Abstract
Growth hormone receptors (GHRs) are especially expressed in the muscle and liver tissues of mandarin fish Siniperca chuatsi. cDNAs of two GHR types (scGHR1 and scGHR2: 1999 and 1963 bp, encoding 638 and 578 amino acids, respectively) have been isolated and characterized in mandarin fish. Both receptors, formed through fish-specific genome duplication, have conserved FGEFS and box1 and 2 motifs. Partial compensatory growth was noted following 10-day re-feeding after 4-week fasting. After 1-week fasting, pituitary GH and muscle scGHR1 and scGHR2 mRNA levels and plasma GH concentrations increased (p < 0.05), while hepatic scGHR1 and scGHR2 mRNA expressions reduced rapidly (p < 0.05). Upon re-feeding, muscle scGHR1 and scGHR2 mRNA expressions decreased, whereas they increased rapidly in the liver, eventually normalizing. Pituitary GH mRNA expression increased to 5.1 times higher than that in the controls 3 days after re-feeding (p < 0.01), and normalized 7 days after re-feeding. The varying expression of scGHR1 and scGHR2 in different tissues suggests their distinct functions. Further, changes in GH, scGHR1, and scGHR2 mRNA expressions suggest their important roles in the growth of mandarin fish. Our results provide comparative insights into evolutionary origins and roles of GHR genes in teleost growth.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Environmental factors such as temperature, photoperiod, and food availability strongly influence growth in fish, which is primarily controlled by the growth hormone (GH)/insulin-like growth factor (IGF) system [1–3]. The classical view holds that pituitary GH stimulates the synthesis and secretion of IGF-1 from the liver and other tissues (e.g., muscle) through growth hormone receptor (GHR). Further, IGF-1 stimulates cell proliferation and differentiation mediated by IGF-1 receptors (IGFR1) [4–6]. In mammals, it has long been proposed that most of the effects of GH are indirect, with IGF1 being the final effector produced primarily in the liver [7]. However, GH also enhances muscle growth directly through receptors on the muscle cells, and this growth is not mediated by liver-derived or muscle-derived IGF1 [8, 9]. Transgenic zebrafish with constitutive overexpression of GH and/or GHR1 display high growth rates [9]. GH affects many physiological functions, including growth, reproduction, immunity, lipid and protein metabolism, osmoregulation, and feeding behavior in fish via GHR signaling pathways [10, 11]. GHRs have been cloned from several teleost species and show differential expression in various tissues [12–17]. Different isoforms of GHR have been identified in gilthead seabream [18], rainbow trout [13], Nile tilapia [14], orange-spotted grouper [15], black seabream [16], and Japanese seabass [19]. GHRs are members of the class I cytokine receptor superfamily composed of an extracellular domain containing a conserved Y/FGEFS motif, a single transmembrane domain, and an intracellular domain containing proline-rich box1 and box2 motifs [1, 4]. The evolution and nomenclature of GHRs is a hot topic of debate [1, 4, 20, 21]. It is reported that fish GHRs are differentially regulated by fasting [21, 22], salinity [22–24], temperature [25], pathogens [26], stress [27], and cortisol and insulin [28, 29]. However, it is still unknown whether the GHRs have similar or distinct functions [4]. Data of the molecular mechanisms by which the GH system modulates skeletal muscle growth in teleosts is limited [2].
The principal environmental regulator of growth is nutritional status [3, 30]. Compensatory growth (CG) has been well studied in teleosts [31] and could be a useful model for studying endocrine regulation during negative, normal, and accelerated growths. The different expression patterns of GH and GHRs have been reported in a few fish species under various nutritional conditions. In catfish, plasma GH concentrations and pituitary GH mRNA increased after 4 weeks of fasting [32]; and in hybrid striped bass, 3 weeks of food deprivation depressed hepatic GHR1/2 mRNA levels, enhanced skeletal muscle expression of GHR2 mRNA, and increased pituitary GH mRNA and plasma GH [33]. In Mozambique tilapia, plasma GH reduced slightly by 4 weeks of fasting, and transcript levels of GHR1 and GHR2 increased in the muscle, but those in the liver were unaffected [22]. Upon re-feeding, fish gained weight at an accelerated rate [31]. Re-feeding for 2 weeks after 4 weeks of fasting in rainbow trout resulted in decreased plasma GH concentrations and increased hepatic GHR1 and GHR2 mRNA levels [34]. In hybrid striped bass, re-feeding resulted in increased expression of GHR in the liver and a return to pre-fasting levels of GHR1 mRNA in muscle [33]. The results were different and somewhat confusing [35]. However, in the fish studied, the sampling frequencies were mostly low. In addition, the time course of the response of GH and GHRs to fasting and re-feeding has been described in detail in few fish species. In mandarin fish, there has been no study of endocrine regulation during variable growth phases, including negative, normal, and accelerated growth.
Siniperca chuatsi is an economically important species for freshwater farming in China and is favored for its large size, meat texture, flavor, and high nutritive value [36, 37]. We have previously studied the sperm ultrastructure of three species in Siniperca [38], the expression of two HSP70 isoforms in mandarin fish [39], and insulin-like growth factor I gene polymorphisms and their effects on growth traits in sinipercid species [40]. In this study, to better understand the roles of GH and GHRs in the growth of mandarin fish, the cDNAs of two GHR subtypes were cloned and characterized. Further, we designed an experiment with different frequencies of sampling and assessed the time course variations in plasma GH, pituitary GH mRNA, and GHR1/2 mRNA levels in the liver and muscle.
Materials and methods
Animals
Mandarin fish was maintained in an indoor circulating water system containing a series of 400-L tanks at Sun Yat-sen University. The fish were allowed to acclimate to the tanks for 1 month prior to the beginning of experiments. Fish were fed twice a day (09:00 and 16:00) to satiety with live fingerlings. The water temperature was maintained at 25–26 °C. The fish used in this study were 5 months old, weighed 145.5 ± 38 g, and were offspring of the same pair. All experiments were conducted in accordance with the guidelines and procedures approved by the Institutional Animal Care and Use Committee at Sun Yat-Sen University.
Sampling
Body weight and length were measured under anesthesia with tricaine methanesulfonate (MS-222, 0.5 g/l) at each sampling time point from the start of the experiment to the end of the experiment. The specific growth rates were calculated as (lnWf − lnWi)/t × 100, where Wf is the final weight (g), Wi is the initial weight (g) at each time interval, and t is the growth time (days). At the time of sampling, six fish from the fed group and six fish from the treated group were netted and anesthetized. Blood was collected from the caudal vasculature using 1-ml heparinized syringes. Plasma was separated by centrifugation at 1000×g for 30 min at 4 °C, and stored at −80 °C for later GH analysis by ELISA. Livers, muscles, and pituitaries were excised, flash-frozen in liquid nitrogen, and stored at −80 °C.
RNA extraction and cDNA preparation
Total RNAs from different mandarin fish tissues were extracted using Trizol Reagent (Invitrogen, USA) according to the manufacturer’s instructions. The integrity of the total RNA was verified by electrophoresis on 1.0 % agarose gel. The concentration and purity of total RNA were measured by a UV spectrophotometer (Nanodrop 2000c, Thermo, USA). Total RNA of 1 µg from each sample was digested with 2 µg RNase-free DNase I (New England Biolabs, USA) and then used for synthesis of the first-strand cDNA with M-MLV Reverse Transcriptase (Invitrogen, USA) according to the manufacturer’s instructions.
Cloning of scGHR1 and scGHR2
Degenerated PCR primers (Table 1) were designed based on the alignment of known GHR1 or GHR2 cDNA sequences and highly conserved regions of amino acid sequences in bony fish (EF052273.1, EF052274.1, AF438176.2, AY573601.2, NM_001279601.1, AY973233.1, KF601961.1, KF770840.1, AB621336.1, AB621337.1). First-strand cDNA synthesis from liver was treated with RNase-free DNase I (New England Biolabs, USA) and used as the template to amplify the fragment cDNAs of scGHR1 and scGHR2. Gene-specific primers (Table 1) were designed based on the partial cDNA sequences identified by RT-PCR, and were used for rapid amplification of cDNA ends with SMARTer RACE cDNA Amplification Kit (Takara, China) according to the user manual. To confirm the composite open reading frame (ORF) sequences, RT-PCR was performed using two pairs of gene-specific primers upstream and downstream of the protein coding sequences. The PCR reactions were as follows: denaturation for 3 min at 94 °C; followed by 40 cycles of 94 °C for 30 s, 55–62 °C for 30 s, and 72 °C for 0.5–3 min; and a final extension step at 72 °C for 10 min. All PCR products were examined by agarose gel electrophoresis, purified using an E.Z.N.A. gel extraction kit (Omega BioTek, USA), and then sub-cloned into a pCR2.1 vector (Invitrogen, USA).
The deduced protein sequence alignments were performed using Clustal X 1.81 software (ClustalWeb: http://www.clustal.org/). The phylogenetic trees of GHR1 and GHR2 were constructed with MEGA 4 by the neighbor-joining method with 1000 replications (MegaWeb: http://www.megasoftware.net/). The transmembrane domains of both GHRs were predicted with the TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). Signal peptides were analyzed with SignalP 4.1 Server (http://www.cbs.dtu.dk/services/SignalP/) and the domains were searched with the Pfam database (PfamWeb: http://pfam.xfam.org/).
Real-time PCR
Real-time PCR was used to quantify the expression of scGH and scGHRs at the transcriptional level. The primers were designed based on the known GH, GHR1 and GHR2 cDNA sequences of S. chuatsi (EF205280.1, KJ477038.1, KJ477039.1). Those used in real-time PCR after screening are shown in Table 1. Real-time PCR was performed with Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen, USA) in a total volume of 10 µl on the Roche Light Cycler 480 Real-Time PCR detection system in 384-well plates according to the manufacturer’s instructions. Each reaction used 1 µl of the cDNA product (20 ng/µl), 0.4 µl of each gene-specific primer (10 µM), 5 µl of Platinum SYBR Green qPCR SuperMix, and 3.2 µl of nuclease-free H2O. Three replicates were set for all reactions, from which mean threshold cycle (Ct) values were calculated. Based on the T m value of primer pairs, the real-time PCR reaction conditions were as follows: UDG incubation at 50 °C for 2 min; initial denaturation at 95 °C for 2 min; and then 40 cycles of 95 °C for 15 s, 57 °C for 15 s, and 72 °C for 30 s. Fluorescence data were collected in the third step of each cycle. When amplification was complete, the melting curves were generated by raising the temperature from 60 to 95 °C in increments of 0.5 °C per 5 s to anneal, and a final denaturation at 95 °C for 10 s.
In a pilot study, expression levels of GAPDH and β-actin in different tissues were not as stable as 18S. The expression levels of 18S were correlated with input RNA concentration, indicating that 18S is a valid reference gene for the experiment. The standard curve for each gene was generated through a tenfold dilution series of plasmids PEASY-T1 (TransGen Biotec, China) containing the target or reference genes. The relative transcript amounts of scGHRs in fish were normalized to the level of 18S rRNA in the same sample by the 2−ΔCt method. The relative expressions of scGHRs in liver and muscle after exposure to fasting and re-feeding were calculated by the comparative CT method (2−ΔΔCt) using the lightcycler 480 Software 1.5 (ROCHE, CH) and indicated as n-fold differences compared to the control group.
Melting curve analyses for all new qPCR assays showed a single peak, confirming the specificity of PCRs. PCR efficiency varied between assays and between runs of a single assay, but was within a reasonable range (90–110 %). No bias was found across the PCR plate.
Tissue distribution of GHR1 and GHR2 transcripts
Three 5-month-old S. chuatsi fish with body weights of 80–120 g were anesthetized using MS-222 and killed by decapitation. Tissue samples of muscle, kidney, gonad, liver, stomach, heart, intestine, brain, gill, and pituitary were collected quickly and flash-frozen in liquid nitrogen and stored at −80 °C until RNA extraction.
Fasting and re-feeding experiments
Eighty-four fish weighing 145.5 ± 38 g, all offspring of the same pair, were randomly divided into 14 tanks representing two groups. The water temperature was maintained between 25 and 26 °C using an air conditioner. The fish were allowed to acclimate to the tanks for 1 month prior to the beginning of the experiment. Following the acclimation period, food was withheld from seven tanks for 4 weeks, while the remaining seven tanks were fed continuously throughout the experiment. After the 4 weeks, fasted and fed fish were fed continuously for the remaining 10 days. Twelve fish, specifically six fed and six fasted/re-fed (from two groups), were terminally sampled in buckets containing MS-222 (0.5 g/l) at seven time points (1–7). Time point 1 (or 2–4) represents fish that had been fasted for 1 (or 2–4) week(s), while 5, 6, and 7 represent 3, 7, and 10 days after re-feeding, respectively. To minimize the effects of crowding on the growth rate, stocking densities were maintained at approximately 3 g/l as fish were removed for sampling. The fish were sampled every week 18:00 after exposure to fasting and every 3 days 19:00 3 h after re-feeding. The liver, pituitary, muscle, and plasma samples of the fish were collected.
Plasma levels of GH by ELISA
Plasma was separated by centrifugation at 1000×g for 30 min at 4 °C, and stored at −20 °C until GH analysis. Plasma GH levels were measured by a GH ELISA Kit (Cusabio, WuHan, China) according to the manufacturer’s instructions. Three replicates were set for all reactions. The standard curve was generated through a dilution series of standard substance (r = 0.99).
Data analysis
Data are expressed as the mean ± SEM. Statistical analysis was performed with SPSS version 18.0 (SPSS Inc., Chicago, IL, USA). Significant differences were identified by a one-way ANOVA followed by Fisher’s least significance difference (LSD) test. A probability of less than 0.05 (p < 0.05) was considered significant.
Results
scGHR1 and scGHR2 sequences
Sequences covering the complete coding regions for GHR1 and GHR2 cDNAs from the liver of mandarin fish were designated as scGHR1 and scGHR2 (GenBank Accession No. KJ477038.1, KJ477039.1). The open reading frame of GHR1 contains 1914 bp and encodes a putative protein of 638 amino acids (Fig. 1), and shares 92 % identity with that of Epinephelus coioides [9] (Fig. 1). GHR1 contains a putative 28 amino acid signal peptide, an erythropoietin receptor (ligand-binding domain) of 104 amino acids, a transmembrane domain of 23 amino acids, and a GHR receptor-binding domain of 296 amino acids (Fig. 1). The open reading frame of GHR2 contains 1734 bp and encodes a putative protein of 578 amino acids (Fig. 2) that has 86 % identity to E. coioides [9]. GHR2 contains a putative 19 amino acid signal peptide, an erythropoietin receptor (ligand-binding domain) of 104 amino acids, a transmembrane domain of 23 amino acids, and a GHR-binding domain of 296 amino acids (Fig. 1). They both contain an extracellular binding FGEFS motif and a cytoplasmic box1 and box2 motif, which are also found in other fish such as E. coioides, Sparus aurata, and Oreochromis niloticus. Several characteristic landmarks of scGHRs have also been identified, including conserved extracellular cysteine residues and the potential N-glycosylation sites. A BLAST search using amino acid sequences of the GHRs revealed that several other conserved domains are also present, such as TFRCRW (at amino acid 56), WKECPEY (89), and PDPP (142) (Fig. 1). scGHR1 has a box1 motif of LLPPVPVP, while scGHR2 has a motif with a V to G replacement (Fig. 1). scGHR1 has a box2 motif of EPWVEFIEVD, while scGHR2 has a box2 motif of DPWVEFIDLD (Fig. 1). scGHR1 has seven extracellular cysteine residues, while scGHR2 has six (Fig. 1). The amino acid sequences of GHR1 and GHR2 in mandarin fish share 40 % identity.
Alignment of amino acid sequences of scGHRs and other vertebrate GHRs. Missed amino acids are marked by dashes. Three GHR protein family signature sequences—FGEFS motif, and conserved box1 and box2—are indicated by shaded regions. The putative signal peptide is in an open rectangle. Transmembrane region is in bold. Extracellular cysteines are double underlined. Potential N-glycosylation sites are in bold. Putative erythropoietin receptor (ligand binding) is underlined with broken line. Growth hormone receptor binding is underlined. The stop codon is denoted by asterisk. The GenBank accession numbers of the GHRs are as follows: Siniperca chuatsi GHR1 (KJ477038.1), Siniperca chuatsi GHR2 (KJ477039.1), Epinephelus coioides GHR1 (EF052273.1), Epinephelus coioides GHR2 (EF052274.1), Sparus aurata GHR1 (AF438176.2) Sparus aurata GHR2 (AY573601.2), Oreochromis niloticus GHR1 (NM_001279601.1), Oreochromis niloticus GHR2 (AY973233.1), Condylura cristata GHR (XM_004678346.1), Gallus gallus GHR (NM_001001293.1), Homo sapiens GHR (NP_000154.1)
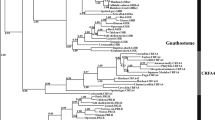
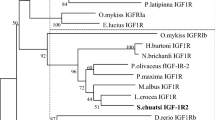
Phylogenetic analysis of teleost GHR1, GHR2 and somatolactin receptor (SLR). The phylogenetic tree was constructed by MEGA 4.0 using neighbor-joining method. Phylogeny test was performed by the bootstrap method with 1000 replicates. The number shown at each branch indicates the bootstrap value (%). GenBank Accession Nos. of fish GHR and SLR used are as follows: Siniperca chuatsi GHR1 (KJ477038.1); Siniperca chuatsi GHR2 (KJ477039.1); Epinephelus coioides GHR2 EF052274.1; Epinephelus coioides GHR1 EF052273.1; Oreochromis urolepis GHR1 EF371466.1; Oreochromis urolepis GHR2 EF371467.1; Sparus aurata GHR1 AF438176.2; Sparus aurata GHR2 AY573601.2; Oncorhynchus mykiss GHR1 NM_001124535.1; Oncorhynchus mykiss GHR2a AAW56611.1; Oncorhynchus mykiss GHR2b AAW27914.1; Danio rerio GHR1 NM_001083578.1; Danio rerio GHR2 NM_001111081.1; Acanthopagrus latus GHR1 JN399057.1; Acanthopagrus latus GHR2 JN399058.1; Anguilla japonica GHR1 AB180476.1; Anguilla japonica GHR2 AB180477.1; Lateolabrax japonicus GHR1 KF601961.1; Lateolabrax japonicus GHR2 KF770840.1; Megalobrama amblycephala GHR1 JN896373.1; Megalobrama amblycephala GHR2 JN896374.1; Pelteobagrus vachellii GHR1 JN392017.1; Pelteobagrus vachellii GHR2 JN392016.1; Takifugu rubripes GHR1 AB621336.1; Takifugu rubripes GHR2 AB621337.1; Oryzias latipes SLR XM011478846.1; Cichlasoma dimerus SLR FJ208943.1; Oreochromis mossambicus GHR1 AB115179.1; Oryzias latipes SLR XP011477148.1; Cichlasoma dimerus SLR ACI42879.1; Oreochromis mossambicus GHR1 BAD83668.1; Paralichthys adspersus GHR1 ABS29325.1; Cynoglossus semilaevis GHR1 AEO97318.1; Cynoglossus semilaevis GHR2 AEO97315.1; Salmo salar SLR ABY76179.1; Oncorhynchus masou SLR BAD51998.1; Mus musculus GHR EDL03404.1; Pseudopodoces humilis GHR XP_005533357.1; Pelodiscus sinensis GHR XP_006139558.1; Oncorhynchus kisutch GHR1 AAK95624.1; Oncorhynchus kisutch GHR2 AAK95625.1
Based on amino acid sequences, the neighbor-joining phylogenetic tree of the fish was constructed to illustrate the phylogenetic relationships of scGHR1 and scGHR2 to their counterparts in other known fish (Fig. 2). This tree places scGHR1 in the GHR1 clade and scGHR2 in the GHR2 clade.
A comparison of the amino acid sequences of mandarin fish GHRs with those from other fish (Table 2) shows that generally the sequence identities between GHR1 and GHR2 within a given species were low (below 39 %), with the exception of Oncorhynchus mykiss GHR1 (omGHR1) and omGHR2, which share 85.9 % identity, while the sequence identities with GHRs of other species were higher. S. chuatsi, E. coioides, Oreochromis urolepis, and S. aurata all belong to the Perciformes, and the identities of GHR1 and GHR2 between these species were all above 70 %.
Tissue distribution of receptors
The mRNA expression profiles of GHR1 and GHR2 in various tissues of mandarin fish were examined by real-time PCR (Fig. 3). Both GHR1 and GHR2 were detected in all tissues tested. For both GHR1 and GHR2, the highest mRNA abundance was observed in skeletal muscle, liver, and heart. When comparing GHR1 and GHR2 mRNA abundance, GHR1 was expressed at higher levels in muscle, brain, and gill than GHR2 (p < 0.05), while GHR2 mRNA was more abundant in liver, heart, gonad, pituitary gland, and kidney than GHR1 (P < 0.05). The lowest GHR1 and GHR2 mRNA abundance was found in the stomach and pituitary gland.
Relative expression of GHR1 and GHR2 mRNA in various tissues of Siniperca chuatsi. mRNA of 18S, GHR1, and GHR2 were quantified by real-time PCR (SYBR Green I), using 18S as a housekeeping gene. All data are represented as the mean ± SEM (n = 3). Different letters indicate significant differences at p < 0.05
Effects of fasting and re-feeding
Throughout the experiment, the body weight of fish in the fed group increased gradually, while that in the fasting group decreased over the 4-week fasting period (Fig. 4a). When re-feeding over the following 10 days, the body weight of previously fasted fish increased quickly, although it was still less than the mean body weight of the fed group. The specific growth rate (SGR) decreased from −0.33 ± 0.07 to −0.44 ± 0.06 in the fasted group over the 4 weeks of fasting. When re-fed, growth rate increased significantly to 1.57 ± 0.16, which was much higher than the continuously fed controls (0.8 ± 0.12, Fig. 4b).
Changes in body weight (a), specific growth rate (b), during 4 weeks of fasting and 10 days of re-feeding in Siniperca chuatsi. Vertical bars indicate ± SEM (body weight, specific growth rate, n = 6). Times of sampling: times 1, 2, 3, 4 mean 1, 2, 3, 4 weeks after fasting, respectively, times 5–7 means 3, 7, 10 days after re-feeding
In this experiment, we investigated the mRNA expression levels of GH in pituitary and GHR1 and GHR2 in liver and muscle of S. chuatsi during feeding, fasting, and re-feeding.
Expression of hepatic scGHR1 and scGHR2 mRNA initially decreased significantly to a lower level than that of the controls after 1 week of fasting (p < 0.05). Over the following 3 weeks of fasting, the levels of hepatic scGHR1 and scGHR2 mRNA were still lower than those of the controls (p < 0.05) (Fig. 5a, b). In muscle, scGHR1 mRNA levels increased to 1.22 times that of the controls after 1 week of fasting and significantly increased to 2.67 times after 4 weeks of fasting (p < 0.01, Fig. 5c). scGHR2 mRNA expression levels in the muscle were always more than those in the continuously fed controls during fasting (p < 0.05, Fig. 5d). GH mRNA in the pituitary of mandarin fish increased significantly during the first 3 weeks of fasting (p < 0.05) and decreased to a normal level after 4 weeks of fasting (Fig. 5e).
Effects of fasting and re-feeding on pituitary, liver and muscle expression of GH, GHR1, GHR2 in Siniperca chuatsi. After acclimated for a month, fish were divided into two groups (n = 42/group). As for fed or re-fed group, fish were fed or fasted for 38 days, and at the end of experiment, fish were fed or re-fed for 6 h before sampling; and in the fasted group, fish had been fasted before sampling. The mRNA expression levels of GHR1, GHR2 in liver (a, b), muscle (c, d), and GH in pituitary (e) of mandarin fish were quantified by real-time PCR and normalized against 18S transcripts. Times of sampling: times 1, 2, 3, 4 mean 1, 2, 3, 4 weeks of fasting, respectively; times 5–7 mean 3, 7, 10 days after re-feeding. Each bar represents the mean ± SEM. (n = 6). Different letters indicate significant differences at p < 0.05
During re-feeding, scGHR1 mRNA expression levels in the liver increased rapidly and were not significantly different from those of the continuously fed controls. scGHR2 initially increased to a level higher than in the controls after 3 days of re-feeding (p < 0.05), and then recovered to a normal level (Fig. 5a, b). There are no significant differences in muscle scGHR1 and scGHR2 mRNA expression during periods 5, 6, and 7 (re-fed period) between fed and re-fed fish (Fig. 5c, d). In addition, no significant differences were observed in scGHR1 and scGHR2 mRNA expression in the fed controls during re-feeding. The mRNA expression of GH in the pituitary increased to a level 5.1 times higher than the controls 3 days after re-feeding (p < 0.01) and decreased to a normal level 7 days after re-feeding (Fig. 5e).
Plasma levels of GH
Plasma GH concentrations in fasted fish over the 4 weeks were significantly higher than in the continuously fed controls (p > 0.05), but when re-fed, plasma concentration of GH decreased to a normal level similar to that of the continuously fed controls (Fig. 6).
Effects of fasting and re-feeding on plasma levels of GH. Vertical bars indicate mean ± SEM (n = 6). Significant differences from the corresponding fed control at the p < 0.05 level are indicated by different letters. Times of sampling: times 1, 2, 3, 4 mean 1, 2, 3, 4 weeks of fasting, respectively, times 5–7 mean 3, 7, 10 days after re-feeding
Discussion
In this study, the cloning, characterization, and expression analysis of two distinct mandarin fish GHR isoforms, namely scGHR1 and scGHR2, are reported for the first time. According to the deduced amino acid sequences, scGHR1 and scGHR2 both have a single short transmembrane domain, an extracellular erythropoietin receptor (ligand-binding) domain located at the C-terminus of the protein, and an intracellular GHR-binding domain, including an FGEFS motif and a box1 and box2 motif, as are characteristic of GHRs also found in Lateolabrax japonicus, rainbow trout, grouper, Nile tilapia, and seabream. The FGEFS motif is important for ligand-binding and signal transduction, as mutations of this motif in humans result in GH insensitivity [41]. The proline-rich box1 takes part in the JAK2–STAT signaling pathway, and internalization of the receptor is mediated by box2 [1]. All of the motifs were highly conserved in mandarin fish compared with the other fish (Fig. 1). Extracellular cysteine residues are involved in disulfide bonds and are structurally important in GHRs [7, 21]. In vertebrates, GHR1 has seven Cys residues with some variability at the last position; GHR2 mostly has five Cys residues with none in the fifth or sixth positions as GHR1 [21] (Fig. 1). The extracellular Cys residues are also highly conserved in scGHR1, while scGHR2 has one more Cys residue not found in other teleosts. On the other hand, the sequence identities between GHR1 and GHR2 within a given species were generally low (below 39 %), with the exception of omGHR1 and omGHR2, which share 85.9 % identity (Table 2). This suggests that there was a high rate of evolution from an ancestral GHR to GHR1 and GHR2 in teleosts.
Two distinct phylogenetic isoforms of GHRs were also identified in rainbow trout [13], Nile tilapia [14], orange-spotted grouper [15], black seabream [16], gilthead seabream [17], and Japanese seabass [19]. In the phylogenetic tree, scGHR1 was grouped with GHR1 and the somatolactin receptor (SLR) of other fish, while scGHR2 was grouped with GHR2 of other fish (Fig. 2). It is now known that descendants of the early sarcopterygians have only one copy of the GHR gene, as seen in the tetrapods, while the early actinopterygians underwent the fish-specific genome duplication (FSGD) and subsequently two distinct clades of GHRs within the teleosts appeared [21]. The exception that omGHR1 and omGHR2 share 85.9 % identity was caused by a more recent duplication (4R) event in several lineages of teleosts, including the salmonids. The tetraploid salmonids had duplicated GHR1/2 genes. Recently, a nomenclature system was proposed by Ellens et al. in which different numbers were used for genes derived from the FSGD; then, different letters for paralogs derived from a recent duplication and SLR should be classified to GHR1 [21]. This nomenclature is also confirmed by analysis of the evolution of receptors for GH and somatolactin in fish and land vertebrates by Fukamachi and Meyer [20]. Our results also support the nomenclature (Fig. 1, 2). Thus, by applying it, omGHR1 and omGHR2 should be designated as omGHR2a and omGHR2b (Table 2). To avoid confusion, the existing names in several species require revision.
A wide range of GHR1 and GHR2 expression levels have been reported in teleost species, including rainbow trout [13], Nile tilapia [14], orange-spotted grouper [15], black seabream [16], gilthead seabream [17], etc. In this experiment, both scGHR1 and scGHR2 were detected in all of the tissues tested, which were in agreement with the various functions of GH in growth, metabolism, reproduction, immunity, and osmoregulation [11, 42]. However, the amounts of scGHR mRNA varied by tissue and the highest expression levels of scGHR mRNA were found in liver and muscle (Fig. 3). Similar patterns have been found in rainbow trout [13], Japanese flounder [43], salmon [44], and black seabream [45]. It appears that GHRs play important roles in the growth and metabolism of fish [21]. Different expressions of GHRs mRNA in tissues have been reported in several teleosts, such as gilthead sea bream [17], Nile tilapia [14], and rainbow trout [13]. In gilthead sea bream, the expression level of GHR2 was significantly lower than GHR1 in liver and adipose tissue, but no significant difference was observed in muscle [17]. In Nile tilapia, GHR2 transcripts were significantly higher than GHR1 in most tissues but lower in muscle [14]. In mandarin fish, scGHR1 mRNA was expressed at higher levels than scGHR2 in muscle, brain, and gill, while the expression of scGHR2 mRNA was higher than that of scGHR1 in liver, heart, gonad, pituitary gland, and kidney. Although the reason why expressions of GHR1 and GHR2 between and/or within tissues in different species were different is not known, it might be ascribed to their long evolution history and to environmental and physiological conditions.
Nutritional state has a strong effect on the expression of GHRs in teleosts. The influences are different among fish species. In masu salmon, the expression of hepatic GHR1 mRNA was reduced in fish fasted for 3 weeks [44]. Skeletal muscle expression of GHR2 mRNA was significantly increased in hybrid striped bass after 3 weeks of complete food restriction, and hepatic GHR1 and GHR2 mRNA levels both declined during food deprivation [33], while the liver of fasting tilapia shows no changes in GHR1 transcript levels [22]. These different results might be due to the different tissues in various fish species and the sampling times. However, decrease of hepatic GHR1/2 mRNA levels and increase of muscle GHR1/2 mRNA levels under fasting conditions have been reported in more teleosts, such as black seabream [45], tilapia [22], and channel catfish [46]. The results in mandarin fish are in agreement with these species. In mandarin fish, the expression of scGHR1 and scGHR2 mRNA in liver decreased during fasting, and in the muscle, the expression of both mRNAs increased (Fig. 5a, b). The reduced expression of hepatic GHR would bring down GH sensitivity, which results in reduced IGF production and growth inhibition. Moreover, the high expression of muscle GHR might promote mobilization of lipids [4]. Thus, as a result of the consumption of lipids, emaciation was observed in mandarin fish during fasting (Fig. 4).
Re-feeding generally restored the fasting-associated changes in fish. Plasma GH, hepatic GHR mRNAs, and muscle GHR mRNAs recovered to pre-fasting levels [1]. In tilapia, muscle GHR1 mRNA declined to below control levels upon re-feeding for 1 and 2 weeks, but recovered 2 weeks after re-feeding, and there was no effect of re-feeding observed in the expression of GH receptor in the liver [47]. In hybrid striped bass, hepatic GHR1 mRNA increased after 10 days of re-feeding and recovered after 40 days of re-feeding. Further, there were no significant differences in muscle GHR1 mRNA expression between or within groups [33]. Although the changes in hepatic GHR mRNA and muscle GHR mRNA during re-feeding in mandarin fish were different, CG was observed and they supported the “restore” mechanism by recovering to normal levels during re-feeding.
GH is the main regulator that controls somatic growth in vertebrates [7, 48]. Changes in GH concentrations during fasting and re-feeding have been studied in many fish. In black seabream, serum GH levels increased progressively during starvation. After 30 days of starvation, serum GH levels were over three times those of the fed controls [45]. In channel catfish, pituitary GH mRNA increased while IGF-I mRNA and plasma IGF-I decreased when fasted for 14 or 28 days [49]. Plasma GH and pituitary GH mRNA levels were significantly increased in seawater-acclimated tilapia fasting for 4 weeks [50]. Two weeks of fasting produced a significant reduction in plasma IGF-I, but not in plasma GH in tilapia Oreochromis mossambicus acclimated to fresh water [51]. Together, these findings indicate that fasting increases plasma GH and pituitary GH mRNA levels in fish. It is believed that the starvation-induced increase in plasma GH could be due to a reduced feedback on the pituitary from the reduction of IGF-I level, in turn resulting from a reduction in hepatic GHR [52]. In mandarin fish, GH mRNAs in the pituitary and plasma GH concentrations both increased after fasting. This result is also in agreement with studies in salmonids [25, 34], tilapia [51], and catfish [49]. The abrupt increase in pituitary GH mRNAs at period 5 in mandarin fish might be ascribed to recovery of metabolism after the long-term starvation and 3 days of re-feeding. The different expression patterns of GH would be affected by environmental conditions and growth phases of different fish species during fasting and re-feeding [25]. Further studies are needed.
It has also been reported that 1 week of re-feeding can restore the pituitary GH mRNA back to its normal levels in E. coioides [53]. In rabbitfish, GH mRNA levels also returned to normal after 6 days of re-feeding [54]. The mRNA expression of GH in the pituitary of mandarin fish returned to a normal level 7 days after re-feeding. These results are in agreement with studies in E. coioides and rabbitfish [53, 54]. In mandarin fish, when the fish were re-fed, the plasma concentration of GH decreased to the same level as that of the continuously fed controls (Fig. 6). In rainbow trout, re-feeding for 2 weeks following 4 weeks of fasting also decreased plasma GH concentrations [25]. In isolated rainbow trout hepatocytes, GH treatment significantly increased the expression of GHR1 and GHR2 mRNA [55], while different results in vivo were found in this study (Fig. 5a, b, e). The increase in plasma GH concentration was accompanied by a decrease in hepatic scGHR1 and scGHR2 mRNA levels. It might be due to the complex regulatory systems in vivo [2, 4].
In summary, cDNAs of two distinct isoforms of GHRs (scGHR1 and scGHR2) have been isolated and characterized in mandarin fish. Both receptors, which arose through FSGD, have conserved the FGEFS motif and box1 and box2 motifs, as characteristic of GHRs also found in other species. They were most highly expressed in muscle and liver. The different expression profiles of scGHR1 and scGHR2 suggest distinct functions in different tissues. There was partial CG during 10 days of re-feeding after 4 weeks of fasting in mandarin fish. The response of GH and GHRs to variable growth phases, including negative, normal, and accelerated growth, has been described in detail with different sampling frequencies along the time course. The changes in GH, scGHR1, and scGHR2 mRNA expression during fasting and re-feeding suggest that they play important roles in the growth of mandarin fish. It has been reported that polymorphisms of GH and GHR genes are associated with growth traits in Boer goat breeds and could be a molecular marker for growth traits [56], so they could also be used as candidate genes for identifying growth-related molecular markers in the selective breeding of mandarin fish. The results provide comparative insights into the roles of these genes in the growth of teleost.
References
Walock CN, Kittilson JD, Sheridan MA (2014) Characterization of a novel growth hormone receptor-encoding cDNA in rainbow trout and regulation of its expression by nutritional state. Gene 533:286–294
Fuentes EN, Valdés JA, Molina A, Bjornsson BT (2013) Regulation of skeletal muscle growth in fish by the growth hormone-Insulin-like growth factor system. Gen Comp Endocrinol 192:136–148
Duan C (1998) Nutritional and developmental regulation of insulin-like growth factors in fish. J Nutr 128:306S–314S
Reindl KM, Sheridan MA (2012) Peripheral regulation of the growth hormone insulin-like growth factor system in fish and other vertebrates. Comp Biochem Physiol A: Mol Integr Physiol 163:231–245
Laviola L, Natalicchio A, Giorgino F (2007) The IGF-I signaling pathway. Curr Pharm Des 13:663–669
Wood AW, Duan C, Bern HA (2005) Insulin-like growth factor signaling in fish. Int Rev Cytol 243:215–285
LeRoith D, Bondy C, Yakar S, Liu JL, Butler A (2001) The somatomedin hypothesis. Endocr Rev 22:53–74
Devlin RH, Yesaki TY, Donaldson EM, Du SJ, Hew CL (1995) Production of germline transgenic Pacific salmonids with dramatically increased growth performance. Can J Fish Aquat Sci 52:1376–1384
Ahmed AS, Xiong F, Pang SC, He MD, Waters MJ, Zhu ZY, Sun YH (2011) Activation of GH signaling and GH-independent stimulation of growth in zebrafish by introduction of a constitutively activated GHR construct. Transgenic Res 20:557–567
Reinecke M, Björnsson BT, Dickhoff WW (2005) Growth hormone and insulin-like growth factors in fish: where we are and where to go. Gen Comp Endocrinol 142:20–24
Bjornsson BT, Johansson V, Benedet S, Einarsdottir IE, Hildahl J, Agustsson T, Jonsson E (2004) Growth hormone endocrinology of salmonids: regulatory mechanisms and mode of action. Fish Physiol Biochem 27:227–242
Pérez-Sánchez J, Calduch-Giner JA, Mingarro M, Vega-Rubín de Celis S, Gómez-Requeni P, Saera-Vila A, Astola A, Valdivia MM (2002) Overview of fish growth hormone family. New insights in genomic organization and heterogeneity of growth hormone receptors. Fish Physiol Biochem 27:243–258
Very NM, Kittilson JD, Norbeck LA, Sheridan MA (2005) Isolation, characterization, and distribution of two cDNAs encoding for growth hormone receptor in rainbow trout (Oncorhynchus mykiss). Comp Biochem Physiol 140B:615–628
Ma XL, Liu XC, Zhang Y, Zhu P, Ye W, Lin HR (2007) Two growth hormone receptors in Nile tilapia (Oreochromis niloticus): molecular characterization, tissue distribution and expression profiles in the gonad during the reproductive cycle. Comp Biochem Physiol 147B:325–339
Li Y, Liu X, Zhang Y, Zhu P, Lin H (2007) Molecular cloning, characterization and distribution of two types of growth hormone receptor in orange-spotted grouper (Epinephelus coioides). Gen Comp Endocrinol 152:111–122
Jiao B, Huang X, Chan CB, Zhang L, Wang D, Cheng CH (2006) The co-existence of two growth hormone receptors in teleost fish and their differential signal transduction, tissue distribution and hormonal regulation of expression in seabream. J Mol Endocrinol 36:23–40
Saera-Vila A, Calduch-Giner JA, Pérez-Sánchez J (2007) Co-expression of IGFs and GH receptors (GHRs) in gilthead sea bream (Sparus aurata L.): sequence analysis of the GHR-flanking region. J Endocrinol 194:361–372
Saera-Vila A, Calduch-Giner JA, Pérez-Sánchez J (2005) Duplication of growth hormone receptor (GHR) in fish genome: gene organization and transcriptional regulation of GHR type I and II in gilthead sea bream (Sparus aurata). Gen Comp Endocrinol 142:193–203
Zhang P, Chi ML, Wen HS, Qian K, Ni M, Zhang YC, Huang ZJ, Song ZF, Chai SH (2015) Cloning of growth hormone receptor and salinity effects on the expression of its related genes in seabass Lateolabrax maculatus. Oceanol Limnol Sin 46:446–453
Fukamachi S, Meyer A (2007) Evolution of receptors for growth hormone and somatolactin in fish and land vertebrates: lessons from the lungfish and sturgeon orthologues. J Mol Evol 65:359–372
Ellens ER, Kittilson JD, Hall JA, Sower SA, Sheridan MA (2013) Evolutionary origin and divergence of the growth hormone receptor family: insight from studies on sea lamprey. Gen Comp Endocrinol 192:222–236
Pierce AL, Fox BK, Davis LK, Visitacion N, Kitahashi T, Hirano T, Grau EG (2007) Prolactin receptor, growth hormone receptor, and putative somatolactin receptor in Mozambique tilapia: tissue specific expression and differential regulation by salinity and fasting. Gen Comp Endocrinol 154:31–40
Shimizu M, Fukada H, Hara A, Dickhoff W (2007) Response of the salmon somatotropic axis to growth hormone administration under two different salinities. Aquaculture 273:320–328
Poppinga J, Kittilson J, McCormick SD, Sheridan MA (2007) Effects of somatostastin on the growth hormone insulin-like growth factors axis and seawater adaptation of rainbow trout (Oncorhynchus mykiss). Aquaculture 273:312–319
Gabillard JC, Yao K, Vandeputte M, Gutierrez J, Le Bail PY (2006) Differential expression of two GH receptor mRNAs following temperature change in rainbow trout (Oncorhynchus mykiss). J Endocrinol 190:29–37
Sitjá-Bobadilla A, Calduch-Giner J, Saera-Vila A, Palenzuela O, Alvarez-Pellitero P, Pérez-Sánchez J (2008) Chronic exposure to the parasite Enteromyxum leei (Myxozoa: Myxosporea) modulates the immune response and the expression of growth, redox and immune relevant genes in gilthead sea bream, Sparus aurata L. Fish Shellfish Immunol 24:610–619
Saera-Vila A, Calduch-Giner JA, Prunet P, Pérez-Sánchez J (2009) Dynamics of liver GH/IGF axis and selected stress markers in juvenile gilthead sea bream (Sparus aurata) exposed to acute confinement: differential stress response of growth hormone receptors. Comp Biochem Physiol 154A:197–203
Norbeck LA, Sheridan MA (2010) Regulation of the growth hormone-insulin-like growth factor system by cortisol and thyroxine in rainbow trout. Annual Meeting of the Society for Integrative and Comparative Biology January, pp 3–7
Birzniece V, Sata A, Ho KK (2009) Growth hormone receptor modulators. Rev Endocr Metab Disord 10:145–156
Pierce AL, Shimizu M, Beckman BR, Baker DM, Dickhoff WW (2005) Time course of the GH/IGF axis response to fasting and increased ration in Chinook salmon (Oncorhynchus tshawytscha). Gen Comp Endocrinol 140:192–202
Ali M, Nicieza A, Wootton RJ (2003) Compensatory growth in fishes: a response to growth depression. Fish Fisheries 4:147–190
Peterson BC, Bilodeau-Burgeois AL, Small BC (2009) Response of the somatotropic axis to alterations in feed intake of channel catfish (Ictalurus punctatus). Comp Biochem Physiol 153A:457–463
Picha ME, Turano MJ, Tipsmark CK, Borski RJ (2008) Regulation of endocrine and paracrine sources of igfs and gh receptor during compensatory growth in hybrid striped sea bass (Morone chrysops × Morone saxatilis). J Endocrinol 199:81–94
Norbeck LA, Kittilson JD, Sheridan MA (2007) Resolving the growth-promoting and metabolic effects of growth hormone: differential regulation of GH-IGF-I system components. Gen Comp Endocrinol 151:332–341
Picha ME, Turano MJ, Beckman BR, Borski RR (2008) Endocrine biomarkers of growth and applications to aquaculture: a mini review of growth hormone, insulin-like growth factor (IGF)-I, and IGF-binding proteins and potential growth indicators in fish. N Am J Aquacult 70:196–211
Liang XF, Lin XT, Li SQ, Liu JK (2008) Impact of enviromental and innate factors on the food habit of Chinese perch Siniperca chuatsi (Basilewsky) (Percichthyidae). Aquacult Res 39:150–157
Chu WY, Fu GH, Chen J, Chen DG, Meng T, Zhou RX, Xia XJ, Zhang JS (2010) Gene expression profiling in muscle tissues of the commercially important teleost, Siniperca chuatsi. Aquacult Int 18:667–678
Luo D, Sun JJ, Lu X, Liu LZ, Chen SJ, Li GF (2011) Comparative sperm ultrastructure of three species in Siniperca (Teleostei:Perciformes: Sinipercidae). Micron 42:884–891
Wang PF, Zeng S, Xu P, Zhou L, Zeng L, Lu X, Wang HF, Li GF (2014) Identification and expression analysis of two HSP70 isoforms in mandarin fish Siniperca chuatsi. Fish Sci 80:803–817
Wang HF, Sun JJ, Lu X, Wang PF, Xu P, Zeng L, Yu DG, Li GF (2013) Identification of insulin-like growth factor I gene polymorphisms using high-resolution melting and its effect on growth traits in sinipercid species. Fish Sci 79:439–446
Langenheim JF, Tan D, Walker AM, Chen WY (2006) Two wrongs can make a right: dimers of prolactin and growth hormone receptor antagonists behave as agonists. Mol Endocrinol 20:661–674
Møller N, Jørgensen JO (2009) Effects of growth hormone on glucose, lipid, and protein metabolism in human subjects. Endocr Rev 30:152–177
Nakao N, Higashimoto Y, Ohkubo T, Yoshizato H, Nakai N, Nakashima K, Tanaka M (2004) Characterization of structure and expression of the growth hormone receptor gene of the Japanese flounder (Paralichtys olivaceus). J Endocrinol 182:157–164
Fukada H, Ozaki Y, Pierce AL, Adachi S, Yamauchi K, Hara A, Swanson P, Dickhoff WW (2004) Salmon growth hormone receptor: molecular cloning, ligand specificity, and response to fasting. Gen Comp Endocrinol 139:67–71
Deng L, Zhang WM, Lin HR, Cheng CH (2004) Effects of food deprivation on expression of growth hormone receptor and proximate composition in liver of black seabream Acanthopagrus schlegeli. Comp Biochem Physiol 137B:421–432
Small BC, Murdock CA, Waldbieser GC, Peterson BC (2006) Reduction in channel catfish hepatic growth hormone receptor expression in response to food deprivation and exogenous cortisol. Domest Anim Endocrinol 31:340–356
Fox BK, Breves JP, Davis LK, Pierce AL, Hirano T, Grau EG (2010) Tissue-specific regulation of the growth hormone/insulin-like growth factor axis during fasting and re-feeding: importance of muscle expression of IGF-I and IGF-II mRNA in the tilapia. Gen Comp Endocrinol 166:573–580
Björnsson BT, Johansson V, Benedet S, Einarsdottir IE, Hildahl J, Agustsson T (2002) Growth hormone endocrinology of salmonids: regulatory mechanisms and mode of action. Fish Physiol Biochem 27:227–242
Small BC, Peterson BC (2005) Establishment of a time-resolved fluoroimmunoassay for measuring plasma insulin-like growth factor I (IGF-I) in fish: effect of fasting on plasma concentrations and tissue mRNA expression of IGF-I and growth hormone (GH) in channel catfish (Ictalurus punctatus). Domest Anim Endocrinol 28:202–215
Fox BK, Riley LG, Hirano T, Grau EG (2006) Effects of fasting on growth hormone, growth hormone receptor, and insulin-like growth factor-I axis in seawater-acclimated tilapia, Oreochromis mossambicus. Gen Comp Endocrinol 148:340–347
Uchida K, Kajimura S, Riley LG, Hirano T, Aida K, Grau EG (2003) Effects of fasting on growth hormone/insulin-like growth factor-I axis in the tilapia, Oreochromis mossambicus. Comp Biochem Physiol 134A:429–439
Duan C, Plisetskaya EM (1993) Nutritional regulation of insulin-like growth factor-I mRNA expression in salmon tissues. J Endocrinol 139:243–252
Pedroso FL, de Jesus-Ayson EG, Cortado HH, Hyodo S, Ayson FG (2006) Changes in mRNA expression of grouper (Epinephelus coioides) growth hormone and insulin-like growth factor I in response to nutritional status. Gen Comp Endocrinol 145:237–246
Ayson FG, de Jesus-Ayson EG, Takemura A (2007) mRNA expression patterns for GH, PRL, SL, IGF-I and IGF-II during altered feeding status in rabbitfish. Gen Comp Endocrinol 150:196–204
Very NM, Sheridan MA (2007) Somatostatin regulates hepatic growth hormone sensitivity by internalizing growth hormone receptors and by decreasing transcription of growth hormone receptor mRNAs. Am J Physiol 292:R1956–R1962
An X, Wang L, Hou J, Li G, Song Y, Wang J, Yang M, Cui Y, Cao B (2011) Novel polymorphisms of goat growth hormone and growth hormone receptor genes and their effects on growth traits. Mol Biol Rep 38:4037–4043
Acknowledgments
This work was supported by the Educational Commission of Guangdong Province, China (No. cxzd1104), the Foshan Innovative and Entepreneurial Research Team Program (No. 2014IT100122), the Special Fund for Agro-scientific Research in the Public Interest (No. 201303048), the Science and Technology Planning Project of Guangdong Province (No. 2007A020300001-1; No. 2012A020800001; No. 2008A020100003), the Cooperative Project of Guangdong Province (No. 2011B090400179) and the Agriculture Science Technology Achievement Transformation project (No.2 012GB2E000338).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lu, X., Gu, Y., Hou, X. et al. Molecular characterization, tissue distribution, and expression regulation from fasting and re-feeding of two growth hormone receptors in mandarin fish Siniperca chuatsi . Fish Sci 82, 155–169 (2016). https://doi.org/10.1007/s12562-015-0941-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12562-015-0941-0