Abstract
This study aims to screen for potential probiotic lactic acid bacteria from the intestines of meat-type pigeon squabs. Ligilactobacillus salivarius YZU37 was identified as the best comprehensive performed strain. Being acid- and bile salt-tolerant, it displayed growth-inhibition activities against Staphylococcus aureus ATCC25923, Escherichia coli ATCC25922, and Salmonella typhimurium SL1344, exhibited sensitivity to 6 commonly used antibiotics, and endowed with good cell surface hydrophobicity, auto-aggregation property, and anti-oxidant activities. Results of in vitro experiments indicated that the bacteriostatic effects of this strain were related to the production of proteinaceous substances that depend on acidic conditions. Whole-genome sequencing of L. salivarius YZU37 was performed to elucidate the genetic basis underlying its probiotic potential. Pangenome analysis of L. salivarius YZU37 and other 212 L. salivarius strains available on NCBI database revealed a pigeon-unique gene coding choloylglycine hydrolase (CGH), which had higher enzyme-substrate binding affinity than that of the common CGH shared by L. salivarius strains of other sources. Annotation of the functional genes in the genome of L. salivarius YZU37 revealed genes involved in responses to acid, bile salt, heat, cold, heavy metal, and oxidative stresses. The whole genome analysis also revealed the absence of virulence and toxin genes and the presence of 65 genes distributed under 4 CAZymes classes, 2 CRISPR-cas regions, and 3 enterolysin A clusters which may confer the acid-dependent antimicrobial potential of L. salivarius YZU37. Altogether, our results highlighted the probiotic potential of L. salivarius YZU37. Further in vivo investigations are required to elucidate its beneficial effects on pigeons.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Probiotics are defined as living microorganisms that offer health benefits to the host when consumed in an adequate quantity [1]. Among the probiotic microorganisms, lactic acid bacteria (LAB) are the most widely used probiotic bacteria. Their beneficial effects can be summarized as normalization of the intestinal microflora, function as alternatives or supplements of antibiotics, regulation of the immune system, inactivation of toxic xenobiotics, and production of nutrients essential for hosts [2]. Due to their favorable effects on the health and performance of hosts, probiotics have been widely used in the poultry industry.
In the past two decades, the restricted prophylactic use of antibiotics in food-producing animals prompted extensive research on substitutes for antibiotics. In broiler chickens, probiotic supplements have demonstrated effective control of microbial infections caused by Salmonella enteritidis [3], Campylobacter jejuni [4], and Eimeria tenella [5]. Additionally, improved feed efficiency (including increased body weight, feed nutritional value, and feed conversion rate), enhanced bone characteristics, improved intestinal morphology, and promoted immune responses were observed in broilers supplemented with probiotic LAB or spore-forming Bacillus bacteria [6, 7]. Probiotic supplementation was also demonstrated to be efficacious in alleviating the adverse effects of heat in broilers exposed to heat stress [8], increasing productive performance and nutrient digestibility in low-protein-fed broilers [9], and improving carcass characteristics in broilers co-administrated with cowpea seeds [10]. Furthermore, improved growth performance and animal behavioral welfare in broiler chickens could be achieved by the combination of probiotics supplementation and a digital poultry system [11].
Accumulated research results obtained in hens at different stages of their growth and development also evidenced various favorable effects associated with in-feed supplementation of probiotics. For example, in ovo and in-feed probiotic supplementation promoted layer embryo and pullet growth [12]. Implementation of probiotics in laying hens improved laying performance (including increased egg production and egg quality) and slowed down the reproductive aging of hens [13]. Significantly increased feed efficiency, improved eggshell quality, and optimized lipid metabolism were documented in the late laying period [14]. In addition, administration of probiotics to laying hens helped reduce the occurrence of injurious behavior, improve animal welfare [15], reduce the cecal load of pathogenic bacteria (Clostridium perfringens, Salmonella spp., and Escherichia coli) [16], and mitigate harmful excretion of nitrogen and phosphorus from the manure [17, 18]. Therefore, the use of probiotics in laying hens has been proved to enhance production performance, improve animal welfare, and promote the sustainable and green development of the industry.
Similarly, beneficial effects of dietary probiotics supplementation on ducks, geese, and quails were reported in terms of enhancing egg quality, increasing performance and reducing mortality, improving meat quality and cecal health, and alleviating pathogen-induced intestinal flora dysbiosis, besides stress amelioration, pathogen control, and immune responses enhancement (recent research results are summarized in supplemental Table S1). Based on the numerous benefits of probiotics for poultry, research on supplemental probiotic products has become one of the fastest growing ventures in poultry industry.
In China, the meat-type pigeon industry is now the fourth largest poultry industry after chicken, duck, and goose industries. Squabs (meat-type pigeon around 4-week old), valued for their high nutritional value and delicate taste of the meat, are the main output of the pigeon industry. As one of the fastest-growing segments of the poultry sector in China, the meat-type pigeon industry produces around 680 million squabs each year, accounting for about 80% of the global production [19]. Given the recorded safe use and recognized beneficial effects of probiotics on chickens, ducks and geese, it can be predicted that probiotics will also play important positive roles in the production of squabs.
However, despite the rapid development of the meat-type pigeon industry and the increasing consumption demand of squabs in the market, few studies have been conducted in meat-type pigeons with the aim to exploit the beneficial effects of probiotics on promoting squab production. As early as 2019, dietary administration of probiotic Enterococcus faecium and Lactobacillus acidophilus pellets was reported to induce temporary shifts in the fecal microbiome composition in adult domesticated Birmingham Roller pigeons [20]. In 2020, research in White King meat-type pigeons revealed that dietary supplementation with Bifidobacterium alone or in combination with mannan oligosaccharides increased body weights and improved immune functions [21]. In 2021, probiotic supplementation of Bacillus velezensis demonstrated effective enhancement of immune responses in circovirus-challenged pigeons [22]. In 2022, a synbiotic product containing chitosan oligosaccharide and Clostridium butyricum showed positive effects of alleviating early-weaned stress and maintaining intestinal health in White King pigeon squabs [23]. Recent studies in 2024 reported that supplementation of drinking water with Enterococcus faecium and Bacillus subtilis improved immunoglobulin levels in pigeon milk [24]. While the lack of intestinal Lactobacillus could be associated with diarrhea in pigeons, the application of Lactobacillus salivarius SNK-6 in health sand proved effective on the treatment and prevention of diarrhea in White Carneau pigeons [25]. Altogether, probiotics-related research in meat-type pigeons is lagging far behind that performed in chickens, ducks, and geese.
Thus, the purpose of this study was to isolate and characterize endogenous LAB within the intestines of meat-type pigeon squabs, with a view to screening promising probiotic strains that had the potential to be applied in the preparation of probiotic supplements targeted specifically for application in meat-type pigeon industry.
Materials and Methods
Sampling
Six healthy (28-day old) meat-type pigeon squabs (the White King breed) used in this study were obtained from Jiangsu Cuigu Pigeon Industry Co. Ltd. (Nanjing, China). The details of pigeon feeding management in this company have been described in a previous article [21]. Briefly, paired pigeons were housed in an aviary provided with nests and perches. Parent pigeons were fed with a free-choice feeding system and received a basal diet mainly consisted of corn, pea, and wheat (the detailed composition and nutrient level of the basal diet was listed in supplemental Table S2). Water, feed, and health sand were supplied ad libitum. Each pair of parent pigeons reared two squabs for nearly 28 days from hatching. Birds were slaughtered in facilities operated by Yangzhou University (permit no. 202,202,169) by cervical dislocation. The intestinal contents of each bird were squeezed aseptically into a 10-mL sterile tube. The attachments were rinsed with 0.5 mL sterile saline and collected in the same tube. Samples from 6 birds were pooled together, homogenized, and used for subsequent isolation of LAB.
Isolation of LAB
The homogenized sample was allowed to stand at 4 °C for 30 min. The supernatants were 10-fold sequentially (from 10−1 to 10−6) diluted with sterile saline. From each diluted sample, an aliquot of 100 µL was plated onto the de Man, Rogosa, and Sharpe (MRS) agar plates (90 mm in diameter, solidified with 1.5% agar) containing 1% CaCO3 and incubated at 37 °C for 24–72 h. Colonies with transparent haloes and different morphologies were selected, and each of them was re-streaked 4 times on MRS agar plates to obtain a pure and single colony.
The preliminarily screened bacterial strains were further morphologically identified via Gram staining. Gram-positive strains were then subjected to the catalase test using 3% H2O2 [26]. The appearance of oxygen bubbles within half a minute indicated the production of catalase, and the test strain was defined as catalase-positive. In all, 28 LAB strains, which were gram-positive and catalase-negative, were selected for subsequent assays. Stock cultures were conserved in MRS broth containing 50% glycerol and stored at −80 °C.
Molecular Identification of Isolated LAB Strains
Genomic DNA was extracted from the isolated LAB strains using the Bacterial Genomic DNA Extraction Kit (G-Clone Biotechnology Co. Ltd., Beijing, China) according to the manufacturer’s protocol. The 16S rDNA sequence was amplified by using the universal primers of 27F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5’-GGTTACCTTGTTACGACTT-3’). PCR products were sent to Sangon Biotech Co., Ltd. (Shanghai, China) and sequenced from both directions. The obtained sequences were compared with known sequences in the rRNA database using the BLAST tool at the NCBI website (http://www.ncbi.nlm.nih.gov/BLAST).
In vitro Tests of LAB Isolates as Potential Probiotics
Acid and Bile Salt Tolerance
Firstly, stocked bacterial isolates were rejuvenated by streaking onto MRS agar plates and incubated at 37 °C for 24 h. For each LAB isolate, a single colony was transferred to a tube containing 1 mL MRS broth. After an overnight incubation at 37 °C, 1 mL bacterial cultures (adjusted to 109 CFU/mL using MRS broth) were centrifuged at 8000 rpm for 5 min. The obtained pellets were washed with phosphate-buffer saline (PBS, 0.01 M, pH7.2) and re-suspended in 10 mL MRS broth which had been acidified to pH 2.5 with 1 N hydrochloric acid or supplemented with 0.3% bile salt (w/v) (Oxgall, B3883, Sigma). After being thoroughly mixed, bacterial cultures were incubated at 37 °C. An aliquot of 100 µL bacterial culture was collected at 0 h, 2 h, and 4 h post-incubation and spotted onto MRS agar plates (3 replicates for each bacterial isolate) after a 10−4 dilution was performed using the MRS broth. The plates were then inverted and incubated at 37 °C for 24 h to monitor the change in total viable count. The tolerance of isolated bacterial cells to acid and bile salt was evaluated by enumerating the viable bacteria colonies grown on MRS agar plates. The survival rate of each isolated bacteria was expressed as the percentage of CFU compared to the initial bacterial viable count [27].
Antagonistic Activities of Isolated LAB Against Pathogenic Bacteria
LAB isolates were investigated regarding their antimicrobial activities against three pathogenic bacteria (Gram-positive Staphylococcus aureus ATCC25923, Gram-negative Escherichia coli ATCC25922 and Salmonella typhimurium SL1344) which were obtained from Jiangsu Key Laboratory of Zoonosis (Yangzhou, China). The LAB isolates were streaked on MRS agar plates (90 mm in diameter) and incubated at 37˚C overnight. A suspension of individual pathogenic bacterium, which was grown at 37 °C for 18 h in LB (Luria-Bertani) broth, was prepared and adjusted to 0.5 McFarland standard (1.5 × 108 CFU/mL). An aliquot of 100 µL suspensions of pathogenic bacteria were inoculated into 8 mL of molten MRS broth containing 0.7% agar, which was overlaid on top of the streaked MRS plates. After solidification, plates were incubated at 37˚C overnight. The diameters of the inhibition zones on the MRS plates were measured. The antimicrobial activity of the LAB strains against three pathogenic bacteria was classified according to the diameter of inhibition zone (D) as follows: D < 10 mm, none (−); 10 ≤ D < 15 mm, weak (+); 15 ≤ D < 20 mm, middle (++); 20 ≤ D < 25 mm, strong (+++); D > 25 mm, very strong (++++) inhibition [28]. The experiment was performed in triplicate.
In vitro Antibiotic Susceptibility Tests
Acid-bile-resistant LAB isolates that displayed growth-inhibitory effects on tested pathogenic bacteria were assessed for their antibiotic susceptibility profiles against 10 commonly used antimicrobials by Kirby-Bauer standard disc diffusion method. Briefly, after an overnight incubation in MRS broth, LAB isolates were pelleted by centrifugation at 8000 rpm for 5 min, washed thrice with PBS, re-suspended in PBS, and adjusted to 0.5 McFarland standard (1.5 × 108 CFU/mL). The obtained bacterial cultures (a volume of 100 µL) were then swabbed evenly as a lawn onto the surface of MRS agar plates (90 mm in diameter), which were allowed to dry at room temperature. The commercial standard antibiotic paper discs (6 mm in diameter, Hangzhou Microbiological Reagents Co., Ltd., Hangzhou, China) were applied onto the solidified agar surface using a disc dispenser. After being left aside for 30 min at 4 °C for the diffusion of antibiotics, the plates were incubated at 37 °C for 24 h and the diameters of resulting inhibition halos were measured. The assay was performed in triplicates. Antibiotics used in this assay belonged to different categories in order to maximize the identification of resistance genotypes, which included the inhibitors of cell wall synthesis: amoxicillin (20 µg), penicillin G (10 U), cefoperazone (75 µg), and vancomycin (30 µg) and the inhibitors of protein synthesis: clindamycin (2 µg), erythromycin (15 µg), kanamycin (30 µg), gentamicin (10 µg), streptomycin (10 µg), and chloramphenicol (30 µg). Results were categorized as susceptibility, moderate susceptibility, or resistance according to the interpretative standards provided by the manufacture which were based on the guidelines of the Clinical and Laboratory Standards Institute M100-S25 (CLSI 2017).
Tests for Cell-Surface Hydrophobicity, Auto-Aggregation, and Antioxidant Activities
LAB isolates and three pathogenic bacteria (Staphylococcus aureus ATCC25923, E. coli ATCC 25,922, and Salmonella typhimurium SL1344) grown in MRS and LB broth, respectively, at 37 °C for 18–20 h were centrifuged at 8000 rpm for 5 min. The obtained precipitate was washed thrice with PBS and re-suspended in PBS. The absorbance of the bacterial suspensions at 600 nm (OD600) was adjusted to 0.6 ± 0.02 in order to standardize the concentration of bacteria (around 108 CFU/mL).
The cell surface hydrophobicity of the LAB isolates was determined by measuring the affinity of bacterial cells for xylene [29]. Equal volumes (2 mL) of LAB suspensions and xylene were vortexed for 30 s and left undisturbed at 37 °C for 1 h to allow phase separation. The lower aqueous phase was carefully collected, and its absorbance at 600 nm was determined (A1). The decrease in the absorbance at 600 nm due to the partitioning of bacterial cells into the xylene phase was taken as a measure of cell surface hydrophobicity. Hydrophobicity (H) was calculated from three replicates according to the following equation: H% = 100 × [1 - A1/A0], where A0 and A1 denoted the OD600 of the bacterial suspension before and after extraction with xylene, respectively. Auto-aggregation assay was performed according to the previously described method [30]. The LAB suspensions (4 mL) were allowed to stand at 37 °C for 5 h. Then, a volume of 1 mL was taken from the upper layer of the bacterial suspension, and its absorbance at 600 nm was determined. The percentage of auto-aggregation (calculated from 3 replicates) was determined according to the following equation: Auto-aggregation (%) = 100 × [1- (At/A0)], where At represented the OD600 of bacterial suspensions after 5-h incubation and A0 was the initial absorbance at 600 nm. Intact cell (IC) and intracellular cell free extraction (CFE) of LAB isolates were prepared according to previous descriptions [31]. Anti-oxidative activities in respects of DPPH and ABTS radical scavenging activities were assayed using the reported method [32].
Analyses of Antibacterial Substances
Evaluation of antibacterial substances was performed in accordance with previous descriptions [31] with slight modifications. Briefly, the LAB bacterial culture broth was adjusted to an OD600 of 0.6, inoculated into MRS liquid medium (2% v/v), incubated at 37 °C for 24 h, thoroughly mixed, and centrifuged at 8000 × g for 10 min at 4 °C. The collected supernatant was sterilized using a 0.22 μm filter to obtain cell-free supernatant (CFS). The pH of the CFS was adjusted to the optimal pH for catalase (7.0), pepsin (2.0), and trypsin (8.0), respectively. After the addition of the protease into the CFS (at the final concentration of 1 mg/mL), the mixture was incubated at 37 °C for 2 h followed by an adjustment of its pH to the initial value (pH 6.5). The Oxford cup agar diffusion method was adopted to determine the growth-inhibition effect of the treated CFS on S. typhimurium SL1344. Firstly, the S. typhimurium SL1344 bacterial culture was washed with PBS twice, adjusted to the concentration of 1 × 108 CFU/mL, added to the semi-solid LB agar at a ratio of 1:100 (v/v), and spread it on a Petri dish to make a plate, on which sterile Oxford cups were gently placed at equal distances. Secondly, 100 µL of the treated CFS was transferred into the cups which were allowed to stand for 2 h and then incubated at 37 °C for 12 h. The diameter of the bacteriostatic zone was measured. The untreated CFS was taken as a control to determine the effects of various pretreatments on the antibacterial activities of the CFS.
Whole Genome Sequencing, Assembly, and Annotation
Genomic DNA was isolated from the 4 L. salivarius strains (designated as YZU37, YZU38, YZU40, and YZU41) using the Bacterial Genomic DNA Extraction Kit (G-Clone Biotechnology Co. Ltd., Beijing, China) according to the manufacturer’s protocol. The obtained DNA extracts were quantified and sequenced using Illumina Novaseq 6000 platform at Tianjin Novogene Bioinformatic Technology Co., Ltd. (Tianjin, China) to generate paired-end 150-bp reads. Clean data were generated after removing adapter sequences and low-quality reads using the Trimmomatic tool v0.38.1 (http://www.usadellab.org/cms/?page=trimmomatic). The quality of the sequences was assessed using the Quast v5.2.0 tool. Clean reads generated in this study have been submitted to NCBI SRA database (accession number SUB13999939). De novo assembly was performed with SPAdes under default parameters by excluding contigs shorter than 100 bp. The draft genomic DNA sequences were annotated using the software tool Prokka [33]. Circular genomic maps were constructed from the resultant genome using the Proksee server (https://proksee.ca) [34].
The species of L. salivarius strains was confirmed by calculating the genome similarity indices, ANI, using the JSpecies server [35] followed by a further phylogenomic analysis using genome-genome comparisons in the TYGS server (https://tygs.dsmz.de). Functional annotation of the L. salivarius YZU37 strain was carried out using RAST-SEED server (Rapid Annotation Subsystem Technology) (https://rast.nmmpdr.org). Analysis of the clusters of orthologous groups (COG) functional groups was performed using EggNOG mapper v2.1.12 (http://eggnog-mapper.embl.de).
Pangenome Comparison and In Silico Molecular Docking Analyses
A comparative pangenome analysis was performed using Roary v3.12.0 [36]. Data used for genome assembly included the whole genome sequence of 212 strains downloaded from the NCBI database and that of the 4 L. salivarius strains (YZU37, YZU 38, YZU 40, and YZU 41) isolated in the current study.
For molecular docking analyses, three-dimensional (3D) conformation of choloylglycine hydrolase (CGH) was predicted using the homology modeling network synthesis server (https://swissmodel.expasy.org/) with gene A0A0F7Q0L6_9LACO from L. salivarius str Ren as the template. The optimization of the CGH structure was performed using AutoDock Tools (v1.5.7), including adding polar hydrogen, calculating the distribution of atomic charges by computing the Gasteiger, and assigning the AD4 type to atoms on CGH. The 3D structures of the 6 substrates of CGH, including glycocholic acid (GCA), glycodeoxycholic acid (GDCA), glycochenodeoxycholic acid (GCDCA), taurocholic acid (TCA), taurodeoxycholic acid (TDCA), and taurochenodeoxycholic acid (TCDCA), were downloaded from the website of Chemical Entities of Biological Interest (ChEBI)(https://www.ebi.ac.uk/chebi/) in the format of mol2, which were then converted to the .pdb format using OpenBabel (v2.4.1). The active site of CGH (of L. salivarius origin) involved in the enzyme-substrate binding was documented by Rani et al. [37]. The docking simulation, docking flexible ligands (substrates) to rigid receptor protein (CGH), was performed by using AutoDock Vina. Results of the substrate-protein interactions with the strongest binding affinities were visualized using PyMOL (v2.5.5).
Identification of CRISPR-Cas System, Antimicrobial Resistance, and Bacteriocin-Encoding Genes
The CRISPR Finder tool was used to detect CRISPR direct repeats and spacers [38]. The Comprehensive Antibiotic Resistance Database (CARD) (https://card.mcmaster.ca/analyze/blast) online server was used to search for antimicrobial resistance genes. The BAGEL4 program (http://bagel4.molgenrug.nl) was used to search for genes associated with bacteriocins.
Results and Discussion
In this study, we isolated 28 gram-positive and catalase-negative LAB from the intestines of 6 meat-type pigeon squabs. All these LAB isolates could hydrolyze CaCO3 and formed transparent zones around the colonies on MRS agar plates. They were firstly identified at the species level using the 16 S rRNA gene PCR-sequencing method. Then, their probiotic features regarding their tolerance to low pH and bile salt, as well as their antagonistic abilities against three pathogenic bacteria, were evaluated. LAB isolates, which demonstrated acid- and bile salt-resistance and growth-inhibition effects on tested pathogenic bacteria, were investigated for their susceptibility profiles to 10 commonly used antibiotics. Based on our results, L. salivarius YZU37 strain was the most promising probiotic candidate. We further characterized its cell surface hydrophobicity, auto-aggregation properties, and antioxidant activities. Its antibacterial substances were also analyzed. Its whole genome was sequenced, and in silico investigations about its metabolic and functional feature were performed to explore its fundamental properties for potential probiotics.
Identification of LAB Isolates by 16 S rRNA Sequencing
Results of sequence comparisons by searching the GenBank 16 S rRNA sequences database revealed that the 28 LAB isolates exhibited a similarity higher than 98% to 3 different LAB species, which included 23 strains of Enterococcus faecium (NR_114742.1, Enterococcus faecium DSM 20,477), 4 strains (designated as YZU37, YZU38, YZU40, YZU41) of L. salivarius (NR_112759.1, L. salivarius JCM 1231), and one strain (YZU39) Lactobacillus johnsonii (NR_117574.1, Lactobacillus johnsonii CIP 103,620).
Tolerance to Acid and Bile Salt
Survival in the acidic and bile salt-containing environment is one of the essential prerequisites for potential probiotic bacteria to proliferate and perform beneficial functions in the hosts’ intestinal tracts.
In the in vitro simulation studies, the unchanged pH (2–3) is a widely used drastic condition to simulate the acidic environment of the stomach [39]. It was generally accepted that isolates capable of tolerating pH 3 for 3 h showed comparably high resistance to acidity and could be considered as acid-resistant strains with promising probiotic properties [40]. Our current results showed that the degrees of acid tolerance varied among the tested strains. After being exposed to low pH (pH 2.5) for 2 h and 4 h, the 28 isolated LAB strains presented an average survival rate of 80.1% and 72.7%, respectively. A total of 17 and 12 strains grew well after 2-h and 4-h incubation, respectively, in acidified MRS broth (pH 2.5) with a survival rate over 80%.
For the bile tolerance assay, most researchers used 0.3% (0.1−0.5%) as a critical concentration for the selection of resistant strains [41]. Our results showed that the majority of the isolated LAB strains exhibited high level of bile tolerance, which was consistent with their intestinal origin. After a 2-h and 4-h exposure to 0.3% bile salt, the 28 isolated LAB strains presented an average survival rate of 96.8% and 90.1%, respectively. In total, 58.6% and 41.4% LAB isolates retained a survival rate above 80% after being incubated in MRS broth containing 0.3% bile salt for 2 h and 4 h, respectively.
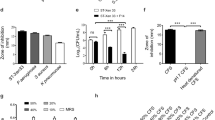
Comparing the impacts of acid and bile salt on the growth of LAB isolates (supplemental File S1), 11 isolated strains (designated as YZU12, 14, 15, 18, 19, 22, 23, 25, 26, 37, and 38) conferred good resistance to both low pH and bile salt (Fig. 1). These acid- and bile-tolerant strains, when being ingested, could be expected to survive and proliferate in the hosts’ intestinal tracts.
Antimicrobial Activity
Inhibitory activity against the growth of pathogenic bacteria is another desirable feature of probiotic strains. This property is closely associated with their probiotic performance of preventing the colonization and infection by pathogenic organisms. In this study, the antibacterial activities of 28 LAB isolates against 3 tested pathogenic bacteria showed disparities (supplemental File S2). Strains YZU37, YZU38, and YZU41 displayed good antibacterial activity against 3 tested pathogenic bacteria (Fig. 2). Strains YZU17, 21, 23, and 36 were more effective against gram-negative S. typhimurium SL 1344, whereas strains YZU13, 19, 30, 31, 33, and 40 exhibited antagonistic activities against gram-positive S. aureus ATCC 25,923. Strain YZU39 was least effective against 3 tested pathogens and displayed the smallest diameters of the inhibition zones.
Antibiotic Susceptibility Test
An important requisite for probiotic strains to be considered safe for animal consumption is that they are antibiotic susceptible (i.e., being able to be eliminated if necessary) or lack acquired antimicrobial resistance properties (i.e., being unable to transmit the antibiotic resistance to pathogenic or potentially pathogenic microorganisms). To address the biosafety concerns, acid- and bile salt-tolerant L. salivarius strains YZU37 and YZU38, which demonstrated the most effective growth-inhibition effects against tested pathogens, together with 2 other L. salivarius strains (YZU40 and YZU41), 2 E. faecium strains (YZU19 and YZU23), and 1 Lactobacillus johnsonii strain YZU39, were tested for their antibiotic susceptibility patterns to 10 most commonly used antibiotics.
LAB isolates displayed a species-specific pattern in their antibiotic susceptibility profiles (Table 1). Four L. salivarius strains displayed similar antibiotic susceptibility profiles. They all were resistant to vancomycin and presented at least medium resistance to aminoglycosides antibiotics (i.e., kanamycin, streptomycin, and gentamicin), while sensitive to the other 6 antibiotics tested.
Here, our results corroborate previous investigations indicating that the occurrence of aminoglycoside and vancomycin resistance was widely observed in LAB.
Aminoglycosides, endowed with polycationic properties, can penetrate into the periplasmic space through binding to the anionic compounds on the bacterial surface. With the participation of a functional electron transport system, the aminoglycoside molecules reach the cytoplasm, bind to the 30 S ribosome subunit, and lead to the production of mistranslated proteins [42, 43]. Most Lactobacillus species are intrinsically resistant to aminoglycosides (mainly gentamicin, kanamycin, streptomycin, and neomycin) [44]. A high natural resistance to the antibiotic group of aminoglycoside was widely reported in LAB strains isolated from various resources [45]. This resistance phenotype observed in LAB has been ascribed to their low permeability of bacterial cell surface for aminoglycosides, especially at low pH, and the absence of cytochrome-mediated electron transport elements [46]. Therefore, resistance to aminoglycosides in these microbes is usually intrinsic, not acquired, and does not constitute a safety concern in itself [47].
As for vancomycin, it is a glycopeptide and the inhibitor of peptidoglycan synthesis. It binds with high affinity to the D-Ala-D-Ala terminus, the peptidoglycan precursors on the bacterial cell wall envelope, and interferes with the maturation process of the peptidoglycan layer [48]. The replacement of the regular D-Ala-D-Ala by dipeptide D-Ala-D-lac or D-Ala-D-Ser, to which vancomycin had low affinity, rendered many Lactobacillus strains intrinsically resistant to vancomycin [49]. Besides Lactobacillus, the vancomycin resistance in Pediococcus, Leuconostoc, and Lactococcus species was also considered to be intrinsic and chromosomally encoded [50]. This intrinsic character, gendered by the substitution of residues in the muramyl pentapeptide cell wall, is non-transferable and presents a low risk of horizontal dissemination to pathogenic and opportunistic bacteria [51]. Therefore, the absence of intrinsic aminoglycoside and vancomycin resistance was not applied as criteria for the exclusion of isolates as probiotic LAB candidates.
Analyses of Other Probiotic Properties
In this study, 7 LAB isolates (designated as YZU37, 38, 40, 41, 19, 23, and 39), which were tested for antibiotic susceptibility, were further analyzed for their characterizations of other probiotic features, including cell surface hydrophobicity, auto-aggregation properties, anti-oxidant activities, and their antimicrobial substances.
Cell surface hydrophobicity, reflecting the capacity of bacteria to adhere to hydrocarbons non-specifically, is correlated to the attachment of bacteria to the epithelium along the digestive tract [52]. It is commonly used to evaluate bacterial strains’ colonization potential [53]. Auto-aggregation, determining the non-specific interactions among bacteria themselves, measures the ability of bacteria to adhere and multiply in the gastrointestinal tracts [54]. Among the 7 strains, the hydrophobicity and auto-aggregation level ranged 13.56−38.65% and 14.85−48.33%, respectively. Strain YZU37 demonstrated a good hydrophobicity of 38.65 ± 3.43% for xylene and an auto-aggregation rate of 32.05 ± 0.62%, which was comparable to previous results obtained in lactobacilli strains [55].
The antioxidant activity of LAB is another important probiotic feature. Our results showed that Lactobacillus johnsonii strain YZU41 and the 4 L. salivarius strains displayed better DPPH and ABTS radical-scavenging activities than the 2 E. faecium strains (supplemental Table S3). Of the 4 L. salivarius strains, YZU37 and YZU38 had equivalent anti-oxidant activities. The DPPH scavenging rates of their CFE and ICS were significantly higher than that of the other two strains (p < 0.05). The ABTS-radical scavenging rates of the CFE of YZU37 and YZU38 were also significantly higher than that of YZU40 and YZU41 (p < 0.05).
Analyses of antibacterial substances revealed complete loss of antimicrobial activity in neutralized CFS of all tested 7 LAB isolates, which indicated that the inhibitory effects these isolates exerted on the growth of pathogenic bacteria were dependent on the acidic environment. In the meantime, significantly reduced antimicrobial activities were observed after the treatments of pepsin, trypsin, and catalase to the CFS, which implied that the antibacterial components were protein-like and, at least, partly were the result of the production of hydrogen peroxides (Supplemental Table S4).
Collectively, strain YZU37 was able to resist pH 2.5 for 4 h without loss of viable cells, retained a survival rate of more than 80% after a 4-h-exposure to 0.3% bile salt, exhibited strong antagonistic effects against the growth of pathogenic Salmonella typhimurium, Staphylococcus aureus, and E. coli strains, was susceptible to the majority antibiotics tested, displayed good hydrophobic and auto-aggregation features, as well as radical scavenging capacities. We concluded strain L. salivarius YZU37 was a putative probiotic candidate and further analyzed its whole genome sequence in order to shed more light on the mechanisms by which it exerted its actions.
Genome Characteristics of L. salivarius YZU37
The complete genome of YZU37 contains a single circular chromosome of 2,159,518 bp with a guanine-cytosine (GC) content of 33.4% (supplemental Fig. S1). Its size (around 2.1 Mb) is similar to the genome size of other L. salivarius strains previously uploaded to the Genome database in NCBI by other researchers.
Species Confirmation
Average nucleotide identity (ANI), which provides a quantitative measure regarding the genetic relatedness of different bacterial strains, was calculated between the YZU37 genome and genome sequences available in the JSpecies server [35]. The ANI values larger than 95−96% were often used as the criterion to confirm the bacterial species. Our results revealed that YZU37 strain showed the highest ANI values for L. salivarius DSM 20,555 (ANIb 97.50% and ANIm 98.06%, respectively) and L. salivarius CECT 5713 (Tetra 0.9884). Phylogenomic analysis using genome-genome comparisons in TYGS revealed that the YZU37 strain was most closely related to L. salivarius DSM 20,555 (Fig. 3). These results confirmed that the YZU37 strain belonged to L. salivarius.
Phylogenetic comparison of Ligilactobacillus salivarius YZU37 with representative complete genomes of other Ligilactobacillus strains carried out in TYGS webserver. (The tree was inferred with FASTME 2.1.6.1. from GBDP distances calculated from 16 S rDNA gene sequences. The bootstrap support value next to each node represents the confidence degree of each branch)
Pangenome Analysis
A comparative pangenome analysis of 216 L. salivarius strains was performed. Of which, genomes of 4 pigeon-origin L. salivarius strains (YZU37, YZU38, YZU40, and YZU41) were newly sequenced in this study and the other 212 genomes were publicly available in the NCBI Genome database. Results from pangenome analysis revealed 14,323 sets of genes. Among them, 7.4% genes were grouped into core (hard core and soft core) genome and 92.6% genes into accessory genome. Nearly 83% (11,889) genes were strain-specific genes (cloud genes), indicating the high genomic heterogeneity among these L. salivarius strains (Fig. 4). Strains isolated from the same species were closely clustered. Of the 4 L. salivarius strains isolated from pigeons in this study, YZU37, YZU38, and YZU41 were clustered into one cluster together with 3 other strains isolated from the feces of wild boar (geographic location: Spain), while YZU40 was in a neighboring cluster with 9 strains isolated from the feces of badgers (geographic location: China). Pigeon- and chicken-origin strains did not group into one cluster as expected. We speculated that the genetic characteristics of intestinal bacterial strains isolated from pigeons (mainly consume raw grains) were more similar to that of strains isolated from wild animals, rather than to those isolated from broilers and laying hens (primarily fed on commercial complete feeds).
By analyzing the presence and absence of genes among the genomes of the 216 strains (Supplemental File S3), two protein-coding genes (dedA_1 and group_13775), annotated as protein DedA and choloylglycine hydrolase, respectively, were uniquely identified in the 4 pigeon-originated strains. DedA family proteins are integral membrane proteins widely found in bacteria and function as undecaprenyl-phosphate (UndP) flippases [56]. Their distinct physiological roles are still enigmatic. At present, it is known that bacterial DedA family members are involved in the maintenance of membrane integrity, colistin resistance, cell division, and pH sensitivity [57]. This dedA_1 gene, uniquely identified in the genomes of the 4 L. salivarius strains derived from pigeons, needs to be further investigated to determine its specific roles.
As for the other protein, choloylglycine hydrolase (CGH, E.C. 3.5.1.24), it is commonly termed as bile salt hydrolase (BSH) or conjugated bile acid hydrolase (CBAH). It hydrolyzes the amide bond in conjugated (glycine- or taurine-conjugated) bile acids, converting them into deconjugated bile acids and releasing free amino acids [58]. Besides its function in bile detoxification, which improves bacterial survivability in the bile-acids-containing intestinal environment, the enzymatic activity of CGH is also associated with bacterial colonization in the gut [59], cholesterol reduction in pigs, dogs, mice, and human [60], and alleviation of Clostridioides difficile infection in human [61]. Due to these beneficial effects on hosts, bacterial CGH activity was considered by some researchers as a functional probiotic biomarker [37].
Pangenome analysis revealed that in the genomes of the 4 pigeon-origin L. salivarius strains, there was a unique CGH-encoding gene which was different from the common one shared by the other 208 L. salivarius strains isolated from human, pig, chicken, badger, and wild boar. Using molecular docking (MD) analysis, we further investigated the interactions between CGH and its 6 substrates, including GCA, GDCA, GCDCA, TCA, TDCA, and TCDCA. MD results showed that the binding affinity of pigeon-origin CGH to 6 conjugated bile acids was generally lower than that of the common CGH shared by other L. salivarius strains of other sources (Fig. 5A and B), indicating a more stable receptor-ligand binding. Detailed non-covalent interactions between CGH (the receptor) and GCA (the ligand), which exhibited the largest difference in the receptor-ligand binding affinity, are shown in Fig. 5C. Compared with the common CGH of other sources, pigeon-origin CGH formed more hydrogen bonds and hydrophobic interactions with its substrate GCA. As far as we know, this is the first report of a unique CGH-encoding gene identified in the genomes of L. salivarius strains isolated from pigeons. Characterized by stronger enzyme-substrate interactions, this pigeon-origin CGH may stably bind to conjugated bile acids and lead to a higher catalytic efficiency. Further studies need to be performed before the specific roles this unique CGH plays in the microbial metabolism of the pigeon’s intestines can be elucidated.
Results of the pangenome comparison analysis. (A, the matrix depicts the results of the comparison of the genomes of the 216 Ligilactobacillus salivarius strains. Of which, 4 strains were isolated from meat-type pigeons and sequenced in this study, while the whole genome sequence of the other 212 strains of different sources were obtained from the Genome database on the NCBI website; B, a pie chart depicting the number of core, soft core, shell, and cloud genes)
MD results showing the interactions of choloylglycine hydrolase (CGH) to its substrates. (A and B, interactions between CGH and its 6 substrates; GCA, glycocholic acid (CHEBI: 17,687); GDCA, glycodeoxycholic acid (CHEBI: 27,471); GCDCA, glycochenodeoxycholic acid (CHEBI: 36,274); TCA, taurocholic acid (CHEBI: 28,865); TDCA, taurodeoxycholic acid (CHEBI: 9410); TCDCA, taurochenodeoxycholic acid (CHEBI: 16,525); C, detailed non-covalent interactions between CGH and GCA)
Annotation and Functional Prediction
Genome annotation using Prokka predicted a total of 2110 genes, including 2036 protein-coding sequences (CDS), 66 tRNAs, 7 rRNAs, and 1 tmRNA. No plasmid sequences were found in the genome. Of the predicted CDS, 1197 genes (58.8%) were functional, and 839 genes (41.2%) were hypothetical or unknown. The 66 tRNA sequences correspond to 20 amino acids: Arg and Gly (6 sequences); Glu and Leu (5 sequences); Gln, Lys, Ser, Thr, Met, and Val (4 sequences); Asp, Asn, and Pro (3 sequences); Cys, Ile, Phe, and Tyr (2 sequences); Ala, Trp, and His (1 sequence).
Analysis of the YZU37 genome on RAST provided an overview of the coded biological features with a subsystem coverage of 27%, distributed in 840 SEED subsystems (Fig. 6). The distribution of different functional groups showed a predominance of genes involved in general processes related to carbohydrates (147), protein metabolism (125), amino acids and derivatives (99), nucleosides and nucleotides (76), and DNA metabolism (70). Fifty-two genes involved in the metabolism of cofactors, vitamins, prosthetic groups, and pigments, including genes involved in the biosynthesis of coenzyme A (7), NAD and NADP (7), riboflavin, FMN, and FAD (7), folate (6), pyridoxine (6), biotin (3), thiamin (3), and lipoic acid (1). The ability of the YZU37 strain to synthesize the B group of vitamins would be a desirable trait for its potential probiotic effects on the hosts.
General overview of biological subsystem distribution of the genes annotated using RAST-SEED server (https://rast.nmmpdr.org)
Analysis of COG functional groups using EggNOGmapper v2 assigned 66.63% genes into 18 clusters (Fig. 7). The highest number was sorted under the function unknown group (S), which contained several loci of interest, including phage-related proteins, enzyme involved in biosynthesis of extracellular polysaccharides, protein involved in the maturation of the 30 S and 50 S ribosomal subunit, and biofilm formation stimulator. The remaining proteins were mostly categorized under functional groups associated with genetic information and processing (J, K, and L), followed by the transport and metabolism-related groups (E, F, G), and the cell wall, membrane, envelope biogenesis group (M).
The KEGG functional annotation by BLASTKOALA assigned approximately half of the genes (53.5%, 1089 genes) into 22 different functional categories (Table 2). Similar to the results of COG analysis, the majority of them were related to protein families (30.84%): genetic information processing (16.80%), signaling and cellular processes (11.29%), and metabolism (2.75%), followed by carbohydrate metabolism (12.86%), nucleotide metabolism (4.87%), amino acid metabolism (4.41%), and environmental information processing (5.51%).
Of note, we identified 8 genes closely associated with the acid-resistant function of YZU37, which included atpB, atpF, and atpE (encoding the integral membrane F0 portion of F0F1 ATP synthase, i.e., subunit A, B, and C, respectively), atpA, atpD, atpG, atpH, and atpC (encoding the catalytic F1 portion of F0F1 ATP synthase, i.e., subunit alpha, beta, gamma, delta, and epsilon, respectively). The F0F1 ATP synthase is one of the main mechanisms adopted by gram-positive bacteria for protection against acidic conditions [62]. Studies in Lactobacillus strains revealed that the cluster of F0F1 ATP synthase subunits served as a main regulator of cytoplasmic pH and improves bacterial tolerance to low pH [63, 64]. Additionally, 3 Na+/H+ antiporters genes (nhaC, napA4, nhaP3), which had been proved to maintain pH and Na+ homeostasis [65], and CadA (lysine decarboxylase 1), which provided cells with protection against mild acidic conditions [66], were also identified in the YZU37 genome. The presence of these proteins may enforce the YZU37 strain’s ability to tolerate an acidic environment.
In terms of bile salt resistance, the presence of choloylglycine hydrolase family protein and inorganic pyrophosphatase (encoded by cbah and ppaC, respectively) endowed the YZU37 strain with superior bile salt tolerance due to the key roles these two proteins played in hydrolyzing non-peptide C-N bonds in conjugated bile acids and maintaining surface tension of the membrane to keep the membrane intact, respectively [67].
We also identified a series of genes involved in the oxidative stress resistance (trxA, trxA1, trxA2, trxB1, and tpx), heavy metal stress resistance (corA, zurR, znuB, znuC, copA), heat stress resistance (hslO, hslV, hslU, dnaK, dnaJ, clpB, clpC, grpE, hrcA), and cold stress resistance (cshB and cspC) [68]. The dltA, dltB, dltC, and dltD genes encoding for proteins with immune-modulatory activities [69] were also identified in YZU37. Moreover, genes encoding cell-surface proteins, such as mapA (maltose phosphorylase), lspA (lipoprotein signal peptidase), and tuf (elongation factor Tu), and other bacterial adhesion activity-related genes, including srtA (sortase A), epsB (protein-tyrosine kinase), eno (enolase), pgi (glucose-6-phosphate isomerase), FbpA (fibronectin-binding protein A N-terminus), and scpB (segregation and condensation protein B), were found in the genome. These results, summarized in Table 3, suggested that YZU37 strain was capable to resist multiple stressful conditions and perform well in terms of adhesion. These properties helped YZU37 adapt to the intestinal environment, survive, reproduction, and exert its probiotic effects on the host.
Carbohydrate-Active Enzymes (CAZymes)
CAZymes are responsible for catalyzing the breakdown, biosynthesis, or modification of carbohydrates and glycoconjugates [70]. Analysis of the YZU37 genome in the dbCAN3 webserver using the predicted amino acid sequences as input revealed that the YZU37 genome comprised 65 CAZymes genes, including 38 glycosyltransferases (GTs) genes, 18 glycoside hydrolases (GHs) genes, 2 carbohydrate esterases (CEs) genes, and 7 carbohydrate-binding modules (CBMs) genes. GHs and GTs are the key enzymes responsible for the hydrolysis/rearrangement and the formation of glycosidic bonds, respectively [71]. The largest family in the YZU37 genome was GTs, which was clustered into 12 families. Among them, GT2 (11) and GT4 (12) were the most abundant. They were mainly involved in the synthesis of extracellular polysaccharide (EPS), which was considered as an immune-related factor associated with the beneficial anti-inflammatory effects of Lactobacillus strains [72]. Based on these results, we concluded that YZU37 strain was capable of utilizing complex carbohydrates to extract energy and nutrients from the environment and enhancing host immune function as well.
CRISPR-Cas System in YZU37
The presence of the CRISPR-Cas system was investigated in the genome of YZU37 to explore its acquired immunity. We identified two CRISPR arrays in the YZU37 genome using the CRISPR-CasFinder. Only one (start at 306,422, 36-bp repeat length, 11 repeats) matched a consensus sequence with evidence level 4 (Supplemental Table S5). Within this array, two accessory CRISPR-associated protein of the CAS system (Cas2_TypeI-II-III and Cas6_Type I-III), one Cas-Type II system (cas1), and five Cas-Type III systems (including csm6, csm4, csm3, csm2, and cas10) were predicted. The CRISPR-CAS system is an immune system acquired by prokaryotes to defend them against foreign genetic elements (plasmids, transposons, viruses, and insertion sequences) and maintain genome fidelity [73]. Research in Enterococci strains revealed an inverse relationship between CRISPR-Cas and antibiotic resistance [74]. The presence of CRISPR-Cas systems in the YZU37 strain implicated its ability to invade and digest foreign DNA, therefore, may limit the horizontal transfer of antibiotic resistance genes [75] and increase the safety of YZU37 as a potential probiotic strain.
Safety-Associated Genes
The profound analysis using IslandViewer 4 did not detect virulence factors or pathogen-associated genes in the genome of YZU37. Under default parameters (perfect and strict hits, only), only one vanT gene in the vanG glycopeptide resistance gene cluster was detected in the YZU37 genome in the CARD database search, which was consistent with our previous phenotypic observation of bacterial resistance to vancomycin. This intrinsic antimicrobial resistance that is common within a single species will not present a safety issue [76].
Bacteriocin-Encoding Genes
The bacteriocin operon in the genome of YZU37 was predicted using the bacteriocin mining tool BAGEL4. Three putative bacteriocinogenic genetic clusters were identified as areas of interest (AOI) at (i) Node 11 length 66,912 cov 217665460.23 (start at 53,441 and end 66,912); (ii) Node 18 length 37,822 cov 291463638.17 (start at 27,222 and end at 37,821); (iii) Node 22 length 26,625 cov 274750904.27 (start at13,223 and end 26,625), which encoded bacteriocin of enterolysin A with levels of identity of 37.11% (E-value = 4.24E-27), 39.74% (E-value = 1.45E-25), and 32.93% (E-value = 1.60E-24), respectively (Fig. 8).
Enterolysin A, belonging to class III bacteriocin, is a heat-labile extracellular peptidoglycan hydrolase [77]. Its presence has been identified in other potential probiotic LAB strains, including Enterococcus faecalis [78], Enterococcus durans [79], Lactobacillus mucosae [80], Pediococcus acidilactici [81], Pediococcus pentosaceus and Lactococcus lactis [82], Limosilactobacillus fermentum [83], and Weissella cibaria [84]. In the present study, the inhibitory effects L. salivarius YZU37 exerted on the 3 tested pathogenic bacteria may be attributed to its cell wall degradation property.
Conclusion
Results obtained in this study led to the identification strain L. salivarius YZU37 from the intestines of meat-type pigeon squabs with the most promising probiotic potential, which met the criteria for probiotic candidates in respects of acid- and bile salt-resistance, anti-pathogenic capacities and antibiotic resistance profile besides good hydrophobicity, co-aggregation ability, and anti-oxidant activities. The genome of YZU37 revealed the presence of various genes related to stress adaption, carbohydrates transports and metabolisms, CRISPR-cas regions, putative bacteriocin clusters, and the absence of virulence and toxin genes. It also harbored a pigeon-unique CGH with stronger binding affinity to conjugated bile salts. Altogether, our current results highlighted YZU37’s probiotic characteristics. Further in vitro and in vivo research is required regarding the safety and technological performance before this pigeon-originated strain may be served as a promising strain for probiotic supplements.
Data Availability
Clean reads generated in this study have been submitted to NCBI SRA database (accession number SUB13999939).
References
Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, Calder PC, Sanders ME (2014) Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol 11(8):506–514. https://doi.org/10.1038/nrgastro.2014.66
Plaza-Diaz J, Ruiz-Ojeda FJ, Gil-Campos M, Gil A (2019) Mechanisms of action of probiotics. Adv Nutr (Bethesda Md) 10(suppl1):S49–S66. https://doi.org/10.1093/advances/nmy063
Khan I, Nawaz M, Anjum AA, Ahmed MU, Tanvir R, Sarwar N, Ashraf MA, Mehmood A, Aqib M (2023) Limosilactobacillus Fermentum IKP 111 reduces pathogen load and improves immunity of broilers when challenged with Salmonella enteritidis. Microb Pathog 185:106401. https://doi.org/10.1016/j.micpath.2023.106401
Ghareeb K, Awad WA, Mohnl M, Porta R, Biarnés M, Böhm J, Schatzmayr G (2012) Evaluating the efficacy of an avian-specific probiotic to reduce the colonization of Campylobacter jejuni in broiler chickens. Poult Sci 91(8):1825–1832. https://doi.org/10.3382/ps.2012-02168
Pan X, Kong R, Liu Q, Jia Z, Bai B, Chen H, Zhi W, Wang B, Ma C, Ma D (2023) Probiotic Enterococcus faecalis surface-delivering key domain of EtMIC3 proteins: immunoprotective efficacies against Eimeria tenella infection in chickens. Microbiol Spectr 11(6):e02455–02423. https://doi.org/10.1128/spectrum.02455-23
Neveling DP, Dicks LMT (2021) Probiotics: an antibiotic replacement strategy for healthy broilers and productive rearing. Probiotics Antimicro 13(1):1–11. https://doi.org/10.1007/s12602-020-09640-z
Popov IV, Algburi A, Prazdnova EV, Mazanko MS, Elisashvili V, Bren AB, Chistyakov VA, Tkacheva EV, Trukhachev VI, Donnik IM, Ivanov YA, Rudoy D, Ermakov AM, Weeks RM, Chikindas ML (2021) A review of the effects and production of spore-forming probiotics for poultry. Anim (Basel) 11(7):1941. https://doi.org/10.3390/ani11071941
Elbaz AM, Ashmawy ES, Ali SM, Mourad DM, El-Samahy HS, Badri FB, Thabet HA (2023) Effectiveness of probiotics and clove essential oils in improving growth performance, immuno-antioxidant status, ileum morphometric, and microbial community structure for heat-stressed broilers. Sci Rep 13(1):18846. https://doi.org/10.1038/s41598-023-45868-9
Imari ZK, Alnajm HR, Zamil SJ (2023) Impact of different levels of probiotic on productive performance, nutrient retention of broiler chickens fed low protein diets. J Adv Vet Anim Res 10(3):395–402. https://doi.org/10.5455/javar.2023.j692
Lefter NA, Gheorghe A, Habeanu M, Ciurescu G, Dumitru M, Untea AE, Vlaicu PA (2023) Assessing the effects of microencapsulated Lactobacillus salivarius and cowpea seed supplementation on broiler chicken growth and health status. Front Vet Sci 10:1279819. https://doi.org/10.3389/fvets.2023.1279819
Zammit VA, Park SO (2023) Impact of the combination of probiotics and digital poultry system on behavior, welfare parameters, and growth performance in broiler chicken. Microorganisms 11(9):2345. https://doi.org/10.3390/microorganisms11092345
Muyyarikkandy MS, Mathew E, Kuttappan D, Amalaradjou MA (2023) Research note: in ovo and in-feed probiotic supplementation improves layer embryo and pullet growth. Poult Sci 102(12):103092. https://doi.org/10.1016/j.psj.2023.103092
Prazdnova EV, Mazanko MS, Chistyakov VA, Denisenko YV, Makarenko MS, Usatov AV, Bren AB, Tutelyan AV, Komarova ZB, Gorlov IF, Weeks R, Chikindas ML (2019) Effect of Bacillus subtilis KATMIRA1933 and Bacillus amyloliquefaciens B-1895 on the productivity, reproductive aging, and physiological characteristics of hens and roosters. Benef Microbes 10(4):395–412. https://doi.org/10.3920/bm2018.0149
Liu L, Zhang G, Qu G, Liu B, Zhang X, Li G, Jin N, Li C, Bai J, Zhao C (2023) Effects of dietary Lactobacillus rhamnosus GG supplementation on the production performance, egg quality, eggshell ultrastructure, and lipid metabolism of late-phase laying hens. BMC Vet Res 19(1):150. https://doi.org/10.1186/s12917-023-03719-9
Jiang S, Hu JY, Cheng HW (2022) The impact of probiotic Bacillus subtilis on injurious behavior in laying hens. Anim (Basel) 12(7):870. https://doi.org/10.3390/ani12070870
Sharma MK, White DL, Singh AK, Liu H, Tan Z, Peng X, Kim WK (2022) Effect of dietary supplementation of probiotic aspergillus Niger on performance and cecal microbiota in hy-line w-36 laying hens. Anim (Basel) 12(18):2406. https://doi.org/10.3390/ani12182406
Alagawany M, Abd El-Hack ME, Arif M, Ashour EA (2016) Individual and combined effects of crude protein, methionine, and probiotic levels on laying hen productive performance and nitrogen pollution in the manure. Environ Sci Pollut Res Int 23(22):22906–22913. https://doi.org/10.1007/s11356-016-7511-6
Abd El-Hack ME, Mahgoub SA, Alagawany M, Ashour EA (2017) Improving productive performance and mitigating harmful emissions from laying hen excreta via feeding on graded levels of corn DDGS with or without Bacillus subtilis probiotic. J Anim Physiol Anim Nutr (Berl) 101(5):904–913. https://doi.org/10.1111/jpn.12522
Jiang SG, Pan NX, Chen MJ, Wang XQ, Yan HC, Gao CQ (2019) Effects of dietary supplementation with dl-methionine and dl-methionyl-dl-methionine in breeding pigeons on the carcass characteristics, meat quality and antioxidant activity of squabs. Antioxid (Basel) 8(10):435. https://doi.org/10.3390/antiox8100435
Grond K, Perreau JM, Loo WT, Spring AJ, Cavanaugh CM, Hird SM (2019) Longitudinal microbiome profiling reveals impermanence of probiotic bacteria in domestic pigeons. PLoS ONE 14(6):e0217804. https://doi.org/10.1371/journal.pone.0217804
Ge B, Yang H, Meng J, Chen X, Wang Z (2020) Effects of Mannan oligosaccharides and/or bifidobacterium on growth and immunity in domestic pigeon (Columba livia Domestica). J Poult Sci 57(4):277–283. https://doi.org/10.2141/jpsa.0190100
Tsai CY, Hu SY, Santos HM, Catulin GEM, Tayo LL, Chuang KP (2021) Probiotic supplementation containing Bacillus velezensis enhances expression of immune regulatory genes against pigeon circovirus in pigeons (Columba livia). J Appl Microbiol 130(5):1695–1704. https://doi.org/10.1111/jam.14893
Wen J, Zhao W, Li J, Hu C, Zou X, Dong X (2022) Dietary supplementation of chitosan oligosaccharide-Clostridium butyricum synbiotic relieved early-weaned stress by improving intestinal health on pigeon squabs (Columba livia). Front Immunol 13:926162. https://doi.org/10.3389/fimmu.2022.926162
Ma H, Li YL, Han PM, Zhang R, Yuan JW, Sun YY, Li JH, Chen JL (2024) Effects of supplementing drinking water of parental pigeons with Enterococcus faecium and Bacillus subtilis on antibody levels and microbiomes in squabs. Anim (Basel) 14(2):178. https://doi.org/10.3390/ani14020178
Fan W, Zhu Y, Hou H, Yao J, Zhu L, Liu H, Yan H (2024) Treatment and prevention of pigeon diarrhea through the application of Lactobacillus SNK-6. Poult Sci 103(4):103476. https://doi.org/10.1016/j.psj.2024.103476
Mulaw G, Sisay Tessema T, Muleta D, Tesfaye A (2019) In vitro evaluation of probiotic properties of lactic acid bacteria isolated from some traditionally fermented Ethiopian food products. Int J Microbiol 2019:7179514. https://doi.org/10.1155/2019/7179514
Rajoka MSR, Hayat HF, Sarwar S, Mehwish HM, Ahmad F, Hussain N, Shah SZH, Khurshid M, Siddiqu M, Shi J (2018) Isolation and evaluation of probiotic potential of lactic acid bacteria isolated from poultry intestine. Microbiology 87(1):116–126. https://doi.org/10.1134/S0026261718010150
Lin CF, Lin MY, Lin CN, Chiou MT, Chen JW, Yang KC, Wu MC (2020) Potential probiotic of Lactobacillus strains isolated from the intestinal tracts of pigs and feces of dogs with antibacterial activity against multidrug-resistant pathogenic bacteria. Arch Microbiol 202(7):1849–1860. https://doi.org/10.1007/s00203-020-01908-w
Ekmekci H, Aslim B, Ozturk S (2009) Characterization of vaginal lactobacilli coaggregation ability with Escherichia coli. Microbiol Immunol 53(2):59–65. https://doi.org/10.1111/j.1348-0421.2009.00115.x
Soemarie YB, Milanda T, Barliana MI (2022) Isolation, characterization, and identification candidate of probiotic bacteria isolated from wadi papuyu (Anabas testudineus Bloch.) a fermented fish product from central Kalimantan. Indonesia. Int J Food Sci 2022:4241531. https://doi.org/10.1155/2022/4241531
Liu Z, Xu C, Tian R, Wang W, Ma J, Gu L, Liu F, Jiang Z, Hou J (2021) Screening beneficial bacteriostatic lactic acid bacteria in the intestine and studies of bacteriostatic substances. J Zhejiang Univ Sci B 22(7):533–547. https://doi.org/10.1631/jzus.B2000602
Shi Y, Cui X, Gu S, Yan X, Li R, Xia S, Chen H, Ge J (2019) Antioxidative and probiotic activities of lactic acid bacteria isolated from traditional artisanal milk cheese from Northeast China. Probiotics Antimicro 11(4):1086–1099. https://doi.org/10.1007/s12602-018-9452-5
Seemann T (2014) Prokka: rapid prokaryotic genome annotation. Bioinf (Oxford England) 30(14):2068–2069. https://doi.org/10.1093/bioinformatics/btu153
Grant JR, Enns E, Marinier E, Mandal A, Herman EK, Chen CY, Graham M, Van Domselaar G, Stothard P (2023) Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res 51(W1):W484–w492. https://doi.org/10.1093/nar/gkad326
Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J (2016) JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinf (Oxford England) 32(6):929–931. https://doi.org/10.1093/bioinformatics/btv681
Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Fookes M, Falush D, Keane JA, Parkhill J (2015) Roary: rapid large-scale prokaryote pan genome analysis. Bioinf (Oxford England) 31(22):3691–3693. https://doi.org/10.1093/bioinformatics/btv421
Rani RP, Anandharaj M, Ravindran AD (2017) Characterization of bile salt hydrolase from Lactobacillus gasseri FR4 and demonstration of its substrate specificity and inhibitory mechanism using molecular docking analysis. Front Microbiol 8:1004. https://doi.org/10.3389/fmicb.2017.01004
Grissa I, Vergnaud G, Pourcel C (2007) CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res 35(Web Server issue):W52-57. https://doi.org/10.1093/nar/gkm360
Kobierecka PA, Wyszyńska AK, Aleksandrzak-Piekarczyk T, Kuczkowski M, Tuzimek A, Piotrowska W, Górecki A, Adamska I, Wieliczko A, Bardowski J, Jagusztyn-Krynicka EK (2017) In vitro characteristics of Lactobacillus spp. strains isolated from the chicken digestive tract and their role in the inhibition of Campylobacter colonization. MicrobiologyOpen 6(5):e00512. https://doi.org/10.1002/mbo3.512
Guo XH, Kim JM, Nam HM, Park SY, Kim JM (2010) Screening lactic acid bacteria from swine origins for multistrain probiotics based on in vitro functional properties. Anaerobe 16(4):321–326. https://doi.org/10.1016/j.anaerobe.2010.03.006
Salehizadeh M, Modarressi MH, Mousavi SN, Tajabadi Ebrahimi M (2020) Evaluation of lactic acid bacteria isolated from poultry feces as potential probiotic and its in vitro competitive activity against Salmonella typhimurium. Vet Res Forum 11(1):67–75. https://doi.org/10.30466/vrf.2018.84395.2110
Ramirez MS, Tolmasky ME (2017) Amikacin: uses, resistance, and prospects for inhibition. Molecules 22(12):2267. https://doi.org/10.3390/molecules22122267
Ramirez MS, Tolmasky ME (2010) Aminoglycoside modifying enzymes. Drug Resist Updat 13(6):151–171. https://doi.org/10.1016/j.drup.2010.08.003
Campedelli I, Mathur H, Salvetti E, Clarke S, Rea MC, Torriani S, Ross RP, Hill C, O’toole PW (2019) Genus-wide assessment of antibiotic resistance in Lactobacillus spp. Appl Environ Microbiol 85(1):e01738–e01718. https://doi.org/10.1128/aem.01738-18
Duche RT, Singh A, Wandhare AG, Sangwan V, Sihag MK, Nwagu TNT, Panwar H, Ezeogu LI (2023) Antibiotic resistance in potential probiotic lactic acid bacteria of fermented foods and human origin from Nigeria. BMC Microbiol 23(1):142. https://doi.org/10.1186/s12866-023-02883-0
Elkins CA, Mullis LB (2004) Bile-mediated aminoglycoside sensitivity in Lactobacillus species likely results from increased membrane permeability attributable to cholic acid. Appl Environ Microbiol 70(12):7200–7209. https://doi.org/10.1128/aem.70.12.7200-7209.2004
Gueimonde M, Sánchez B, C GDLR-G, Margolles A (2013) Antibiotic resistance in probiotic bacteria. Front Microbiol 4:202. https://doi.org/10.3389/fmicb.2013.00202
Stogios PJ, Savchenko A (2020) Molecular mechanisms of Vancomycin resistance. Protein Sci 29(3):654–669. https://doi.org/10.1002/pro.3819
Ammor MS, Flórez AB, Van Hoek AH, De Los Reyes-Gavilán CG, Aarts HJ, Margolles A, Mayo B (2008) Molecular characterization of intrinsic and acquired antibiotic resistance in lactic acid bacteria and bifidobacteria. J Mol Microbiol Biotechnol 14(1–3):6–15. https://doi.org/10.1159/000106077
Mathur S, Singh R (2005) Antibiotic resistance in food lactic acid bacteria-a review. Int J Food Microbiol 105(3):281–295. https://doi.org/10.1016/j.ijfoodmicro.2005.03.008
Zhang S, Oh JH, Alexander LM, Özçam M, Van Pijkeren JP (2018) D-Alanyl-D-Alanine ligase as a broad-host-range counterselection marker in Vancomycin-resistant lactic acid bacteria. J Bacteriol 200(13):e00607–e00617. https://doi.org/10.1128/jb.00607-17
Kos B, Susković J, Vuković S, Simpraga M, Frece J, Matosić S (2003) Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J Appl Microbiol 94(6):981–987. https://doi.org/10.1046/j.1365-2672.2003.01915.x
Taheri HR, Moravej H, Tabandeh F, Zaghari M, Shivazad M (2009) Screening of lactic acid bacteria toward their selection as a source of chicken probiotic. Poult Sci 88(8):1586–1593. https://doi.org/10.3382/ps.2009-00041
Del Re B, Sgorbati B, Miglioli M, Palenzona D (2000) Adhesion, autoaggregation and hydrophobicity of 13 strains of Bifidobacterium longum. Lett Appl Microbiol 31(6):438–442. https://doi.org/10.1046/j.1365-2672.2000.00845.x
Peres CM, Alves M, Hernández-Mendoza A, Liliana, Moreira, Silva SL, Bronze MDR, Vilas-Boas LF, Peres C, Malcata FX (2014) Novel isolates of lactobacilli from fermented Portuguese olive as potential probiotics. Lwt-Food Sci Technol 59(1):234–246. https://doi.org/10.1016/j.lwt.2014.03.003
Scarsbrook HL, Urban R, Streather BR, Moores A, Mulligan C (2021) Topological analysis of a bacterial DedA protein associated with alkaline tolerance and antimicrobial resistance. Microbiology 167(12):001125. https://doi.org/10.1099/mic.0.001125
Todor H, Herrera N, Gross CA (2023) Three bacterial DedA subfamilies with distinct functions and phylogenetic distribution. mBio 14(2):e0002823. https://doi.org/10.1128/mbio.00028-23
Geng W, Lin J (2016) Bacterial bile salt hydrolase: an intestinal microbiome target for enhanced animal health. Anim Health Res Rev 17(2):148–158. https://doi.org/10.1017/s1466252316000153
Bustos AY, Font De Valdez G, Fadda S, Taranto MP (2018) New insights into bacterial bile resistance mechanisms: the role of bile salt hydrolase and its impact on human health. Food Res Int 112:250–262. https://doi.org/10.1016/j.foodres.2018.06.035
Dong Z, Lee BH (2018) Bile salt hydrolases: structure and function, substrate preference, and inhibitor development. Protein Sci 27(10):1742–1754. https://doi.org/10.1002/pro.3484
Mullish BH, Mcdonald JK, Pechlivanis A, Allegretti JR, Kao D, Barker GF, Kapila D, Petrof EO, Joyce SA, Gahan CGM, Glegola-Madejska I, Williams HRT, Holmes E, Clarke TB, Thursz MR, Marchesi JR (2019) Microbial bile salt hydrolases mediate the efficacy of faecal microbiota transplant in the treatment of recurrent Clostridioides difficile infection. Gut 68(10):1791–1800. https://doi.org/10.1136/gutjnl-2018-317842
Cotter PD, Hill C (2003) Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol Mol Biol Rev 67(3):429–453. https://doi.org/10.1128/mmbr.67.3.429-453.2003
Corcoran BM, Stanton C, Fitzgerald GF, Ross RP (2005) Survival of probiotic lactobacilli in acidic environments is enhanced in the presence of metabolizable sugars. Appl Environ Microbiol 71(6):3060–3067. https://doi.org/10.1128/aem.71.6.3060-3067.2005
Sun Y, Zhang S, Li H, Zhu J, Liu Z, Hu X, Yi J (2022) Assessments of probiotic potentials of lactiplantibacillus plantarum strains isolated from Chinese traditional fermented food: phenotypic and genomic analysis. Front Microbiol 13:895132. https://doi.org/10.3389/fmicb.2022.895132
D’souza S, Garcia-Cabado A, Yu F, Teter K, Lukacs G, Skorecki K, Moore HP, Orlowski J, Grinstein S (1998) The epithelial sodium-hydrogen antiporter Na+/H+ exchanger 3 accumulates and is functional in recycling endosomes. J Biol Chem 273(4):2035–2043. https://doi.org/10.1074/jbc.273.4.2035
Kanjee U, Gutsche I, Alexopoulos E, Zhao B, El Bakkouri M, Thibault G, Liu K, Ramachandran S, Snider J, Pai EF, Houry WA (2011) Linkage between the bacterial acid stress and stringent responses: the structure of the inducible lysine decarboxylase. EMBO J 30(5):931–944. https://doi.org/10.1038/emboj.2011.5
Zhang L, Ma H, Kulyar MF, Pan H, Li K, Li A, Mo Q, Wang Y, Dong H, Bao Y, Li J (2022) Complete genome analysis of Lactobacillus fermentum YLF016 and its probiotic characteristics. Microb Pathog 162:105212. https://doi.org/10.1016/j.micpath.2021.105212
Wu YP, Liu DM, Zhao S, Huang YY, Yu JJ, Zhou QY (2022) Assessing the safety and probiotic characteristics of Bacillus coagulans 13002 based on complete genome and phenotype analysis. Lwt-Food Sci Technol 155:112847. https://doi.org/10.1016/j.lwt.2021.112847
Kandasamy S, Yoo J, Yun J, Lee KH, Kang HB, Kim JE, Oh MH, Ham JS (2022) Probiogenomic in-silico analysis and safety assessment of lactiplantibacillus plantarum DJF10 strain isolated from Korean raw milk. Int J Mol Sci 23(22):14494. https://doi.org/10.3390/ijms232214494
Benini S (2020) Carbohydrate-active enzymes: structure, activity, and reaction products. Int J Mol Sci 21(8):2727. https://doi.org/10.3390/ijms21082727
Ye K, Li P, Gu Q (2020) Complete genome sequence analysis of a strain Lactobacillus pentosus ZFM94 and its probiotic characteristics. Genomics 112(5):3142–3149. https://doi.org/10.1016/j.ygeno.2020.05.015
Ryan PM, Stolte EH, London LEE, Wells JM, Long SL, Joyce SA, Gahan CGM, Fitzgerald GF, Ross RP, Caplice NM, Stanton C (2019) Lactobacillus mucosae DPC 6426 as a bile-modifying and immunomodulatory microbe. BMC Microbiol 19(1):33. https://doi.org/10.1186/s12866-019-1403-0
Makarova KS, Koonin EV (2015) Annotation and classification of CRISPR-Cas systems. Methods Mol Biol 1311:47–75. https://doi.org/10.1007/978-1-4939-2687-9_4
Palmer KL, Gilmore MS (2010) Multidrug-resistant enterococci lack CRISPR-cas. mBio 1(4):e00227. 10
Dos Santos BA, De Oliveira JDS, Parmanhani-Da-Silva BM, Ribeiro RL, Teixeira LM, Neves FPG (2020) CRISPR elements and their association with antimicrobial resistance and virulence genes among Vancomycin-resistant and Vancomycin-susceptible enterococci recovered from human and food sources. Infect Genet Evol 80:104183. https://doi.org/10.1016/j.meegid.2020.104183
EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP), Rychen G, Aquilina G, Azimonti G, Bampidis V, Bastos ML, Bories G, Chesson A, Cocconcelli PS, Flachowsky G, Gropp J, Kolar B, Kouba M, López-Alonso M, López Puente S, Mantovani A, Mayo B, Ramos F, Saarela M, Villa RE, Wallace RJ, Wester P, Glandorf B, Herman L, Kärenlampi S, Aguilera J, Anguita M, Brozzi R, Galobart J (2018) Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J 16(3):e05206. https://doi.org/10.2903/j.efsa.2018.5206
Malinicova L, Dubikova K, Piknova M, Pristas P, Javorsky P (2012) Peptidoglycan hydrolase enterolysin a recognizes lipoteichoic acid chains in the cell walls of sensitive bacteria. Protein Pept Lett 19(9):924–929. https://doi.org/10.2174/092986612802084410
Khan H, Flint SH, Yu PL (2013) Determination of the mode of action of enterolysin A, produced by Enterococcus faecalis B9510. J Appl Microbiol 115(2):484–494. https://doi.org/10.1111/jam.12240
Enuh BM, Gedikli S, Aytar Çelik P, Çabuk A (2023) Genome sequence and probiotic potential of newly isolated Enterococcus durans strain MN187066. Lett Appl Microbiol 76(3):ovad035. https://doi.org/10.1093/lambio/ovad035
Jia Y, Yang B, Ross P, Stanton C, Zhang H, Zhao J, Chen W (2020) Comparative genomics analysis of Lactobacillus mucosae from different niches. Genes (Basel) 11(1):95. https://doi.org/10.3390/genes11010095
Li Z, Song Q, Wang M, Ren J, Liu S, Zhao S (2021) Comparative genomics analysis of Pediococcus acidilactici species. J Microbiol 59(6):573–583. https://doi.org/10.1007/s12275-021-0618-6
Mileriene J, Aksomaitiene J, Kondrotiene K, Asledottir T, Vegarud GE, Serniene L, Malakauskas M (2023) Whole-genome sequence of Lactococcus lactis subsp. lactis LL16 confirms safety, probiotic potential, and reveals functional traits. Microorganisms 11(4):1034. https://doi.org/10.3390/microorganisms11041034
Dos Santos CI, Campos CDL, Nunes-Neto WR, Do Carmo MS, Nogueira FB, Ferreira RM, Costa EPS, Gonzaga LF, Araújo JMM, Monteiro JM, Monteiro C, Platner FS, Figueiredo IFS, Holanda RA, Monteiro SG, Fernandes ES, Monteiro AS, Monteiro-Neto V (2021) Genomic analysis of limosilactobacillus fermentum ATCC 23271, a potential probiotic strain with anti-candida activity. J Fungi (Basel) 7(10):794. https://doi.org/10.3390/jof7100794
Tenea GN, Hurtado P, Ortega C (2020) A novel Weissella cibaria strain UTNGt21O isolated from wild Solanum quitoense fruit: genome sequence and characterization of a peptide with highly inhibitory potential toward gram-negative bacteria. Foods 9(9):1242. https://doi.org/10.3390/foods9091242
Funding
This research was funded by the Open Project Program of Jiangsu Key Laboratory of Zoonosis (No. R2110) and the Postgraduate Research & Practice Innovation Programs of Jiangsu Province (Yangzhou University) (No. SJCX22_1770 and No. SJCX23_1960).
Author information
Authors and Affiliations
Contributions
Shaoqi Tian and Manhong Ye wrote the main manuscript. Shaoqi Tian and Yinhong Jiang performed most of the lab work and prepared Figs. 1 and 2 and Table 1. Qiannan Han, Chuang Meng, and Bin Zhou participated in the lab work and prepared Figs. 3, 4, 6, 7, and 8 and Tables 2 and 3 in bioinformatics analysis. Feng Ji and Bin Zhou prepared Fig. 5 in molecular docking analysis. Manhong Ye participated in the whole lab work, bioinformatics, and molecular docking analyses. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tian, S., Jiang, Y., Han, Q. et al. Putative Probiotic Ligilactobacillus salivarius Strains Isolated from the Intestines of Meat-Type Pigeon Squabs. Probiotics & Antimicro. Prot. (2024). https://doi.org/10.1007/s12602-024-10289-1
Accepted:
Published:
DOI: https://doi.org/10.1007/s12602-024-10289-1