Abstract
Alpha-synuclein (SNCA) oligomers have been reported to inhibit autophagy. Aminochrome-induced SNCA oligomers are neurotoxic, but the flavoenzyme DT-diaphorase prevents both their formation and their neurotoxicity. However, the possible protective role of DT-diaphorase against autophagy impairment by aminochrome-induced SNCA oligomers remains unclear. To test this idea, we used the cell line RCSN-3NQ7SNCA, with constitutive expression of a siRNA against DT-diaphorase and overexpression SNCA, and RCSN-3 as control cells. A significant increase in LC3-II expression was observed in RCSN-3 cells treated with 20 μM aminochrome and 10 μM rapamycin followed by a decrease in cell death compared to RCSN-3 cells incubated with 20 μM aminochrome alone. The incubation of RCSN-3NQ7SNCA cells with 20 μM aminochrome and 10 μM rapamycin does not change the expression of LC3-II in comparison with RCSN-3NQ7SNCA cells incubated with 20 μM aminochrome alone. The incubation of both cell lines preincubated with 100 nM bafilomycin and 20 μM aminochrome increases the level of LC3-II. Under the same conditions, cell death increases in both cell lines in comparison with cells incubated with 20 μM aminochrome. These results support the protective role of DT-diaphorase against SNCA oligomers-induced autophagy inhibition.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
α-Synuclein (SNCA) belongs to the synuclein family that includes β and ƴ synuclein. All members of this family are predominantly neuronal proteins that physiological conditions are preferentially located in the presynaptic terminal (George 2002). These proteins have N-terminal sequence with different repeats, although they differ at their C-terminus (Bellucci et al. 2012). The sequence of SNCA protein is constituted by N-terminal rich in lysine and plays a crucial role in the modulation and interaction with membranes. On the other hand, it has a disordered C-terminal that has been implicated in the regulation of its nuclear localization and interaction with metals, small molecules, and proteins (Ulmer and Bax 2005; Masliah et al. 2005). This sequence is divided into three major regions: N-terminal region (residues 1–60) contains 4 imperfect repeats of 11 amino acids with a highly conserved hexamer motif (KTKEGV), which has been involved in the formation of amphipatic α–helices, essential for membrane binding (Clayton and George 1998). This portion includes sites for SNCA variants related to familial PE: A30P, A53T, and E46K (Bellucci et al. 2012). The central region (residues 61–95) constitutes the non-β-amyloid component (NAC), an indispensable region for aggregation; the deionization of long segments of this region decreases the oligomerization of SNCA and fibrillogenesis in vitro (El-Agnaf et al. 1998, Giasson et al. 2001). The C-terminal region (residues 96–140 amino acids) contains tyrosine residues which, upon being mutated, abolish the fibrillation capacity of SNCA. This part mediates the interaction with oxidized catechols as aminochrome. Oxidative modifications on ASYN by dopamine metabolites have been considered to be responsible for the selective vulnerability of dopaminergic neurons (Xu et al. 1998; Conway et al. 2001). The function of SNCA is not completely clear. It is related to synaptic function and vesicle trafficking.
Mutations in SNCA gene have been associated to a familial form of Parkinson’s disease such as alpha-synuclein mutant (Petrucci et al. 2016). SNCA mutations induce its aggregation to cytotoxic oligomers. SNCA aggregates to a fibrillary beta-pleated sheet structure in Lewy bodies and related SNCApathies (Spillantini et al. 1998; Domert et al. 2016), where SNCA is the major constituent of Lewy bodies (Emamzadeh 2016). Evidence from studies in vitro and in vivo has shown that a prerequisite of SNCA pathology is its oligomerization into cytotoxic oligomers (Conway et al. 2001; Volles and And Lansbury 2003; Ingelsson 2016). The intracellular formation of SNCA oligomers is induced by dopamine oxidation products, such as dopaminochrome, aminochrome, or 5,6-indolequinone (Norris et al. 2005; Bisaglia et al. 2010). Dopamine oxidation to neuromelanin is a normal pathway because neuromelanin pigment can be observed in the substantia nigra of healthy seniors (Segura-Aguilar et al. 2014, 2016; Herrera et al. 2017). Aminochrome, the most stable o-quinone formed during dopamine oxidation, can be neurotoxic by forming adducts with proteins such as SNCA (Muñoz et al. 2015) or by one-electron reduction by flavoenzymes which transfer one electron (Arriagada et al. 2004). However, the flavoenzymes DT-diaphorase (EC.1.6.99.2) prevents aminochrome neurotoxicity by catalyzing the two-electron reduction of aminochrome to leukoaminochrome (Arriagada et al. 2004; Lozano et al. 2010; Paris et al. 2011, 2010; Muñoz et al. 2012; Zafar et al. 2006). Recently, we reported that DT-diaphorase prevents the formation of aminochrome-induced oligomers and its neurotoxicity (Muñoz et al. 2015). However, it remains unclear whether DT-diaphorase prevents autophagy dysfunction as a consequence of aminochrome-induced SNCA oligomer formation. To test this hypothesis, we used the RCSN-3 wild-type cell line and RCSN-3NQ7SNCA (expressing both a siRNA against DT-diaphorase and pLv21ASYN plasmid encoding a SNCA to increase its expression in this cell line).
Materials and Methods
Chemicals
Dopamine, CM-Sephadex C50-100, tyrosinase (EC 1.14.18.1), and dopamine from mushroom were from Sigma-Aldrich (St. Louis, MO, USA (TH (A-1)): sc-374047).
Synthesis and Purification of Aminochrome
Dopamine (7.5 mmol) and 15 ng of tyrosinase were incubated in 1.5 ml of 25 mM phosphate buffer pH 6.0, for 10 min at 25 °C. To purify the aminochrome formed, the incubation solution was loaded on a column 17–0.7 cm resin CM-Sephadex C-50-120 which was eluted first with 30 ml of 25 mM phosphate buffer pH 6.0. The eluate from the column was recovered in 500 ml aliquots. Each aliquot was monitored spectrophotometrically by absorbance at 280, 478, and 600 nm wavelengths. The recovered pure aminochrome (5–7 ml) was eluted with 25 mM of phosphate buffer pH 6.0; dopamine was recovered only after 5 ml of 25 mM 4-Morpholineethanesulfonic acid sodium salt (MES) pH 6.0 (Paris et al. 2010).
Cell Culture
The RCSN-3 cell line was grown in monolayers, with a doubling time of 52 h, a plating efficiency of 21% and a saturation density of 56,000 cells/cm2 in normal growth media composed of DME/HAM-F12 (1:1), 10% bovine serum and 2.5% fetal bovine serum. The cultures were kept in an incubator at 37 °C with 100% humidity, and the cells grew well in atmospheres of both 5 and 10% CO2 (Lozano et al. 2010; Paris et al. 2008). RCSN-3 cells are derived from the substantia nigra of an adult rat. The cell line does not require differentiation to express catecholaminergic traits such as tyrosine hydroxylase, dopamine transport, VMAT-2, norepinephrine transport, dopamine release, monoamine oxidase (MAO)-A expression (but not MAO-B); the formation of neuromelanin and DT-diaphorase is responsible for 94% of the total quinone reductase activity catalyzed by flavoenzymes (Paris et al. 2008).
Transfection of RCSN-3 and RCSN-3Nq7 Cells
RCSN-3 and RCSN-3NQ7 cells were transduced with lentiviral plasmid pLvGFP-SNCA constructed in our laboratory in order to overexpress SNCA. Lentiviral particles were obtained by transfecting HEK-293 T cells with a mixture of 6 mg DNA of pLvGFP-SNCA coding for GFP-IRES-SNCA, 6 mg DNA of packing plasmid pMDG and FUGENE HD (Roche) in a relationship (8:2 DNA:FUGENE HD) in 300-ml cell culture medium without serum. After 1 h, serum was added to the cells and was incubated for 24, 48, and 72 h after which the supernatant was collected and filtered with a 0.45-mm filter. As a control, pRetrosuper without DNA insert and pMDG were transfected using FUGENE HD as explained above. Transduced cells were incubated in the presence of 40 mg/l gentamicin sulfate to select the cells that have the plasmid with a gene resistant to gentamicin sulfate.
Western Blot
The samples of LC3I/II were separated by SDS-PAGE (12% wt/vol.). The separated proteins were then transferred electrophoretically to a 0.2-mm nitrocellulose membrane. After blocking with a solution of 0.5% skim milk in 10 mM Tris-HCl pH 7.6, 150 mM NaCl, and 0.025% Tween 20 for at least 4 h, the membrane was incubated with anti-MAP1LC3B N(20) goat sc-16755 antibody (Santa Cruz Biotechnology), diluted 1:500 overnight at room temperature in the same buffer. The membrane was washed three times for 15 min with a solution of 10 mM Tris-HCl pH 7.6, 150 mM NaCl, and 0.025% Tween 20 and incubated for 2 h with an anti-goat alkaline phosphatase-linked antibody. After a final washing, LC3I/II was detected using BCIP/NBT (Zymed laboratories Inc.)
Cell Death
Cell death was determined using a LIVE/DEAD Viability/Cytotoxicity kit (Molecular Probes, L-3224) after incubating the cells in a culture medium for 24 h in the presence of aminochrome, rapamycin, or bafilomycin A1, purified as previously described (Paris et al. 2010).
Statistical Analysis
The data were expressed as the mean ± SD values, and statistical significance was assessed using analysis of variance (ANOVA) for multiple comparisons and Student’s t test.
Results
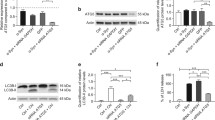
We determined the LC3I/II protein with Western blotting to follow autophagy in RCSN-3 and RCSN-3NQ7SNCA cells treated with 20 μM aminochrome. A significant increase in LC3-II in RCSN-3 cells occurred in the 20 μM of aminochrome and 10 μM rapamycin condition, compared with cells treated with 20 μM aminochrome alone (bars 3 and 5—Fig. 1b, p < 0.05) and also with RCSN-3NQO7SNCA in the same condition (bars 4 and 6—Fig. 1b, p < 0.05). The preincubation of RCSN-3 and RCSN-3NQ7SNCA cells with bafilomycin A1 for 2 h prior to the addition of 20 μM of aminochrome induces a significant LC3-II increase in both cell lines in comparison with cells treated with 20 μM aminochrome alone (bars 3 and 7 and 4 and 8—Fig. 1b, p < 0.05). However, no significant difference in LC3-II expression was observed between RCSN-3 and RCSN-3NQ7SNCA cells treated with 20 μM of aminochrome and 100 nM bafilomycin A1 (bars 7 and 8—Fig. 1b).
The effect of aminochrome on LC3-II expression in RCSN-3NQ7SNCA overexpressing SNCA. Determination of LC3-II expression through Western blot analysis. a Total of 30 μg of RCSN-3 WT and RCSN-3NQ7SNCA untreated cell homogenate (control) was applied in lanes 1 and 8, respectively. The same amount of protein from of RCSN-3 WT and RCSN-3NQ7SNCA cell homogenate treated with 20 μM aminochrome was applied to lanes 2 and 6; cell homogenate treated with 20 μM aminochrome + 10 μM rapamycin was applied to lanes 3 and 7; cell homogenate treated with 20 μM aminochrome + 10 nM Bafilomycin A1 was applied to lanes 4 and 8. b The values represent the means ± SD (n = 3). The statistical significance was assessed using analysis of variance (ANOVA) for multiple comparison and Student’s t test (*p < 0.01)
We incubated the cells with 10 μM rapamycin for 24 h to determine whether an increase in autophagy protects against aminochrome-induced cell death. The incubation of RCSN-3 cells treated with 10 μM rapamycin and 20 μM aminochrome induces a small but not significant decrease in cell death compared to cells incubated with 20 mM aminochrome alone (bars 3 and 5—Fig. 2). No significant effect was observed when RCSN-3NQ7SNCA cells were incubated with 10 μM rapamycin in the presence of 20 μM aminochrome, in comparison with RCSN-3NQ7SNCA cells incubated with 20 mM aminochrome alone (bars 4 and 6—Fig. 2). In addition, rapamycin does not have a protective effect with respect to aminochrome-induced cell death, because of the significant increase in cell death for RCSN-3NQ7SNCA with 10 μM rapamycin and 20 μM aminochrome compared to RCSN-3 cells (bars 5 and 6—Fig. 2, p < 0.001). The preincubation of RCSN-3NQ7SNCA and RCSN-3 cells with 100 nM bafilomycin A1 for 2 h prior to the addition of 20 μM aminochrome induced a significant increase in aminochrome-induced cell death in comparison with these cells treated with 20 μM aminochrome alone (bars 3 and 7 and 4 and 8—Fig. 2, *p < 0.05; **p < 0.005; ***p < 0.001).
The role of autophagy in aminochrome-induced cell death in RCSN-3NQ7SNCA overexpressing SNCA. The role of autophagy in aminochrome-induced cell death was determined. The RCSN-3 and RCSN-3NQ7SNCA cells were incubated with cell culture medium (Co); 20 μM aminochrome (AM 20); 10 nM bafilomycin A1 pretreatment (BF) with 20 μM aminochrome (AM 20 BF) and 20 μM of aminochrome and rapamycin (AM 20 RAP). The role of increased autophagy in aminochrome-induced cell death was determined by incubating the cells with the autophagy-inducer rapamycin 10 μM or by preincubating the cells for 2 h with 10 nM bafilomycin A1 (BF) prior to the addition of 20 μM aminochrome (AM) for 24 h. The values are the means ± SD (n = 3). The statistical significance was assessed using analysis of variance (ANOVA) for multiple comparisons and Student’s t test (*p < 0.05; ***p < 0.0001)
Discussion
The formation of SNCA oligomers seems to be essential for SNCA neurotoxic effects (Gruden et al. 2014; Lashuel et al. 2002; Volles and And Lansbury 2003). In the familial form of Parkinson’s disease, the mutations induce the formation of neurotoxic oligomers but the question is what induces SNCA oligomers in the sporadic form of the disease (Ottolini et al. 2017). Interestingly, o-quinones generated during dopamine oxidation to neuromelanin have been reported to induce the formation of SNCA oligomers. Dopamine oxidation to neuromelanin is a sequential event where several o-quinones are formed (dopamine ➔ dopamine o-quinone ➔ aminochrome ➔ 5,6-indolequinone ➔ neuromelanin). Dopaminochrome induces the formation of SNCA oligomers (Norris et al. 2005), but the structure of this o-quinone generated during dopamine oxidation to neuromelanin remains unclear (Herrera et al. 2017). The compound 5,6-indolequinone also forms adducts with SNCA (Bisaglia et al. 2007). Aminochrome, the most stable and the most studied o-quinone, induces the formation of neurotoxic oligomers (Muñoz et al. 2015). Dopamine o-quinone is very unstable and does not form adducts with SNCA (Van Laar et al. 2009). It has been reported that the majority of SNCA oligomers formed during dopamine oxidation have noncovalent interactions between SNCA and these o-quinones (Bisaglia et al. 2010). Interestingly, DT-diaphorase prevents both the formation of aminochrome-induced SNCA oligomers and their neurotoxicity (Muñoz et al. 2015).
One important factor in the aggregation process of SNCA is its protein levels, which depend on the balance between the synthesis, degradation, and secretion of the protein. The control of the intracellular SNCA level is mediated by two mechanisms, with pathways mediated by proteasomes and lysosomes, respectively (Cuervo 2008; Liu et al. 2003), and might be involved in the degradation of SNCA according to the cell stress level (Abeliovich et al. 2000; Ebrahimi-Fakhari et al. 2011). Consistent evidence points to a role for mainly macroautophagy and chaperone-mediated autophagy (CMA), in the regulation of SNCA levels. Indeed, SNCA has been found in lysosomal compartments, and rapamycin, a well-known macroautophagy-enhancing agent, promotes its lysosomal-mediated degradation. Rapamycin binds to FKBP1A/FKBP12 and inhibits mechanistic target of rapamycin (serine/threonine kinase)-1 (MTORC1); the complex binds to the FRB domain of MTOR and limits its interaction with RPTOR, thus inducing autophagy, but only providing partial MTORC1 inhibition (Klionsky et al. 2016). Our experiments show no difference in the expression of the LC3-II marker of autophagy between RCSN-3 cells with normal expression of SNCA and RCSN-3NQ7SNCA cells with overexpression of SNCA, both in the presence or absence of aminochrome. Interestingly, the incubation of RCSN-3 cells with both aminochrome and rapamycin induces a significant increase in LC3-II expression contrasting with no effect observed in RCSN-3NQ7SNCA cells. A possible explanation for the lack of effect of rapamycin in RCSN-3NQ7SNCA cells may be the distribution of oligomers of SNCA. It has been reported that SNCA oligomer distribution both in the cytosol and the nucleus and the presence of SNCA in the nucleus can disturb LC3-II expression (Emmanouilidou et al. 2010; Mak et al. 2010; Nakamura et al. 2008). A correlation between the increase in LC3-II expression and decrease in cell death was observed in RCSN-3 cells treated with aminochrome and rapamycin. Similarly, a correlation between the lack of effect on LC3-II expression and no change in cell death in RCSN-3NQ7SNCA cells was observed when the cells were treated with aminochrome and rapamycin. The increase in LC3-II expression and cell death observed both in RCSN-3 and RCSN-3NQ7SNCA cells treated with aminochrome and bafilomycin A1 can be explained by the intracellular accumulation of autophagosomes due to autophagy inhibition. In the RCSN-3 cell, the increase in expression is explained by the fact that bafilomicyn A1 is V-ATPase inhibitor that causes an increase in lysosomal/vacuolar pH, and, ultimately, blocks fusion of autophagosomes with the vacuole; the latter may result from inhibition of ATP2A/SERCA with this autophagic flux is often inferred on the basis of LC3-II turnover (Klionsky et al. 2016). In the RCSN-3NQ7SNCA cells, the same condition could be explained by the absence of DT-diaphorase where aminochrome inhibits the autophagy (Huenchuguala et al. 2014; Muñoz et al. 2012). In addition, overexpression of SNCA induces CMA dysfunction (Xilouri et al. 2013) and defects in lysosomal acidification (Crews et al. 2010; Cuervo et al. 2004; Yu et al. 2009). In conclusion, our results support the protective role of DT-diaphorase against aminochrome-induced neurotoxicity. Specifically, DT-diaphorase prevents autophagy inhibition by preventing the formation of aminochrome-induced SNCA oligomers.
References
Abeliovich A, Schmitz Y, Farinas I, Choi-Lundberg D, Ho W, Castillo P, Shinsky N, Verdugo J, Armanini M, Ryan A et al (2000) Mice lacking α-synuclein display functional deficits in the nigrostriatal dopamine system. Neuron 25:239–252
Arriagada C, Paris I, Sanchez de las Matas M, Martinez-Alvarado P, Cardenas S, Castaneda P, Graumann R, Perez-Pastene C, Olea-Azar C, Couve E (2004) On the neurotoxicity mechanism of leukoaminochrome o-semiquinone radical derived from dopamine oxidation: mitochondria damage, necrosis, and hydroxyl radical formation. Neurobiol Dis 16:468–477
Bellucci A, Zaltieri M, Navarria L, Grigoletto J, Missale C, Spano P (2012) From α-synuclein to synaptic dysfunctions: new insights into the pathophysiology of Parkinson’s disease. Brain Res 1476:183–202
Bisaglia M, Mammi S, Bubacco L (2007) Kinetic and structural analysis of the early oxidation products of dopamine: analysis of the interactions with alpha-synuclein. J Biol Chem 282(21):15597–15605
Bisaglia M, Tosatto L, Munari F, Tessari I, de Laureto PP, Mammi S, Bubacco L (2010) Dopamine quinones interact with alpha-synuclein to form unstructured adducts. Biochem Biophys Res Commun 394(2):424–428
Clayton DF, George JM (1998) The synucleins: a family of proteins involved in synaptic function, plasticity, neurodegeneration and disease. Trends Neurosci 21(6):249–254
Conway K, Rochet J, Bieganski R, Lansbury P (2001) Kinetic stabilization of the alpha-synuclein protofibril by a dopamine-alpha-synuclein adduct. Science 294:1346–1349
Crews L, Spencer B, Desplats P, Patrick C, Paulino A, Rockenstein E, Hansen L, Adame A, Galasko D, Masliah E (2010) Selective molecular alterations in the autophagy pathway in patients with Lewy body disease and in models of α-synucleinopathy. PLoS One 5(e):9313
Cuervo AM (2008) Autophagy and aging: keeping that old broom working. Trends Genet 24(12):604–12. doi:10.1016/j.tig.2008.10.002
Cuervo A, Stefanis L, Fredenburg R, Lansbury P, Sulzer D (2004) Impaired degradation of mutant alpha-synuclein by chaperonemediated autophagy. Science 305:1292–1295
Domert J, Sackmann C, Severinsson E, Agholme L, Bergström J, Ingelsson M, Hallbeck M (2016) Aggregated alpha-synuclein transfer efficiently between cultured human neuron-like cells and localize to lysosomes. PLoS One 11(12):e0168700. doi:10.1371/journal.pone.0168700
Ebrahimi-Fakhari D, Cantuti-Castelvetri I, Fan Z, Rockenstein E, Masliah E, Hyman B, McLean P, Unni V (2011) Distinct roles in vivo for the ubiquitin-proteasome system and the autophagy-lysosomal pathway in the degradation of α-synuclein. J Neurosci 31:14508–14520
El-Agnaf OM, Jakes R, Curran MD, Middleton D, Ingenito R, Bianchi E, Pessi A, Neill D, Wallace A (1998) Aggregates from mutant and wild-type alpha-synuclein proteins and NAC peptide induce apoptotic cell death in human neuroblastoma cells by formation of beta-sheet and amyloid-like filaments. FEBS Lett 440(1–2):71–75
Emamzadeh FN (2016) Alpha-synuclein structure, functions, and interactions. J Res Med Sci 21:29
Emmanouilidou E, Stefanis L, Vekrellis K (2010) Cell-produced α-synuclein oligomers are targeted to, and impair, the 26S proteasome. Neurobiol Aging 31:953–968
George JM (2002) The synucleins. Genome Biol 3(1):REVIEWS3002
Giasson BI, Murray IV, Trojanowski JQ, Lee VM (2001) A hydrophobic stretch of 12 amino acid residues in the middle of alpha-synuclein is essential for filament assembly. J Biol Chem 276(4):2380–2386
Gruden MA, Davydova TV, Narkevich VB, Fomina VG, Wang C, Kudrin VS, Morozova-Roche LA, Sewell RD (2014) Intranasal administration of alpha-synuclein aggregates: a Parkinson’s disease model with behavioural and neurochemical correlates. Behav Brain Res 263:158–168
Herrera A, Muñoz P, Steinbusch HW, Segura-Aguilar J (2017) Are dopamine oxidation metabolites involved in the loss of dopaminergic neurons in the nigrostriatal system in Parkinson’s disease? ACS Chem Neurosci. doi:10.1021/ acschemneuro.7b00034
Huenchuguala S, Muñoz P, Zavala P, Villa M, Cuevas C, Ahumada U, Graumann R, Nore B, Couve E, Mannervik B et al (2014) Glutathione transferase mu 2 protects glioblastoma cells against aminochrome toxicity by preventing autophagy and lysosome dysfunction. Autophagy 10:618–630
Ingelsson M (2016) Alpha-synuclein oligomers-neurotoxic molecules in Parkinson’s disease and other Lewy body disorders. Front Neurosci 10:408
Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ (2016) Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12:1–222
Lashuel HA, Hartley D, Petre BM, Walz T, Lansbury PT Jr (2002) Neurodegenerative disease: amyloid pores from pathogenic mutations. Nature 418, 291
Liu C, Corboy M, DeMartino G, Thomas P (2003) Endoproteolytic activity of the proteasome. Science 299:408–411
Lozano J, MuÑoz P, Nore B, Ledoux S, Segura-Aguilar J (2010) Stable expression of short interfering RNA for DTdiaphorase induces neurotoxicity. Chem Res Toxicol 23:1492–1496
Mak S, McCormack A, Manning-Bog A, Cuervo A, Di Monte D (2010) Lysosomal degradation of α-synuclein in vivo. J Biol Chem 285:13621–13629
Masliah E, Rockenstein E, Adame A, Alford M, Crews L, Hashimoto M, Seubert P, Lee M, Goldstein J, Chilcote T, Games D, Schenk D (2005) Effects of alpha-synuclein immunization in a mouse model of Parkinson’s disease. Neuron 46:857–868
Muñoz P, Paris I, Sanders LH, Greenamyre JT, Segura-Aguilar J (2012) Overexpression of VMAT-2 and DT-diaphorase protects substantia nigra-derived cells against aminochrome neurotoxicity. Biochim Biophys Acta 1822:1125–1136
Muñoz P, Cardenas S, Huenchuguala S, Briceño A, Couve E, Paris I, Segura-Aguilar J (2015) DT-diaphorase prevents aminochrome-induced alpha-synuclein oligomer formation and neurotoxicity. Toxicol Sci 145(1):37–47
Nakamura K, Nemani V, Wallender E, Kaehlcke K, Ott M, Edwards R (2008) Optical reporters for the conformation of α-synuclein reveal a specific interaction with mitochondria. J Neurosci 28:12305–12317
Norris E, Giasson B, Hodara R, Xu S, Trojanowski J, Ischiropoulos H, Lee V (2005) Reversible inhibition of alpha-synuclein fibrillization by dopaminochrome-mediated conformational alterations. J Biol Chem 280:21212–21219
Ottolini D, Calí T, Szabò I, Brini M (2017) Alpha-synuclein at the intracellular and the extracellular side: functional and dysfunctional implications. Biol Chem 398(1):77–100
Paris I, Lozano J, Cardenas S, Perez-Pastene C, Saud K, Fuentes P, Caviedes P, Dagnino-Subiabre A, RaismanVozari R, Shimahara T (2008) The catecholaminergic RCSN-3 cell line: a model to study dopamine metabolism. Neurotox Res 13:221–230
Paris I, Perez-Pastene C, Cardenas S, Iturriaga-Vasquez P, Muñoz P, Couve E, Caviedes P, Segura-Aguilar J (2010) Aminochrome induces disruption of actin, alpha-, and beta-tubulin cytoskeleton networks in substantia-nigra-derived cell line. Neurotox Res 18:82–92
Paris I, Muñoz P, Huenchuguala S, Couve E, Sanders LH, Greenamyre JT, Caviedes P, Segura-Aguilar J (2011) Autophagy protects against aminochrome-induced cell death in substantia nigra-derived cell line. Toxicol Sci 121:376–388
Petrucci S, Ginevrino M, Valente EM (2016) Phenotypic spectrum of alpha-synuclein mutations: new insights from patients and cellular models. Parkinsonism Relat Disord 22(Suppl 1):S16–S20
Segura-Aguilar J, Paris I, Muñoz P, Ferrari E, Zecca L, Zucca F (2014) Protective and toxic roles of dopamine in Parkinson’s disease. J Neurochem 129:898–915
Segura-Aguilar J, Muñoz P, Paris I (2016) Aminochrome as new preclinical model to find new pharmacological treatment that stop the development of Parkinson’s disease. Curr Med Chem 23:346–359
Spillantini M, Crowther R, Jakes R, Hasegawa M, Goedert M (1998) Alpha-synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with Lewy bodies. Proc Natl Acad Sci U S A 95:6469–6473
Ulmer TS, Bax A (2005) Comparison of structure and dynamics of micelle-bound human alpha-synuclein and Parkinson disease variants. J Biol Chem 280(52):43179–43187
Van Laar V, Mishizen A, Cascio M, Hastings T (2009) Proteomic identification of dopamine-conjugated proteins from isolated rat brain mitochondria and SH-SY5Y cells. Neurobiol Dis 34:487–500
Volles M, And Lansbury P (2003) Zeroing in on the pathogenic form of alpha-synuclein and its mechanism of neurotoxicity in Parkinson’s disease. Biochemistry 42:7871–7878
Xilouri M, Brekk O, Stefanis L (2013) Alpha-synuclein and protein degradation systems: a reciprocal relationship. Mol Neurobiol 47:537–551
Xu Y, Stokes A, Roskoski R, Vrana K (1998) Dopamine in the presence of tyrosinase covalently modifies and inactivates tyrosinase hydroxylase. J Neurosci Res 54:691–697
Yu W, Dorado B, Figueroa H, Wang L, Planel E, Cookson M, Clark L, Duff K (2009) Metabolic activity determines efficacy of macroautophagic clearance of pathological oligomeric α-synuclein. Am J Pathol 175:736–747
Zafar KS, Siegel D, Ross D (2006) A potential role for cyclized quinones derived from dopamine, DOPA, and 3,4-dihydroxyphenylacetic acid in proteasomal inhibition. Mol Pharmacol 70:1079–1086
Acknowledgements
This study was supported by FONDECYT # 1100165. RCSN-3 cells are made available by P. Caviedes upon request (pcaviede@med.uchile.cl).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Muñoz, P.S., Segura-Aguilar, J. DT-diaphorase Protects Against Autophagy Induced by Aminochrome-Dependent Alpha-Synuclein Oligomers. Neurotox Res 32, 362–367 (2017). https://doi.org/10.1007/s12640-017-9747-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12640-017-9747-4