: Literature review is described in Sect. 2. Section 3 gives detailed discussion of proposed method which includes improved thresholding method, feature extraction and GA based features selection. In Sect. 4, detailed experimental results are presented. Conclusion of the paper is given in Sect. 5.
2 Literature review
Nowadays a number of images data are generated in clinical laboratories which are difficult to segment within the acceptable time. Additionally, manual review of images is difficult, tiring and time consuming (Popuri et al. 2012). Computer based methods for the segmentation and classification of tumor prove to be more effective and desirable. But these methods contain several challenges such as texture, appearance, distinct shape, an association between natural texture and tumor, irregularity, discontinuity and border. Major aim of computer based methods is correct detection and classification of tumor region.
In general, detection and classification of tumor region consist of four major steps including preprocessing or normalization, tumor segmentation, feature extraction and classification. The preprocessing step plays a vital role because the rest of steps such as segmentation and feature extraction are dependent on it. Several artifacts exist in MR images which have effects on segmentation accuracy. These artifacts are noise in images, poor contrast, irregularity and more (Patil and Udupi 2012). Therefore in literature, several methods are proposed to remove these problems. Wu et al. (Wu et al. 2014) presented a method in which noise is removed from MR images using structural knowledge. In (Chaddad et al. 2014), morphological operations are utilized to remove extra artifacts. Local and non local neighborhood spatial information is utilized in (Damodharan and Raghavan 2015) to remove the noise effects.
Moreover, several other filters are used in the processing step to remove noise and other artifacts such as high pass filter (Hamamci et al. 2012), sharpening filter, histogram equalization technique (Senthilkumaran and Thimmiaraja 2014), Gaussian high pass filter (Acunzo et al. 2012) and edge detectors. Furthermore, stripping the skull region is a procedure which is important in brain tumor detection and used to remove the skull region from MR images (Liu et al. 2014). Skull stripping can be done manually or using some morphological operations (Doshi et al. 2013). Abbasi et al. (Abbasi and Tajeripour 2017) implemented local binary pattern (LBP) and histogram of gradient (HOG) based features for tumor detection. Bias filed correction and histogram matching method are used in the preprocessing step. Then Otsu thresholding is applied for tumor detection. Thereafter, HOG and LBP features are extracted and classified by Random Forest (RF) classifier. The introduced method is evaluated on BRAST 2013 data set and shows improved performance. An Artificial Bee Colony (ABC) and fuzzy c-means (FCM) based algorithm is implemented in (Menon and Ramakrishnan 2015) for brain tumor detection. The ABC algorithm is used to search threshold value for fitness function. In order to get a fitness function, the original image is disintegrated by discrete wavelet transform (DWT) method. Lastly, segmented regions are converted into clusters by FCM which help in finding out the tumor region. Ariyo et al.(Ariyo et al. 2017) introduced a fusion of spatial FCM and K-means clustering method for brain tumor segmentation from MR images. The FCM is incorporated with spatial neighborhood to remove the noise effects. Then K-means clustering is employed to sort image pixels. Then these pixels are grouped into a singular cluster. The introduced algorithm results are calculated by comparing their values with ground truth values and shows improved performance as contrasted to existing methods. El Abbadi and Kadhim (2017) described a new tumor classification method based on feature extraction and probabilistic neural network (PNN) approach. The implemented approach includes three primary steps as preprocessing, gray level co-occurence matrix (GLCM) and gray level run length matrix (GLRLM) feature extraction and finally classification by PNN. The implemented method is tested on 50 images and achieved accuracy up to 98%.
Other methods are introduced as well in literature which efficiently detect and classify tumor regions from MR images such as Gabor wavelet and statistical features based approach (Nabizadeh and Kubat 2015), convolutional neural network (CNN) based brain tumor segmentation (Pereira et al. 2016), Gaussian mixture model (GMM) based brain tumor classification (Chaddad 2015), hybrid methods (Sharma et al. 2018), texture features based classification (Soltaninejad et al. 2018) and few more (Hore et al. 2015; Moraru et al. 2016, 2017; Rajinikanth et al. 2018; Rani et al. 2017; Samanta et al. 2015; Tian et al. 2017; Wang et al. 2018). These existing techniques give importance to segmentation and feature extraction techniques for brain tumor diagnosis and classification from MR images. Therefore in this study, focus is on hybrid features and segmentation methods to make the system more efficient.
3 Proposed methodology
A novel brain tumor detection and classification approach is proposed in this article from MR images. The proposed method consists of four major steps including preprocessing, region of interest (ROI) detection, multiple feature extraction for best features selection and classification. After classification, tumor area is detected from tumor images as shown in Fig. 2. The comprehensive explanation of each step is given below.
3.1 Image preprocessing
Image preprocessing is an essential part of several computer vision and image processing applications. In medical image processing, this step gained much attention due to several factors such as image acquisition, illumination, low contrast and complex background. An effective preprocessing step plays its primary role in producing better segmentation results which in turn produces high classification accuracy (Gao et al. 2011). Therefore, a hybrid image preprocessing technique is proposed which follows skull stripping, image de-noising and size normalization. All these steps are essential and are required to facilitate segmentation phase where the salient region/tumor is identified and separated from the background. Here in this research work, primary focus is to identify the tumor which is initially marked using skull stripping method. Later noise is removed from the database images so that infected regions are extracted with greater accuracy. The implemented approach is applied to two selected data sets namely Harvard imaging data set and Private collected images. Both data sets contain some noise and resizing issues, therefore this technique helps to remove these factors. The sample images are shown in Fig. 3.
Let \(~{{\varvec{\Delta}}}\) denotes the selected data sets, \({\text{I}}\left( {{\text{i}},{\text{j}}} \right)\)denotes an input image having dimension \(M \times N\), where \(~I\left( {i,j} \right) \in {{\varvec{\Delta}}}\). In MR images, focus is to detect ROI, where ROI denotes the tumor region. Therefore, initially skull stripping is used. In MR images, a skull is a region which is neither a tumor nor ROI. A manual skull striping method is utilized to detect ROI (Kalavathi and Prasath 2016) which is brain tumor. Therefore, all those regions which belong to brain tumor are of high priority whilst rest of the information is of low priority. The effects of skull stripping on Private and Harvard data sets are shown in Fig. 4.
Afterwards, a Gaussian filter is used to remove noise effects in the skulled images because these images hold a lot of speckle noise. Recently, several techniques are introduced which deal with image de-noising such as (Ashour et al. 2018a, b; Dey 2015 # 37). The Gaussian filter (Deng and Cahill 1993) does not defeat the peak signals and only minimizes variance among raise and fall of signals. According to center limit theorem, noises combined effect tends towards Gaussian. Therefore, Gaussian filter is utilized for speckle noise removal as well as for other minimal noises if they exist. Gaussian filter undoubtedly makes the edges blur but this problem is controlled by using different ranges of std and mean. \(\sigma =0.3\) is initialized in this research. The Gaussian filter is defined as in Eq. (1) and Eq. (2).
where, \(~{I^G}(i,j)\) denotes the de-noising image using Gaussian function, \({\sigma ^2}\) denotes the variance of noisy skulled image, \(i\) and \(j\) are distances from the origin in \(x\) and \(y\) axes and \(X \in (i,j)\) respectively. The effects of de-noising method using Gaussian function are shown in Fig. 5.
After all, the size of de-noising image is normalized based on image intensity range. Size normalization is a process in which images are resized to some standard resolution which is 256 × 256 whereas images from the original database are of resolution 228 × 212. Hence, it is quite clear from the resolution that there is a minimal change in size. This process transforms the gray scale enhanced image \({I^G}\left( {i,j} \right)~\) with multiple dimensions having intensity ranges from \({I^G}\left( {i,j} \right):\left( {X \subseteq ~{{\mathbb{R}}^2}} \right) \to \left\{ {{{\text{{\rm I}}}_{Min}}~to~{{\text{{\rm I}}}_{Max}}} \right\}~\) to a new gray scale image with new intensities as \({I_{New}}\left( {i,j} \right):\left( {X \subseteq ~{{\mathbb{R}}^2}} \right) \to \left\{ {{{\text{{\rm I}}}_{MinN}}~to~{{\text{{\rm I}}}_{MaxN}}} \right\}\). The normalization process is performed linearly and defined as follows in Eq. (3).
Here \({{\text{{\rm I}}}_{Min}}~,{{\text{{\rm I}}}_{Max}}\) are the min and max intensities before normalization and \({{\text{{\rm I}}}_{MinNew}}~,~{{\text{{\rm I}}}_{MaxNew}}\) are the new min and max intensities after normalization. The implementation detail of preprocessing method is given in Algorithm 1. The resized image is passed to segmentation method for the detection of tumor region.

3.2 Tumor segmentation
In MR images, segmentation of tumor region is a difficult and stimulating task due to several factors such as irregularity, shape and similarity with a healthy region. Therefore, to resolve these factors, several methods are introduced in literature for the segmentation of tumor and lesion regions such as FCM based tumor segmentation (Bahadure et al. 2018), principal component analysis (PCA) based brain tumor segmentation from MR images (Kaya et al. 2017), uniform segmentation (Nasir et al. 2018), modified K-means clustering (Kumar and Mathai 2017) and few more (Yasmin et al. 2014). To follow these techniques, a new improved thresholding technique is implemented based on binomial mean, variance and standard deviation. The description of segmentation method is given below:
Let normalized image is \({I_{New}}\left( {i,~j} \right)\) with \(L\) gray levels \(\left( {0,1,2, \ldots ,~L - 1} \right)~\) and total number of pixels \(~N=\mathop \sum \limits_{{i=0}}^{{~L - 1}} {n_i}\), where \({n_i}\) are pixels at certain gray level. The probability of each level is computed as follows by Eq. (4).
The output of proposed improved thresholding method is in logical form containing two classes \({C_{j=1}} \in \{ 0,1,2, \ldots {t_1}\}\) and \({C_{j=2}} \in \{ {t_1}+1,~ \ldots L - 1\}\) with binomial mean value \(\mu ({t_1})\) and binomial variance \(~\sigma _{k}^{2}\left( {{t_1}} \right)\) respectively. Then the cumulative probability \({P_k}\left( {{t_1}} \right)~\) is calculated as below in Eq. (5) and Eq. (6).
where, \(\tilde {k}\) represents class \(~k \ne 1\). Binomial mean and variance for each class are calculated through Eq. (7) to Eq. (13).
Thereafter, these values are substituted into a threshold function to get final threshold values from Eq. (14) and Eq. (15).
where, \(\mu =\mathop \sum \limits_{{i=1}}^{N} \frac{{p({n_i})}}{N}~\) which denotes the mean value. Hence, the final threshold is selected as in Eq. (16) and Eq. (17).
where, \({\tilde {\phi }^T}\) is a threshold value, \(L\) are gray level pixels and \({I_B}(i,j)\) is binary image. Some morphological operations like clossing and filling are performed on binary image in the last step to optimize the segmentation results. The sample results for both data sets are shown in Figs. 6 and 7 which reflects that the proposed segmentation method performed good for MR images.
3.3 Feature extraction
In the area of machine learning, feature extraction techniques got much attention for several research domains such as human action recognition (Khan et al. 2018; Sharif et al. 2017), signature verification (Sharif et al. 2018), license plate recognition (Khan et al. 2017), medical imaging (Zhou et al. 2018) etc. The effective and more related features always produced best classification results. Therefore, in this step, efficient and robust features are much important because irrelevant and redundant features degrade the system classification accuracy. In the presented technique, nine geometric and four GLCM features are extracted for the classification of healthy and tumor images. The idea behind the geometric and Harlick feature extraction is to catch the information of tumor part in one vector to generate the best classification accuracy. After that, fused features are optimized by GA. GA gives best features based on their fitness function which is later provided to SVM for classification. The flow diagram of feature extraction, fusion and selection is shown in Fig. 8.
3.3.1 Geometric features
Geometric features, also known as shape features, are extracted in the proposed method from the whole data set. In this work, the purpose of geometric features extraction is to measure the shape information of tumor region. Therefore, nine geometric features are extracted (Sharif et al. 2018) such as area, major axis length, filled area, orientation, extent, perimeter, solidity, circularity and minor axis length. These features are computed from segmented tumor regions one by one for all images and combined in one matrix denoted by R. The matrix R includes the extracted geometric features and labels for all images. The computed matrix R is further utilized in the next step of features fusion. The major aim of these features extraction is to obtain the geometric information of tumor and fused in texture features for better classification accuracy. These features are calculated from a final binary image as follows:
Area of image is calculated by total number of pixels in the image using Eq. (18).
Filled Area denoted by ƑA,is calculated by the number of ‘ON’ pixels in the image after filling all the holes, given in Eq. (19). The total number of 1’s in the image corresponds to filled area; it will be 0 in case of healthy images. The value returned by this function is a subset of Area.
Orientation corresponds to angle θ between horizontal axis of the segmented image and major axis of an image. It gives a scalar value in return which is stored in feature vector. The perimeter is total number of boundary pixels in the binary image. Solidity of image can be calculated by dividing the area by its calculated convex area. Major axis length is the longest distance of two points from the foci of an ellipse. It is the distance between two farthest points on the longest diameter of ellipse. Minor axis length is the shortest distance between two points from the foci of an ellipse. It is the distance between two farthest points on the shortest diameter of ellipse. Circularity gives the scalar value and is computed from area divided by the square of perimeter and multiplied by some constant value. These features are calculated as follows from Eq. (20) to Eq. (25).
Here \(\upvarrho\) and \(\varvec{\varsigma}\) represent bounding box and convex hull area respectively, x and y are points on the ellipse and f is focal point of the ellipse, \(~\varvec{\iota}~{\varvec{a}}{\varvec{n}}{\varvec{d}}~\varvec{\omega}\) are the length and width of boundary of the segmented image. Thereafter, these features are stored in one vector R having length 1 × 9 and further fused with Harick texture features.
3.3.2 Texture features
Lately, in the area of computer vision, texture features gain much importance for texture analysis of an image. In the domain of medical imaging, texture features are utilized for the analysis of lesion or tumor because these features are reflected as a surface property of tumor whereas each tumor has its own statistical texture representation. Therefore, they differ from each other (Kalaivani and Chitrakala 2018). In this work, Harlick texture features are extracted for classification. As discussed above that these features lie in the category of texture features to get texture information of an image, therefore these are calculated from the gray scale images and give spatial information and intensity variation. Texture features calculation helps for better classification. To calculate the texture features, two approaches are being used that are structured and statistical. In the presented technique, second order statistical features are extracted.
The texture of an image \(I(i,j)\) is stored in a matrix called GLCM (Albregtsen 2008) represented as \({\xi _t}(s,t|{\delta _i},{\delta _j})\), here \(0 \leqslant s \leqslant {G_l} - 1\) and \(0 \leqslant t \leqslant {G_l} - 1\) and \({G_l}\) represent total number of gray levels in the image. The components of matrix \({\xi _t}\left( {s,t|{\delta _i},{\delta _j}} \right)~\) contain the value of frequency with two pixels having \({G_l}\), where x and y are separated by some distance \(({\delta _i},{{{\varvec{\updelta}}}_j})\). The \(~{\xi _t}(s,t)\) is calculated for one direction only which is θ = 0 and distance δ = 1 as shown in Fig. 9.
As mentioned above, four texture features are extracted such as contrast, correlation, uniformity and homogeneity. One of the primary reasons of selecting only four features is to select most robust information. Contrast is a property of an image that returns intensity contrast of a pixel and its adjacent pixel from the whole image and values are stored in a feature vector. Result of contrast is 0 if there is no variance in the image. Correlation is a property of an image that returns a value which expresses how a pixel in the image is inter related to its adjacent pixel, its value can be 1, − 1 or NaN. Uniformity is a property of an image that returns a value which is sum of square elements of GLCM; it can be 0 or 1. Homogeneity is a property of an image that calculates intimacy of elements’ distribution in texture feature vector, its value is 1 for diagonal GLCM and it is also known as inverse difference moment (IDM).
The features are calculated as follows from Eq. (26) to Eq. (29).
The range of contrast features is [0\(,{({\text{S}}(G(s,t),1) - 1)^2}\)], correlation is [− 1, 1] and other two features is [0, 1]. Thereafter, mean, standard deviation, skewness and kurtosis are computed for each extracted texture feature. GLCM analyzes the pairs of horizontally adjacent matrix. To calculate the skewness and kurtosis, initially GLCM features are calculated from a binary segmented image which analyzes the pairs of horizontally adjacent matrix of size 2 × 2 = 4 and final vector of size 1 × 16. Mathematically, kurtosis and skewness are defined as follows:
where, \(S\) denotes the skewness value, \(KR\) is kurtosis, \(E\) is expected mean value and \({x_i}\) denotes the adjacent scale matrix.
After that, geometric and texture features are fused by a serial based simple concatenation method. The fusion process denotes that two sets of features having distinct pattern information are combined in one vector to get more accurate results. The fusion of these features is computed as follows.
Let \({\zeta _g}\) be the geometric feature vector, \({\xi _t}\) be the texture feature vector and then the fusion vector \({\varphi ^f}\) is given as below in Eq. (30).
where, \(~i~\epsilon ~\left\{ {1, \ldots ,~9} \right\}\) and \(j~\epsilon ~\left\{ {1, \ldots ,~16} \right\}\). Hence the final vector can also be defined as in Eq. (31).
where, m = 1,2,3,..9 and n = 1,2,3,…16. The dimensions for the fused feature vector is 1 × 25 for each image. Afterwards, these fused features are fed to GA for feature optimization. Here an existing GA approach is utilized for feature selection (Sharif et al. 2018) having cost function mean squared error (MSE). The best selected features are fed to SVM for classification.
3.4 Classification
In this step, the selected features are classified by SVM. SVM is a supervised learning classifier mostly used for classification in the domain of machine learning. Several kernels functions are implemented for SVM. Few kernel functions such as linear kernel function, cubic kernel function, quadratic kernel function, polynomial kernel function and radial basis function (RBF) kernel function are utilized for classification. In the proposed method, least square SVM with linear kernel function is selected because of having binary classes.
For classification, first inputs and their corresponding labels \(\left( {{I_{T1}},{{\varvec{\Upsilon}}}} \right){\text{~are~given~to~it}}\), where \({I_{T1}}\) denotes the selected features of given images and \({{\varvec{\Upsilon}}}\) denotes assigned labels provided to the classifier. In the presented work, binary classification is performed for the classification of healthy and tumor images.
Let training data is given by \(\{ \left( {{{\varvec{I}}_{{\varvec{T}}1}},{{\mathbf{\Upsilon }}_1}} \right) \ldots ,~\left( {{{\varvec{I}}_{{\varvec{T}}{\varvec{n}}}},~{{\mathbf{\Upsilon }}_{\varvec{n}}}} \right)\}\) and feature set is defined by \({\varvec{\varphi}^{\varvec{f}}}=\left\{ {\varvec{\varphi}_{1}^{{\varvec{f}}} \ldots ,~\varvec{\varphi}_{{\varvec{n}}}^{{\varvec{f}}}} \right\},\) where \({\varvec{\varphi}^{\varvec{f}}} \in ~{{\mathbb{R}}^{\varvec{n}}}\)and \({\mathbf{\Upsilon }} \in [ - 1,1]~~\) to show healthy or unhealthy images. For this purpose, least squares SVM is used for classification with linear kernel function (Suykens and Vandewalle 1999). The detailed classification results are given in Results section.
4 Experimental results
In this section, proposed algorithm is evaluated on two data sets including Harvard medical Images (Hamamci et al. 2012) and Private dataset. The experiments are performed in two steps. In the first step, segmentation results are discussed in terms of tanimoto coefficient (TC), DOI (JaccardIndex), number of pixels and area. Then in the second step, classification results are calculated for which SVM is used as a leading classifier and its performance is compared with several other classification methods such as linear SVM (LSVM), quadratic SVM (QSVM), cubic SVM (CSVM), k nearest neighbor (KNN), decision tree (DT) and bagged tree (BT). The performance of these classification methods are calculated in terms of sensitivity, specificity, false positive rate (FPR), accuracy and false negative rate (FNR). All experiments are performed in MATLAB 2017b using a personal computer with 16 GB of RAM.
4.1 Tumor segmentation results
In this section, segmentation results are analyzed to check the authenticity of proposed improved thresholding method. Four parameters including TC, DOI (JaccardIndex), number of pixels and area are calculated to examine the performance. These parameters are calculated as follows from Eq. (33) to Eq. (35).
where, \(~\lambda\) denotes TC and \(\xi\) denotes DOI parameters. The value of TC is between 0 and 1, 0 ≤ TC ≤ 1. For segmentation results, 20 brain tumor images are taken from both selected datasets and their results are computed. The selected parameters are calculated by their ground truth images. The ground truth images of Harvard data set are designed by an expert doctor and the ground truth images of Private data set are already shown in Fig. 3. The segmentation results for Private data set are given in Table 1 having average TC as 0.926 and DOI as 0.918. Thereafter, same parameters are calculated for Harvard data set using ground truth images and achieved average TC value as 0.935 and DOI as 0.961 given in Table 2. The proposed segmentation method performed better on Harvard data set as compared to existing method (Vishnuvarthanan et al. 2016) in terms of TC and DOI values as presented in Table 3. Moreover, a comparison is conducted for Private data set with existing methods in terms of average TC and average DOI values as given in Table 4.
4.2 Classification results
The proposed classification results are validated on two publicly available data sets. The first data set is private which consists of total 85 images having 39 healthy and 46 unhealthy. A ratio of 50:50 is chosen for the validation of presented algorithm. All results are calculated using 10-fold cross validation (Desai et al. 2016). Initially, fusion of geometric and texture features results is obtained having maximum accuracy 98.13%, sensitivity 97.99%, specificity 97.09% and FPR as 0.023 on LSVM as given in Table 5. Then the classification results are calculated for feature selection algorithm in two different training/ testing ratios. In the first test, training/ testing ratio 40:60 is selected and achieved classification accuracy 98.29%, sensitivity 98.13%, specificity 98.19 and FPR as 0.045 on LSVM presented in Table 6. In the second test, selection of training/ testing ratio 50:50 is done to achieve classification accuracy 99.99%, sensitivity 99.99%, specificity 99.99% and FPR as 0.001 given in Table 7. The classification results from Table 7 show that the proposed algorithm performed efficiently for feature selection algorithm on ratio 50:50 for training and testing. Moreover, the accuracy results for separate feature extraction method are calculated to show the strength of proposed fused and selection approach. The classification accuracy of each feature extraction type is given in Table 8 which shows the presented system authenticity. Further, a graphical comparison is presented in Fig. 10 to show that the LSVM achieved best classification performance on selection based algorithm.
Next, proposed algorithm is tested on Harvard data set which consists of total 120 MR images including 41 healthy and 79 tumors. For classification results, a ratio of 50:50 for training and testing is selected. It means that 20 healthy and 40 tumor images are utilized for testing. Initially, fused feature vector is applied for classification results and achieved maximum accuracy 97.80%, sensitivity 96.92% and specificity 95.10% on LSVM as given in Table 9. Then classification accuracy is calculated for proposed feature selection method in two different steps. In the first step, 40:60 training/testing approach is used and achieved accuracy 98.90%, sensitivity 97.99% and specificity 98.67% as given in Table 10. Then in the second step, 50:50 ratio is chosen for training/testing and achieved maximum accuracy 99.69%, sensitivity 98.39% and specificity 99.99% as presented in Table 11. From Table 11, it is clear that LSVM performed significantly well for 50:50 approach and produced better results as compared to fused feature vector performance. Moreover, the classification accuracy for each extracted feature is computed as given in Table 12 which makes the proposed system more effective. A graphical comparison for classification performance of fused features and selected features is also given in Fig. 11 which proves the authenticity of proposed method. Finally, the classification accuracy of presented method is calculated on Harvard data set in Table 13 which shows its outperformance as compared to several other existing methods of same domain in literature.
4.3 Discussion
Detection and classification results in both graphical and numerical forms are discussed in this section. The proposed method consists of four major steps as shown in Fig. 2. In tumor segmentation step, an improved segmentation method is implemented which is tested on two data sets and gives efficient performance as shown in Figs. 6 and 7. Moreover, the segmentation qualitative results for both data sets are given in Tables 1 and 2. The comparison of proposed segmentation results is also given in Tables 3 and 4.
Thereafter, classification results are presented in four different steps. In the first step, classification is performed on fusion of feature vector and results are given in Tables 5 and 9. Then in the second step, best features selection method is performed and classification results are obtained on 40:50 training/ testing ratio as given in Tables 6 and 10. It is clear from these tables that the proposed selection method performed better as compared to fused features. In the third step, a 50:50 procedure is adopted and achieved better results as compared to the first two steps as given in Tables 7 and 11. In addition, negative predictive value (NPV) and positive predictive value (PPV) for each classifier are calculated given in Tables 5, 6, 7, 9, 10 and 11.
In order to justify that proposed segmentation algorithm performs well; the proposed method is compared with popular existing techniques such as Otsu, multi-thresholding, Watershed and k-means on selected data sets such as Harvard and Private (Fig. 12). To compare the performance of proposed method with existing techniques, recall rate, precision rate and F-Measure are calculated for Private data set as given in Tables 13 and 14 and Fig. 13 which show that the proposed method outperformed. Moreover, the comparison is also conducted on Harvard data set in terms of recall rate and precision rate. An additional parameter of F-measure is also considered which shows a fair comparison of some of the precision and recall. Achieved average rates using proposed method are 93.79%, 93.91% and 94.29%, respectively. Few numeric results are also plotted in Figs. 14, 15 and 16.
The performance of extracted features is also tested as given in Tables 8 and 12. In Table 8, geometric features achieved classification accuracy 96.13% whereas texture features reached up to 94.21%. But after fusion of geometric and texture features, the classification accuracy is improved and reached up to 98.13% using LSVM. In Table 12, the classification accuracy for geometric features is 97.2% whereas for texture features, best accuracy achieved is 91% on CSVM. After fusion, LSVM performed well and attained accuracy of 97.80%.
A graphical comparison of classification accuracy for both data sets is shown in Figs. 10 and 11. The proposed features selection performance for 10-fold cross validation of ratios 50:50, 40:60 and all fused features set are plotted in Figs. 16 and 17. Figure 16 shows that the classification accuracy range of Clinical dataset on selected classifiers is 84–98% for fused features, 96–98% for proposed method of 40:60 approach and 98–99.90% for 50:50 approach. Similarly in Fig. 18, the classification accuracy range is given for Harvard dataset. Both Figs. 17 and 18 show that the proposed method significantly outperformed on ratio 50:50 and achieved best accuracy. Lastly, classification results of the proposed method for Harvard data set are compared in Table 15. Yang et al. (Yang et al. 2015) achieved classification accuracy 97.78% on Harvard data set which is later improved by Zhang et al. (Zhang et al. 2015). Moreover, Wang et al. (Wang et al. 2016) achieved 98.67% accuracy but presented method performed well and reached up to 99.69%.
Lately, HOG and LBP features are utilized for brain tumor classification. The LBP features also known as texture features are originally introduced for face recognition and HOG features known as shape based features are used for human detection. However, recently several researchers extract LBP and shape features for brain tumor classification such as (Lenz et al. 2018), (Nabizadeh and Kubat 2015) and (Abbasi and Tajeripour 2017). These methods show improved performance in terms of Jaccard Index, sensitivity, specificity and Dice on Brats 2013, real and simulated data sets.
5 Conclusion
Medical imaging applications gained much attention from last few years in the area of computer vision. Brain tumor diagnosis and classification is an important step in this domain and recently through computer vision techniques, it may be helpful for doctors in their clinics. In this paper, an improved thresholding and best feature extraction approach for tumor segmentation and classification from MR images is proposed. Preprocessing problem is resolved in the first step by the implementation of a Gaussian filter. Then tumor part is segmented by a novel improved thresholding method which is further optimized by morphological operations. Thereafter, geometric and Harlick features are extracted which are fused by a serial based method. After that, GA is used to select best features from a fused vector which are later on fed to LSVM for classification. The proposed segmentation method is tested on two data sets such as Private images and Harvard Medical School dataset having accuracy 91.8% and 96.1%. Moreover, best classification results are achieved by features selection algorithm with accuracy 99.99% and 99.69% for Private and Harvard datasets, respectively. Experimental results show that segmentation and classification accuracy of proposed technique is significantly well as compared to several existing methods. From the above discussion, it is concluded that accurate tumor segmentation is much important to obtain the best accuracy. Also, the fusion of texture and geometric features along with best features selection algorithm provided good results as compared to individual features. The proposed system has few limitations which will be dealt in future work. The limitations of this system are oversegmentation, classification on large datasets, and selection of best features. The oversegmentation is done when a tumor appears on the border regions whereas, the classification accuracy is affected by irrelevant features.
References
Abbasi S, Tajeripour F (2017) Detection of brain tumor in 3D MRI images using local binary patterns and histogram orientation gradient. Neurocomputing 219:526–535
Acunzo DJ, MacKenzie G, van Rossum MC (2012) Systematic biases in early ERP and ERF components as a result of high-pass filtering. Journal of neuroscience methods 209(1):212–218
Ajaj K, Syed NA (2015) Image processing techniques for automatic detection of tumor in human brain using SVM. Int J Adv Res Comput Commun Eng 4(4):147–171
Albregtsen F (2008) Statistical texture measures computed from gray level coocurrence matrices. Image Processing Laboratory, Department of Informatics, University of Oslo, version 5
Amin J, Sharif M, Yasmin M, Fernandes SL (2018) Big data analysis for brain tumor detection: deep convolutional neural networks. Future Gener Comput Syst 87:290–297
Ariyo O, Zhi-guang Q, Tian L (2017) Brain MR segmentation using a fusion of K-means and spatial fuzzy C-means. In: 2017 International conference on computer science and application engineering (CSAE 2017), pp 863–873
Ashour AS, Beagum S, Dey N, Ashour AS, Pistolla DS, Nguyen GN, Shi F (2018a) Light microscopy image de-noising using optimized LPA-ICI filter. Neural Comput Appl 29(12):1517–1533
Ashour DS, Rayia DMA, Dey N, Ashour AS, Hawas AR, Alotaibi MB (2018b) Schistosomal hepatic fibrosis classification. IJNCR 7(2):1–17
Avina-Cervantes JG, Lindner D, Guerrero-Turrubiates J, Chalopin C (2016) Automatic brain tumor tissue detection based on hierarchical centroid shape descriptor in Tl-weighted MR images. In: Paper presented at the 2016 international conference on electronics, communications and computers (CONIELECOMP)
Bahadure NB, Ray AK, Thethi HP (2018) Comparative approach of MRI-based brain tumor segmentation and classification using genetic algorithm. J Digit Imaging, 1–13
Bauer S, Nolte L-P, Reyes M (2011) Fully automatic segmentation of brain tumor images using support vector machine classification in combination with hierarchical conditional random field regularization. In: Paper presented at the international conference on medical image computing and computer-assisted intervention
Benson CC, Lajish VL, Rajamani K (2016) A novel skull stripping and enhancement algorithm for the improved brain tumor segmentation using mathematical morphology. Int J Image Graph Signal Process 8:59–66
Calabrese C, Poppleton H, Kocak M, Hogg TL, Fuller C, Hamner B,.. . Allen M (2007) A perivascular niche for brain tumor stem cells. Cancer cell 11(1):69–82
Chaddad A (2015) Automated feature extraction in brain tumor by magnetic resonance imaging using gaussian mixture models. Int J Biomed Imaging 2015:868031. https://doi.org/10.1155/2015/868031
Chaddad A, Zinn PO, Colen RR (2014) Brain tumor identification using Gaussian Mixture model features and decision trees classifier. In: Paper presented at the information sciences and systems (CISS), 2014 48th annual conference on
Chua AS, Egorova S, Anderson MC, Polgar-Turcsanyi M, Chitnis T, Weiner HL, Healy BC (2015) Using multiple imputation to efficiently correct cerebral MRI whole brain lesion and atrophy data in patients with multiple sclerosis. NeuroImage 119:81–88
Damodharan S, Raghavan D (2015) Combining tissue segmentation and neural network for brain tumor detection. IAJIT. 12:1
Deng G, Cahill L (1993) An adaptive Gaussian filter for noise reduction and edge detection. In: Paper presented at the nuclear science symposium and medical imaging conference, 1993, 1993 IEEE conference record
Desai U, Martis RJ, Nayak G, Seshikala C, Sarika G, Shetty K (2016) Decision support system for arrhythmia beats using ECG signals with DCT, DWT and EMD methods: a comparative study. J Mech Med Biol 16(01):1640012
Doshi J, Erus G, Ou Y, Gaonkar B, Davatzikos C (2013) Multi-atlas skull-stripping. Acad Radiol 20(12):1566–1576
El Abbadi NK, Kadhim NE (2017) Brain cancer classification based on features and artificial neural network. Brain 6:1
Gao Y, Mas JF, Kerle N, Navarrete Pacheco JA (2011) Optimal region growing segmentation and its effect on classification accuracy. Int J Remote Sens 32(13):3747–3763
Hamamci A, Kucuk N, Karaman K, Engin K, Unal G (2012) Tumor-cut: segmentation of brain tumors on contrast enhanced MR images for radiosurgery applications. IEEE Trans Med Imaging 31(3):790–804
Havaei M, Davy A, Warde-Farley D, Biard A, Courville A, Bengio Y, Larochelle H (2017) Brain tumor segmentation with deep neural networks. Med Image Anal 35:18–31
Hore S, Chakroborty S, Ashour AS, Dey N, Ashour AS, Sifaki-Pistolla D, Chaudhuri S (2015) Finding contours of hippocampus brain cell using microscopic image analysis. J Adv Microsc Res 10(2):93–103
Irum I, Shahid M, Sharif M, Raza M (2015) A review of image denoising methods. J Eng Sci Technol Rev. 8:5
Kadam DB, Gade SS et al (2012) Neural network based brain tumor detection using MR images. Int J Comput Sci Commun 2(2):325–331
Kalaivani A, Chitrakala S (2018) An optimal multi-level backward feature subset selection for object recognition. IETE J Res 2:1–13
Kalavathi P, Prasath VS (2016) Methods on skull stripping of MRI head scan images—a review. J Digit Imaging 29(3):365–379
Kaya IE, Pehlivanlı A, Sekizkardeş EG, Ibrikci T (2017) PCA based clustering for brain tumor segmentation of T1w MRI images. Comput Methods Programs Biomed 140:19–28
Khan MA, Sharif M, Javed MY, Akram T, Yasmin M, Saba T (2017) License number plate recognition system using entropy-based features selection approach with SVM. IET Image Process. https://doi.org/10.1049/iet-ipr.2017.0368
Khan MA, Akram T, Sharif M, Javed MY, Muhammad N, Yasmin M (2018) An implementation of optimized framework for action classification using multilayers neural network on selected fused features. Pat Anal Appl 6:1–21
Kumar R, Mathai KJ (2017) Brain Tumor segmentation by modified K-mean with morphological operations. Brain 6:8
Lenz M, Krug R, Dillmann C, Stroop R, Gerhardt NC, Welp H, Hofmann MR (2018) Automated differentiation between meningioma and healthy brain tissue based on optical coherence tomography ex vivo images using texture features. J Biomed Opt 23(7):071205
Liu J, Li M, Wang J, Wu F, Liu T, Pan Y (2014) A survey of MRI-based brain tumor segmentation methods. Tsinghua Sci Technol 19(6):578–595
Masood S, Sharif M, Yasmin M, Raza M, Mohsin S (2013) Brain image compression: a brief survey. Res J Appl Sci 5(1):49–59
Menon N, Ramakrishnan R (2015) Brain tumor segmentation in MRI images using unsupervised artificial bee colony algorithm and FCM clustering. In: Paper presented at the Communications and Signal Processing (ICCSP), 2015 International Conference on
Moraru L, Moldovanu S, Dimitrievici LT, Ashour AS, Dey N (2016) Texture anisotropy technique in brain degenerative diseases. Neural Computing and Appl 5:1–11
Moraru L, Moldovanu S, Dimitrievici LT, Shi F, Ashour AS, Dey N (2017) Quantitative diffusion tensor magnetic resonance imaging signal characteristics in the human brain: a hemispheres analysis. IEEE Sens J 17(15):4886–4893
Mosquera JM, Sboner A, Zhang L, Chen CL, Sung YS, Chen HW, Singer S (2013) Novel MIR143-NOTCH fusions in benign and malignant glomus tumors. Genes Chromosom Cancer 52(11):1075–1087
Nabizadeh N, Kubat M (2015) Brain tumors detection and segmentation in MR images: Gabor wavelet vs. statistical features. Comput Electr Eng 45:286–301
Nasir M, Attique Khan M, Sharif M, Lali IU, Saba T, Iqbal T (2018) An improved strategy for skin lesion detection and classification using uniform segmentation and feature selection based approach. Microsc Res Tech
Nazir M, Wahid F, Ali Khan S (2015) A simple and intelligent approach for brain MRI classification. J Intell Fuzzy Syst 28(3):1127–1135
Ohgaki H, Kleihues P (2013) The definition of primary and secondary glioblastoma. Clin Cancer Res 19(4):764–772
Patil S, Udupi V (2012) Preprocessing to be considered for MR and CT images containing tumors. IOSR J Electrical Electron Eng 1(4):54–57
Pereira S, Pinto A, Alves V, Silva CA (2016) Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging 35(5):1240–1251
Popuri K, Cobzas D, Murtha A, Jägersand M (2012) 3D variational brain tumor segmentation using Dirichlet priors on a clustered feature set. Int J Comput Assis Radiol Surg 7(4):493–506
Rajinikanth V, Satapathy SC, Dey N, Vijayarajan R (2018) DWT-PCA image fusion technique to improve segmentation accuracy in brain tumor analysis microelectronics. In: Anguera J, Satapathy S, Bhateja V, Sunitha K (eds) Microelectronics, electromagnetics and telecommunications. Lecture notes in electrical engineering, vol 471. Springer, Singapore
Rani J, Kumar R, Talukdar FA, Dey N (2017) The brain tumor segmentation using fuzzy c-means technique: a study. In: Recent advances in applied thermal imaging for industrial applications. IGI Global, pp 40–61
Raza M, Sharif M, Yasmin M, Masood S, Mohsin S (2012) Brain image representation and rendering: a survey. Res J Appl Sci 21:4
Samanta S, Ahmed SS, Salem MA-MM, Nath SS, Dey N, Chowdhury SS (2015) Haralick features based automated glaucoma classification using back propagation neural network. In: Paper presented at the proceedings of the 3rd international conference on frontiers of intelligent computing: theory and applications (FICTA) 2014
Senthilkumaran N, Thimmiaraja J (2014) Histogram equalization for image enhancement using MRI brain images. Paper presented at the Computing and Communication Technologies (WCCCT), 2014 World Congress on
Sharif M, Khan MA, Akram T, Javed MY, Saba T, Rehman A (2017) A framework of human detection and action recognition based on uniform segmentation and combination of Euclidean distance and joint entropy-based features selection. EURASIP J Image Video Proces 2017(1):89
Sharif M, Khan MA, Faisal M, Yasmin M, Fernandes SL (2018) A framework for offline signature verification system: Best features selection approach. Pattern Recogn Lett
Sharma M, Purohit G, Mukherjee S (2018) Information retrieves from brain MRI images for tumor detection using hybrid technique K-means and artificial neural network (KMANN) networking communication and data knowledge engineering. Springer, Heidelberg pp. 145–157
Soltaninejad M, Yang G, Lambrou T, Allinson N, Jones TL, Barrick TR, Ye X (2018) Supervised learning based multimodal MRI brain tumour segmentation using texture features from supervoxels. Comput Method Program Biomed
Suykens JA, Vandewalle J (1999) Least squares support vector machine classifiers. Neural Proces Lett 9(3):293–300
Tian Z, Dey N, Ashour AS, McCauley P, Shi F (2017) Morphological segmenting and neighborhood pixel-based locality preserving projection on brain fMRI dataset for semantic feature extraction: an affective computing study. Neural Comput Appl 5:1–16
Vishnuvarthanan G, Rajasekaran MP, Subbaraj P, Vishnuvarthanan A (2016) An unsupervised learning method with a clustering approach for tumor identification and tissue segmentation in magnetic resonance brain images. Appl Soft Comput 38:190–212
Wang S, Du S, Atangana A, Liu A, Lu Z (2016) Application of stationary wavelet entropy in pathological brain detection. Multimedia Tools Appl 2:1–14
Wang Y, Shi F, Cao L, Dey N, Wu Q, Ashour AS,.. . Wu L (2018) Morphological segmentation analysis and texture-based support vector machines classification on mice liver fibrosis microscopic images. Curr Bioinform
Wu W, Chen AY, Zhao L, Corso JJ (2014) Brain tumor detection and segmentation in a CRF (conditional random fields) framework with pixel-pairwise affinity and superpixel-level features. Int J Comput Assis Radiol Surg 9(2):241–253
Yang G, Zhang Y, Yang J, Ji G, Dong Z, Wang S, Wang Q (2015) Automated classification of brain images using wavelet-energy and biogeography-based optimization. Multimed Tools Appl 1:1–17
Yasmin M, Sharif M, Mohsin S, Azam F (2014) Pathological brain image segmentation and classification: a survey. Curr Med Imaging Rev 10(3):163–177
Zhang YD, Chen S, Wang SH, Yang JF, Phillips P (2015) Magnetic resonance brain image classification based on weighted-type fractional Fourier transform and nonparallel support vector machine. Int J Imaging Syst Technol 25(4):317–327
Zhou M, Scott J, Chaudhury B, Hall L, Goldgof D, Yeom K,.. . Napel S (2018) Radiomics in brain tumor: image assessment, quantitative feature descriptors, and machine-learning approaches. Am J Neuroradiol 39(2):208–216
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sharif, M., Tanvir, U., Munir, E.U. et al. Brain tumor segmentation and classification by improved binomial thresholding and multi-features selection. J Ambient Intell Human Comput 15, 1063–1082 (2024). https://doi.org/10.1007/s12652-018-1075-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12652-018-1075-x