Abstract
Heat shock protein 70 (HSP70) plays important roles in multiple cellular stress responses. Two HSP70 isoforms, ScHSP70a and ScHSP70b, were identified from Siniperca chuatsi in this study. ScHSP70a and ScHSP70b shared all but one of their 639 amino acids and showed a remarkable homology to HSP70s of other species. Expression profile analysis by qRT-PCR revealed that both isoforms were detected throughout embryonic development, and a striking feature of ScHSP70a was its significant up-regulation during the crystal stage. ScHSP70a and ScHSP70b were expressed ubiquitously and at a low level in tissues under non-stressed conditions. However, they were dramatically induced by heat shock at different levels, and their induction was positively correlated with the increasing rate of temperature. Although insensitive to hypoxia in the heart, both genes were greatly induced by hypoxia in the liver, and the induction was returned to the basal level after re-oxygenation for 24 h. Additionally, Aeromonas hydrophila infection also markedly augmented ScHSP70a and ScHSP70b expression in a time-dependent manner in the head kidney and spleen, and the ScHSP70a induction levels were much higher than those of ScHSP70b. These results suggest that ScHSP70a and ScHSP70b contribute differently to embryonic development and protection against damage from high temperature, hypoxia and bacterial infection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Heat shock proteins (HSPs) are a super family of ubiquitous and evolutionarily highly conserved proteins, which function as chaperone molecules to maintain the cellular protein homeostasis under both normal and physiological stress conditions [1, 2]. HSPs modulate the folding and assembly of nascent proteins, refold or degrade damaged proteins, assist in the membrane translocation of organellar and secretory proteins, as well as regulate signal transduction [3, 4]. According to their sequence homology and approximate molecular weights, HSPs are generally classified into several conserved groups: HSP100, HSP90, HSP70, HSP60 and small HSPs [5]. Of these proteins, HSP70s are thought to be the central components of the cellular network of molecular chaperones and folding catalysts [3]. The HSP70 family is composed mainly of two subgroups, the constitutive heat shock cognate 70 proteins (HSC70s) and the inducible HSP70s. HSC70 is actively expressed in non-stressed cells and remains unchanged or only mildly induced upon stress stimulation [6]. It is involved in many cellular functions, such as protein translocation, cell apoptosis and embryonic development [6]. In contrast, HSP70 can be strongly induced from low basal levels by various environmental stresses, such as heat shock, heavy metals, hypoxia and pathogenic infection. It is required for a variety of cellular processes and for the response to environmental changes [7]
In teleosts, genes or cDNAs of HSP70 have been identified in a number of species, such as Oncorhynchus mykiss [8], Danio rerio [9], Oreochromis mossambicus [10], Xiphophorus maculatus [11] and Ctenopharyngodon idella [12], and observations of expression patterns in these studies suggested that HSP70 plays a vital role in enhancing the survival and health of cells or whole organisms upon being stressed. It is noted that there are multiple HSP70 isoforms in mammals [7, 13] and Drosophila melanogaster [14], and these isoforms differ with respect to their expression patterns. Human HSP70 isoforms show different expression profiles in the developmental stages, among different tissues and under stress conditions [13] and D. melanogaster HSP70 isoforms reveal the characteristics of tissue- and development-specific induction after heat shock and during recovery [14]. In teleosts, however, only a few species such as Oryzias latipes, D. rerio, O. mykiss, X. maculatus and Fundulus heteroclitus macrolepidotus have been reported to have two or more HSP70 isoforms [2, 11, 15, 16], and information about different expression patterns between teleost HSP70 isoforms during embryonic development, as well as in different tissues upon being stressed, remains rare.
The mandarin fish, Siniperca chuatsi, is an economically important fish and has been widely cultured in China [17]. With the development of intensive production in the aquaculture industry, various environmental stresses, including high temperature, hypoxia and Aeromonas hydrophila infection, the causative agent of outbreaks of bacterial hemorrhagic septicemia disease, have frequently threatened the health of this fish, causing serious economic losses [18, 19]. Understanding the expression characteristics and roles of HSP70 during stresses will contribute to improving the abilities of fish to tolerate stress and resist disease. However, the HSP70 gene and its response to environmental stresses are basically unknown in S. chuatsi. Therefore, in the present study, we identified and characterized two distinct inducible HSP70 isoforms in S. chuatsi and comparatively assessed their expression during embryonic development, on exposure to heat shock, hypoxia, and an A. hydrophila challenge, using real-time qRT-PCR. This study provides a better understanding of the correlations between the expression of HSP70 isoforms in S. chuatsi and during its embryonic development, as well as different environmental stresses, and lays the foundation for further study of the mechanism of anti-adverseness in S. chuatsi.
Materials and methods
Animals and sampling
Mandarin fish S. chuatsi weighing approximately 120–140 g were obtained from BaiRong Aquatic breeding Co., Ltd (Guangdong, China). The fish were maintained in a circulating water system containing a series of 400-l tanks at 25 °C for at least 2 weeks prior to processing. The fish were fed with fresh juvenile Cirrhinus molitorella at a ratio of approximately 5 % of the total biomass. For sampling, the fish were anesthetized with tricaine methanesulfonate (MS-222) and tissue samples were rapidly removed by team dissection, snap-frozen in liquid nitrogen, and stored at −80 °C until RNA extraction. All experiments were approved by the Animal Research and Ethics Committees of Sun Yat-sen University and performed in accordance with the guidelines of the committee.
Total RNA and genomic DNA isolation
Total RNA was extracted from tissues with EZNA total RNA kit II (Omega Bio-Tek, USA) according to the manufacturer’s instructions. Genomic DNA was isolated from the muscle using a TIANamp Genomic DNA Kit (Tiangen Biotec, China). The concentration and quality of RNA/DNA were verified by spectrophotometer and electrophoresis on 1.0 % agarose gel.
Cloning of full-length cDNAs and genomic DNAs of ScHSP70 isoforms
A schematic diagram of cloning of HSP70 full-length cDNAs from S. chuatsi is shown in Fig. 1. The fish used in cloning was exposed to heat shock at 34 °C for 60 min, and then tissues, including heart, head kidney, liver and muscle were sampled. First-strand cDNA synthesis was performed using 2 μg of DNase I-treated total RNAs with the first-strand cDNA synthesis using M-MLV for RT-PCR kit (Invitrogen) according to the manufacturer’s instructions. The cDNAs of the four tissues were mixed and then used as the template for polymerase chain reaction (PCR). Two couples of degenerate primers, HSP-F1/HSP-R1 and HSP-F2/HSP-R2 (Table 1), based on the highly conserved regions of amino acid sequences of HSP70 family members of other fish, were designed to amplify the HSP70 cDNA fragments from S. chuatsi using the nested PCR.
Cloning strategy of HSP70a and HSP70b full-length cDNAs from Siniperca chuatsi. Arrows represent primers for intermediate fragment, 5′- and 3′-end amplification, and lines under them indicate their amplification sequences. The open reading frames (ORFs) are indicated by dark gray boxes, and the untranslated regions (UTRs) are indicated by light gray boxes
Full-length cDNA sequences of HSP70s were obtained by the technique of rapid amplification of cDNA ends (RACE). The 5′-RACE PCR was performed with the purified first-strand cDNA as the template, which had a poly(C) end added to the 5′ terminal, and nested primers HSP70-R1 and HSP70-R2 (Table 1) as the reverse primers and an abridged anchor primer (AAP) or an abridged universal amplification primer (AUAP) as the forward primers. Based on the two distinct 5′-untranslated regions (UTRs) of HSP70 cDNAs we obtained, forward gene-specific primers for ScHSP70a and ScHSP70b (Table 1) were designed for two rounds of 3′-RACE PCR. The anchor primer (AP) and AUAP were used as the reverse primers in the 1st and 2nd rounds, respectively. In addition, in order to verify the correctness of assembly of the intermediate fragment, a 5′-RACE fragment and a 3′-RACE fragment of each of the cDNA gene-specific primers ORF-HSP70a-F/-R for ScHSP70a and ORF-HSP70b-F/-R for ScHSP70b (Table 1) were designed to amplify their complete ORFs using first-strand cDNA as the template, and the PCR products were sequenced.
Amplifications for cloning the ScHSP70 cDNAs were performed as follows: denaturation at 94 °C for 3 min, followed by 35 cycles at 94 °C for 30 s, 55–60 °C for 30 s and 72 °C for 1–2.5 min, and a final extension of 10 min at 72 °C. The PCR amplification products were purified with a Universal DNA Purification Kit (Tiangen Biotec, China) and cloned into pEASY-T1 cloning vector (TransGen Biotec). Recombinants were identified by blue/white screening and confirmed by PCR, and then positive ones were selected and sequenced.
The genomic DNA sequences coding ScHSP70a and ScHSP70b were amplified from total genomic DNA by gene-specific primers ORF-HSP70a-F/-R and ORF-HSP70b-F/-R, respectively, which were located at the terminus of 5′- and 3′-UTRs of ScHSP70s cDNA sequences. PCR conditions for the genomic DNA sequence cloning were as follows: initial denaturation at 94 °C for 3 min, followed by 38 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s and extension at 72 °C for 3 min, and a final extension of 10 min at 72 °C. The PCR amplification products were cloned into pEASY-T1 cloning vectors (TransGen Biotec, China) and sequenced.
BLAST searches and sequence alignments
Nucleotide and amino acid sequences that are homologous to ScHSP70 were retrieved by the BLAST programs at the National Center for Biotechnology Information (http://www.blast.ncbi.nlm.nih.gov/). Protein sequence alignments were performed by ClustalX 2.1 (http://www.clustal.org), and then used for constructing the phylogenetic tree with MEGA5.2 software according to the neighbor-joining method [20]. The compute pI/MW tool of ExPASy (http://www.expasy.org/proteomics) was used to calculate the molecular weight (MW) and isoelectronic point (pI) of deduced protein of S. chuatsi HSP70s. The program ScanProsite (http://www.us.expasy.org/tools/scanprosite/) was used to search the protein motifs.
Embryos and tissues
Embryos were obtained by natural crossing. Fertilized eggs were hatched in three vertical fish egg incubators. The clean and oxygenated water with a temperature of 27 °C entered incubators continuously under pressure from below and kept the eggs in a suspended state. Embryos at different embryonic stages including fertilized eggs, 16-cell stage, morula, blastula, gastrula, closure of blastopore, appearance of myomere, tail-bud stage, muscle burl stage, crystal stage, blood circulating stage, pre-hatched larvae and 1-day post-hatched larvae (1 dph) were collected and snap-frozen in liquid nitrogen. Thirty embryos or twelve larvae in every stage from each incubator were collected as one sample.
A range of tissues including brain, heart, gill, head kidney, liver, spleen, muscle, stomach, intestines and opisthonephros from the fish (body weight, 150–160 g; body length, 16.0–16.5 cm) reared in non-stressed conditions at 25 °C were sampled and used for analysis the tissue distribution of the two ScHSP70 isoforms by real-time qRT-PCR.
Heat shock treatment
To examine the expression patterns of two S. chuatsi HSP70 isoforms in response to thermal stress, the fish were subjected to three regimes of thermal stress: acute, fast and gradual heat shock. For acute heat shock, 20 fish (mean body weight of 140 ± 20 g) maintained in the water at 25 °C were carefully and directly shifted to a tank of 2,000 l in the circulating water system at a constant temperature of 34 °C. Six fish were randomly collected after each heat treatment (2, 6 and 12 h), and another six individuals maintained at 25 °C served as the control group.
In the other two regimes of thermal stress, fish were transferred to the tank of 2,000 l in the circulating water system (initial temperature: 25 °C), and then the water temperature was elevated from 25 to 38.8 °C (semilethal temperature acquired from our preliminary experiment) at an average speed of 1.2 °C increase per hour for fast heat shock and 0.2 °C increase per hour for gradual heat shock. Six fish were randomly sampled when the temperature reached 34 °C, six more at 38.8 °C, and another six individuals maintained at 25 °C were taken as the control group.
Hypoxia exposure and re-oxygenation
Twenty-four fish (mean body weight of 130 ± 20 g) acclimated to a normal oxygen environment (dissolved oxygen, DO: ≥5.6 mg/l, temperature: 25 °C) for 1 week were randomly and evenly divided into four equal groups (A–D): A, exposure to a low oxygen level of 0.9 ± 0.1 mg/l of DO for 4 h; B, re-oxygenation under normal oxygen for 24 h after the 4-h hypoxia; C and D, exposure to normal oxygen, served as control groups for A and B, respectively. The dissolved oxygen of 0.9 ± 0.1 mg/l was maintained by controlling the water and nitrogen inflows. Dissolved oxygen was monitored continuously using a YSI Model 550A dissolved oxygen meter (Geo Scientific Ltd, USA).
Bacterial challenge
Sixty healthy S. chuatsi (140 ± 15 g, BW) were used for a challenge of virulent bacteria A. hydrophila, provided by MOE Key Laboratory of Aquatic Product Safety, Sun Yat-sen University. Each fish was intraperitoneally injected with a total of 0.5 ml of A. hydrophila (5 × 108 CFU/ml), which was diluted with sterile phosphate buffer solution (PBS, pH 7.4). Another 30 fish injected with the same volume of aseptic PBS were used as the control group. Six individuals from the treatment and control groups were randomly collected and tissues were sampled at 6, 12, 24, 48 and 72 h after injection, respectively.
Real-time qRT-PCR
Quantitative real-time PCR (real-time qRT-PCR) was used to quantify the expression of ScHSP70s at the transcriptional level according to MIQE validation guidelines [21]. 18S rRNA was selected as the reference gene, because it was stably expressed among different tissues and upon stress exposure in the present study. Total RNA (0.9 μg) from each sample was reverse-transcribed using a PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa, Japan). Specific primers (Table 1) were designed to amplify PCR fragments for ScHSP70a, ScHSP70b and S. chuatsi 18S rRNA. The amplification was performed on the LightCycler® 480 II Real-Time PCR System (Roche) using SYBR® Premix Ex Taq™ II (Tli RNaseH Plus) (TaKaRa, Japan) in 384-well plates. The final reaction volume was 10 μl consisting 1.0 μl of cDNA sample (10 ng/μl), 0.4 μl of each gene-specific primer (10 μM), 3.2 μl of nuclease-free H2O and 5 μl of 2× SYBR Premix Ex Taq II. All reactions were carried out in three technical replicates, from which mean threshold cycle (CT) values were calculated. Based on Tm value of primer pairs, cycling conditions were designed as: initial denaturation step at 95 °C for 30 s, followed by 40 cycles of denaturation at 95 °C for 5 s, annealing at 58 °C for 20 s and extension at 72 °C for 30 s, then a dissociation curve step (95 °C for 1 s, 60 °C for 20 s and 95 °C for 1 s) was conducted to verify the PCR specificity.
The standard curve for ScHSP70a, ScHSP70b and S. chuatsi 18S rRNA was generated through a 10-fold dilution series of plasmids pEASY-T1 (TransGen Biotec, China) containing the target or reference genes. Amplification efficiencies (E) of real-time qPCR were determined by employing the equation E = 10(−1/slope) − 1. Here, amplification efficiencies of ScHSP70a, ScHSP70b and S. chuatsi 18S rRNA were 100, 99.4 and 104.6 %, respectively.
The relative transcript amounts of HSP70s of the experimental fish were normalized to the level of 18S rRNA in the same sample by the 2−ΔCt method and the relative expression levels of HSP70s after exposure to heat shock, hypoxia and bacterial challenge were determined by the 2−ΔΔCt method and indicated as n-fold differences relative to the control group.
Statistical analysis
Statistical analyses were performed using SPSS software version 18.0 (SPSS Inc., Chicago, IL, USA). Data presented in this study were analyzed using one-way ANOVA followed by Tukey’s test and expressed as mean ± SD.
Results
Cloning and characterization of two distinct ScHSP70 isoforms
Considering the possibility that more than one isoform of HSP70 exists in S. chuatsi, we carefully designed a RACE strategy to clone their cDNA sequences (Fig. 1). Firstly, a mixture of total cDNAs from multiple tissues of the heat-treated fish was used as the PCR amplification template to avoid certain isoforms being expressed in a tissue-specific manner. Secondly, the primers for intermediate fragment cloning were designed at conserved regions of the HSP70 but with high degeneracy. As a result, two intermediate cDNA fragments with extremely high nucleotide sequence identity (>99 %) were isolated, and their 5′-UTRs were amplified by the same 5′-RACE primers. The sequence differences between their 5′-UTRs allowed us to further design their specific 3′-RACE primers. Finally, two full-length cDNAs were successfully obtained. BLAST analysis of nucleotide and deduced amino acid sequences of the two cDNAs revealed that they belong to the HSP70 family. Sequence alignment showed that they share 99.8 % identity with a single substitution of Arg-Ser at position 623, indicating that these two transcripts represent distinct isoforms of HSP70. Thus, in the present study, they are designated as ScHSP70a and ScHSP70b and deposited in GenBank under the accession numbers JX500752 and KF150224, respectively.
The full length of ScHSP70a cDNA sequence was 2,224 bp [excluding the poly(A) tail], containing an open reading frame (ORF) of 1,920 bp, a 5′-UTR of 104 bp and a 3′-UTR of 200 bp with a polyadenylation signal (AATAAA) located 14 nucleotides upstream from the poly(A) tail. The deduced amino acid sequence encoded a protein of 639 amino acids with a calculated molecular mass of 70.20 kDa and a pI of 5.41. The complete ScHSP70b cDNA was 2,348 bp in length [excluding the poly(A) tail], consisting of a 5′-UTR of 110 bp, and 3′-UTR of 318 bp with a polyadenylation signal (AATAAA) located 16 bases before the poly(A) tail and an ORF of 1,920 bp encoding a protein of 639 amino acids with a calculated MW of 70.13 kDa and a pI of 5.35.
ScHSP70a and ScHSP70b were composed of an N-terminal ATPase domain (amino acids, aa 1–383), a peptide (substrate)-binding domain (aa 387–545) and a C-terminal domain (aa 539–622) [6]. Three signature motifs of the HSP70 family were identified in both ScHSP70a and ScHSP70b sequences (signature 1, IDLGTTYS; signature 2, IFDLGGGTFDVSIL; and signature 3, VVLVGGSTRIPKIQK) (Fig. 2). The other conserved motifs, such as the cytoplasmic characteristic EEVD motif known to be the site of interaction with some co-chaperones, the bipartite nuclear localization signal (KRKHKKDIGQNKRALRRRTACERAKRT), that is involved in the selective translocation of HSP70 into the nucleus, and the non-organellar stress protein motif RARFEEL were also identified in both ScHSP70 isoforms (Fig. 2).
Alignment of amino acid sequences of ScHSP70s and other vertebrate HSP70s. Identical amino acid residues are indicated by dots. Missed amino acids are marked by dashes. Three HSP70 proteins family signature sequences are indicated by shaded regions. The putative bipartite nuclear localization signal is underlined and the non-organellar motif RARFEEL is marked by a dashed underline. The C-terminal EEVD motif is marked by double underlines. The GenBank accession numbers of the HSP70s are as follows: Siniperca chuatsi HSP70a (AFU54391), Siniperca chuatsi HSP70b (AGS83422), Lates calcarifer (AEH27544), Oreochromis niloticus (ACI42865), Lutjanus sanguineus (ADO32584) Sciaenops ocellatus (ADL18372), Fundulus heteroclitus macrolepidotus (ABB17042), Xiphophorus maculatus (BAB72168), Danio rerio (BAB72170), Homo sapiens (NP_005336), Mus musculus (NP_034609)
Using the ORF primers (Table 1) close to the 5′- and 3′-terminals, genomic DNA fragments of ScHSP70a (GenBank accession no. KF500540) and ScHSP70b (GenBank accession no. KF500541) were isolated, with lengths of 2,980 and 2,661 bp, respectively. Genomic DNA structure analysis revealed that both genes contained no introns in their coding regions, which meant the uninterrupted ORF of 1,920 bp encoded the entire protein. However, there were two introns with lengths of 780 and 331 bp within the two 5′-UTRs, respectively (Fig. 3).
Genomic structures of ScHSP70a and ScHSP70b. Wide boxes represent exons, and narrow boxes indicate introns. The ORFs are indicated by dark gray boxes, and the UTRs are indicated by light gray boxes. Boxes between wide and narrow represent the terminus region in the UTRs of the cDNAs which were not amplified in the genomic DNA. E Exon, I intron, UTR untranslated regions
Sequence comparison and phylogenetic analysis
The homology analysis based on the amino acid sequences revealed that the predicted HSP70a and HSP70b protein of S. chuatsi displayed similar amino acid residues to those of other vertebrates (Fig. 2). The highest identity (95 %) was with Lates calcarifer, Oreochromis niloticus and Lutjanus sanguineus. Identities with the other species were 94 % (Sciaenops ocellatus), 93 % (F. h. macrolepidotus), 92 % (X. maculatus) and 87 % (D. rerio). They also shared 87 and 85 % identities with human and mouse HSP70s, respectively.
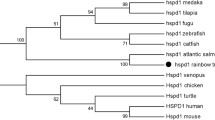
In the phylogenetic tree constructed with HSP70s and HSC70s in vertebrates (Fig. 4), the fish HSP70 family members were clearly clustered into two distinct groups, fish HSC70 group and fish HSP70 group, respectively. And ScHSP70a and ScHSP70b identified in the present study were clustered into the fish HSP70 group, revealing that the ScHSP70a and ScHSP70b were HSP70s rather than HSC70s. In addition, ScHSP70a and ScHSP70b showed distant relationship to the mammalian MHC-linked HSP70 group and mammalian HSP70B′ group.
Phylogenic tree for the HSP70 and HSC70 amino acid sequences of vertebrates. The tree was generated by MEGA 5.2 software using the neighbor-joining method, following ClustalX 2.1. Scale bar indicates an evolutionary distance of 0.02 amino acid substitutions per position in the sequence. Bootstrap values of greater than 50 % are indicated (1,000 replicates). The four clusters of fish HSC70, fish HSP70, mammalian MHC-linked HSP70 and mammalian HSP70B′ are boxed. ScHSP70a and ScHSP70b identified from Siniperca chuatsi in this study are underlined. Sequence database accession numbers in GenBank are indicated in parentheses. HSC70 Heat shock cognate 70 protein, MHC major histocompatibility complex
Expression of ScHSP70a and ScHSP70b during embryonic developmental stages
The mRNA expression levels of ScHSP70a and ScHSP70b were detected throughout the embryonic developmental stages from fertilized eggs to 1-day post-hatched larvae (Fig. 5). During embryonic developmental stages, the expression of both genes showed high values and no significant changes from fertilized eggs to blastula. The expression level of ScHSP70a mRNA decreased from the stage of gastrula to tail-bud stage, significantly increased during the muscle burl stage and crystal stage, and then decreased again and was maintained at low levels until 1 dph. The mRNA expression level of ScHSP70b also decreased from the stage of gastrula to a low level, at which it was maintained until 1 dph except for a moderate increase at pre-hatched larvae stage.
Expression of ScHSP70a a and ScHSP70b b during embryonic developmental stages of Siniperca chuatsi. The mRNA levels were quantified by real-time qRT-PCR and normalized by 18S rRNA. Thirty embryos or 12 larvae in each stage were collected as one sample. Each bar represents the mean ± SD of three samples. Different letters above the bars indicate significant difference (P < 0.05). 1 Fertilized eggs, 2 16-cell stage, 3 morula, 4 blastula, 5 gastrula, 6 closure of blastopore, 7 embryo body stage, 8 tail-bud stage (optic vesicle), 9 muscle burl stage, 10 crystal stage, 11 blood circulating stage, 12 pre-hatched larvae, 13 1 day post-hatched larvae (dph)
Tissue expression of ScHSP70a and ScHSP70b
Under normal physiological conditions, transcripts of ScHSP70a and ScHSP70b were universally detected in brain, heart, gill, head kidney, liver, spleen, muscle, stomach, intestines and opisthonephros (Fig. 6) at substantially different levels. ScHSP70a was expressed at the highest level in the intestines, with lower levels in the head kidney and opisthonephros, and at the lowest level in muscle. ScHSP70b had the highest level in the brain, followed by the intestines, and was almost undetectable in the liver. In general, mRNA expression of ScHSP70a was higher than that of ScHSP70b in all tissues except for the brain in which the expression of the two HSP70 isoforms was nearly equal.
Expression patterns of ScHSP70a and ScHSP70b after heat shock exposure
To investigate the expression patterns of ScHSP70a and ScHSP70b after heat shock exposure, the fish were exposed to three regimes of heat shock: acute, fast and gradual heat shock. The experimental temperatures in the three regimes were based on the fact that S. chuatsi has a natural temperature range of between 15 and 32 °C [22] with an upper lethal limit of 34 °C under acute heat stress, and approximately 39 °C under fast and gradual heat shocks measured in the present study. The expression levels of ScHSP70a and ScHSP70b in heart, liver and head kidney in the time-course of acute heat shock exposure are shown in Fig. 7. The results showed that acute heat shock for 2 h at 34 °C resulted in a strong up-regulation of ScHSP70a by averages of 663.7-, 184.0- and 89.1-fold of that in controls in heart, liver and head kidney, respectively. Meanwhile, ScHSP70b expression was also dramatically increased by averages of 4,782.3-, 2,063.3- and 877.4-fold in heart, liver and head kidney, respectively. Although ScHSP70b showed lower basal expression levels under non-stressed conditions, its increased level was more remarkable than that of ScHSP70a in heart and liver following 2-h heat shock treatment. The time-course results revealed that the expression levels of ScHSP70a and ScHSP70b peaked after 2 h of heat shock exposure, but began to decrease towards control levels thereafter. Compared to ScHSP70b, it took more time for ScHSP70a to return its expression to the control levels.
Expression of ScHSP70a and ScHSP70b during acute heat shock exposure. The mRNA levels in heart (a), liver (b) and head kidney (c) in the time-course were measured by real-time qRT-PCR and normalized by 18S rRNA. Fish that were maintained at 25 °C over the entire experimental period were taken as the control group. Data presented are expressed as mean ± SD (n = 6). Different letters above the bars represent significant difference (P < 0.01)
The expression levels of ScHSP70a and ScHSP70b in heart under fast heat shock (elevating temperature rate: ~1.2 °C/h) and gradual heat shock (elevating temperature rate: ~0.2 °C/h) were also investigated. Results showed that the expression of both genes was significantly up-regulated by the two regimes of heat shock when the temperature elevated from 25 °C to 34 and 38.8 °C (Fig. 8). Under fast heat shock from 25 to 34 °C, the mRNA levels of ScHSP70a and ScHSP70b were increased by averages of 906.3- and 3,240.6-fold, respectively, which continued to be increased with the temperature elevating to 38.8 °C, by averages of 3,708.5- and 18,450.2-fold, respectively. Likewise, gradual heat shock also resulted in up-regulation of ScHSP70a and ScHSP70b, but was much lower than those under fast heat shock, by averages of 208.2- and 108.5-fold, respectively, at 34 °C and by 2,017.1- and 8,031.7-fold, respectively, at 38.8 °C.
Expression of ScHSP70a a and ScHSP70b b under fast and gradual heat shock regimes. The elevating temperature rate in the fast and gradual heat shock regimes were 1.2 and 0.2 °C/h, respectively. The mRNA levels were quantified by real-time qRT-PCR and normalized by 18S rRNA. Data presented are expressed as mean ± SD (n = 6)
Expression of ScHSP70a and ScHSP70b after hypoxia exposure and re-oxygenation
In order to know whether hypoxia influences the expression of ScHSP70a and ScHSP70b, their mRNA levels in heart and liver in response to hypoxia and re-oxygenation were detected and the results are shown in Fig. 9. In heart, ScHSP70a and ScHSP70b mRNA levels tended to increase after hypoxia treatment but this increase did not reach significance. Although ScHSP70a was not significantly induced in the heart by hypoxia, its mRNA level was increased to 2.1-fold during re-oxygenation compared with the control. Meanwhile, the re-oxygenation treatment had no effects on ScHSP70b mRNA expression in the heart. However, the expression of both ScHSP70a and ScHSP70b was greatly increased in liver upon 4-h acute hypoxia exposure, by 286.2- and 1,303.0-fold, respectively, and declined back to their baselines during re-oxygenation over 24 h.
Expression of ScHSP70a and ScHSP70b in response to hypoxia. The mRNA levels in heart (a) and liver (b) were measured by real-time qRT-PCR and normalized by 18S rRNA. Fish were sampled after continuous exposure to severe hypoxia conditions (DO 0.9 ± 0.1 mg/l) for 4 h. After 4 h of hypoxia the DO levels were adjusted back to normoxic levels within 20 min and fish were sampled after 24 h of re-oxygenation. Data presented are expressed as mean ± SD (n = 6). Different letters above the bars represent significant difference (P < 0.05)
Expression of ScHSP70a and ScHSP70b after A. hydrophila infection
The expression patterns of ScHSP70a and ScHSP70b in head kidney and spleen from 6- to 72-h post-infection with A. hydrophila were examined by real-time qRT-PCR, and the results are shown in Fig. 10. Since there were no significant changes in the mRNA levels of both ScHSP70 isoforms during the 72 h after injection with PBS (P > 0.05), their respective average expression level was used as a control. Analysis showed that treatment with A. hydrophila induced ScHSP70a and ScHSP70b mRNA expression from 6- to 24-h post-infection in head kidney and spleen. During the time-course experiment, the ScHSP70a mRNA level was initially markedly increased and reached the highest level of 25.1-fold in the head kidney and 5.4-fold in the spleen at 6 h in comparison to control groups, and gradually decreased to the baselines thereafter; the expression of ScHSP70b was also significantly up-regulated and reached the highest level of 15.4- and 10.7-fold in head kidney and spleen, respectively, at 6 h post-infection compared with control groups, and then fluctuated from 6- to 48-h post-induction until returned to the baselines. In addition, the induction of ScHSP70a mRNA was much higher than that of ScHSP70b at any time point post-infection in both immune tissues.
Expression of ScHSP70a and ScHSP70b after A. hydrophila infection. The mRNA levels in head kidney (a) and spleen (b) in the time-course were quantified by real-time qRT-PCR and normalized by 18S rRNA. Data presented are expressed as mean ± SD (n = 6). Different letters above the bars indicate significant difference among different time points (P < 0.05)
Discussion
The present study is the first report of cloning, characterization and expression analysis of two distinct S. chuatsi HSP70 isoforms, ScHSP70a and ScHSP70b. According to the deduced amino acid sequences, ScHSP70a and ScHSP70b share all but one (Arg-Ser at position 623) of their 639 amino acids, being more than 99 % identical. Their deduced amino acid sequences show a remarkable homology (84–95 %) to HSP70s of other species, as much as 95 % with that of L. calcarifer, O. niloticus and L. sanguineus. They have perfectly conserved N-terminal ATPase and peptide-binding domains, and a less conserved C-terminal domain compared with HSP70s from other species. Their C-terminal EEVD motif, non-organellar RARFEEL motif and bipartite nuclear localization signal indicate they are nuclear-cytosolic HSP70s [23]. Another striking feature of ScHSP70a and ScHSP70b genes is the absence of introns in their coding regions. Such a gene structure is characteristic of inducible HSP70 genes, rather than the constitutively expressed HSC70 genes [24]. It has been suggested that lack of introns in the HSP70 gene allows the mRNA to be rapidly translated into a nascent protein by circumventing the block in RNA splicing upon cellular stress [25, 26]. Real-time qRT-PCR experiments revealed that ScHSP70a and ScHSP70b mRNAs are expressed at low levels in various tissues under non-stressed conditions, but are dramatically induced after heat shock. Taken together, these observations strongly suggest that ScHSP70a and ScHSP70b sequences identified in the present study correspond to the inducible nuclear-cytosolic HSP70.
It has been suggested that HSP70 isoforms differ with respect to their expression patterns during developmental stages, in different tissues and upon stresses. For instance, human HSP70-1a and HSP70-1b genes are expressed at their highest levels in juvenile tissues while HSP70-6 is most expressed in the neonate [7]. HSP70-1a and HSP70-1b are heat-inducible while HSP70-1t is not induced by heat stress and HSP70-6 is only induced after severe stress insults [7]. In RTG-2 cells of O. mykiss, HSP70a and HSP70b displayed similar profiles in both the temperature-shift and time-course heat shock experiments, but at different expression levels [15]. Cadmium slightly induced the expression of HSP70-1 in platyfish cultured tailfin cells, but it did not alter the HSP70-2 expression [11]. To investigate the expression patterns of the ScHSP70a and ScHSP70b identified in the present study, their mRNA levels during embryonic development, in different tissues and under different stress conditions, including heat shock, hypoxia and A. hydrophila infection were determined by real-time qRT-PCR.
During embryonic development, ScHSP70a and ScHSP70b showed similar expression patterns in the early embryonic developmental stages from fertilized eggs to the tail-bud stage. Their mRNAs, maintained at relatively high levels from fertilized eggs until the blastula stage, are considered as maternal origin according to the demonstration that zygotic transcription is activated until the midblastula transition (MBT) period [27]. Thereafter, their mRNA levels decreased, and were maintained at low levels until the tail-bud stage. At the later embryonic developmental stages, however, the expression patterns of ScHSP70a and ScHSP70b differed obviously, from the muscle burl stage to crystal stage in particular. The low mRNA expression level of ScHSP70b was maintained until 1 dph except for a moderate increase at pre-hatched larvae stage, while the expression of ScHSP70a was significantly increased from the muscle burl stage to crystal stage, suggesting that ScHSP70a plays a particular role during these stages. This speculation can be supported by the observation that zebrafish HSP70 is required for embryonic lens formation [28, 29].
ScHSP70a and ScHSP70b showed different expression patterns in tissue distribution and after exposure of heat shock, hypoxia and A. hydrophila infection. Both genes were expressed at low levels in all tissues tested, while in most cases ScHSP70a levels were much higher than ScHSP70b levels. Although expressed at low levels under non-stressed conditions, both ScHSP70a and ScHSP70b were strongly heat-inducible. Their mRNA levels were dramatically increased in heart, liver and head kidney after exposure to 34 °C for 2 h. The quick and high expression of ScHSP70a and ScHSP70b suggests that they are directly involved in the resistance of organisms to heat stress. In the time-course experiments, ScHSP70a and ScHSP70b revealed very similar expression patterns. Their mRNA induction levels were initially markedly increased and then decreased gradually towards their control levels following the non-lethal acute heat shock. This tendency is generally consistent with the induction pattern of HSP70a and HSP70b in RTG-2 cells of rainbow trout upon heat stress [15]. Another finding was that ScHSP70b had much higher increasing rates than ScHSP70a in all the three tissues, indicating the induction of ScHSP70b is more sensitive than that of ScHSP70a on exposure to heat shock. It is clear that transcriptional induction of HSP70 requires the binding of activated heat shock transcription factors (HSFs) to heat shock elements (HSEs) located within the HSP70 gene promoter regions [30, 31]. Therefore, it can be speculated that the different induction levels between ScHSP70a and ScHSP70b may be related to the characteristics of their HSEs. Meanwhile, their induction varies among tissues. Both ScHSP70a and ScHSP70b are more stongly expressed in heart than in liver and head kidney after heat shock, which may be associated with the different HSF activities in these tissues.
The expression patterns of ScHSP70a and ScHSP70b in heart under fast and gradual heat shock regimes were further investigated. Their expression levels up-regulated gradually with the increasing temperature under both temperature regimes; however, their induction rate during gradual heat shock was slower than during fast heat shock. These data indicate that the induction of ScHSP70s is positively correlated with the elevated temperature as well as the increasing rate of temperature.
It is well established that HSP70 protects mammalian cells against ischemic and hypoxic injury [32, 33]. However, in fish, less is known about the relationship between HSP70 gene expression and hypoxic stress [34]. Previous reports showed that hypoxia significantly induced the expression of HSP70 mRNA in embryos of D. rerio [35], as well as in gill, spleen and liver of Sebastes schlegeli [36]. The combined expression of HSP70/HSC70 protein in blood, brain and muscle of juvenile O. niloticus was also highly induced during hypoxia [37]. These findings suggest that a particular level of HSP70 expression is required during hypoxia [34]. Similarly, mRNA expression of both ScHSP70a and ScHSP70b was also elevated in liver of S. chuatsi after hypoxia treatment in this study, which indicates that ScHSP70a and ScHSP70b may contribute to the resistance of S. chuatsi for hypoxic injury. However, no significant changes of ScHSP70a and ScHSP70b expression were observed in heart after hypoxia exposure compared to those under normoxic conditions, revealing their expression upon hypoxia is in a tissue-specific manner. Stenslokken et al. [34] demonstrated that distinct HSP70 isoforms in Carassius carassius showed different expression profiles under hypoxia, which was also found in our study. The mRNA accumulation of ScHSP70a in liver was far greater than that of ScHSP70b following hypoxia exposure. In heart, the ScHSP70a level was significantly enhanced after re-oxygenation for 24 h, while ScHSP70b expression was still maintained at the control level. These differences between ScHSP70a and ScHSP70b expression under hypoxia/re-oxygenation suggest that ScHSP70a may play more roles in cytoprotection against hypoxia injury. It is documented that HSP70-2 in human hepatocellular carcinoma (HCC) and hepatoma (HepG2) cells was strongly increased under hypoxic conditions, and HSP70-2 up-regulation is due to the direct binding of hypoxia-inducible factor to hypoxia-responsive elements (HREs) in HSP70-2 promoter [38]. However, whether this mechanism is true for teleost fish still needs further elucidation.
In mammals, HSP70 is demonstrated to protect the organism against pathogens not only by functioning as a molecular chaperone, but by mediating the innate immune responses [39]. Enhanced HSP70 can protect cells by inhibiting the production of pro-inflammatory cytokines and promoting the expression of anti-inflammatory cytokines [40, 41]. HSP70 is released into extracellular compartments and functions as an endogenous “danger signal” to promote immune responses that are involved in protection of the cytoplasmic components against bacterial infection [42, 43]. Moreover, HSP70 serves as a ligand for Toll-like receptors to modulate pathogen-associated molecular pattern (PAMP)-induced Toll-like receptor (TLR) signaling, which is beneficial to stimulation of innate immunity and elimination of pathogens [39]. In fish, organisms responding to pathogens by altering HSP70 mRNA synthesis has also been reported. Infection with Vibrio harveyi increased L. sanguineus HSP70 mRNA level in the head kidney [44]. HSP70 mRNA expression was augmented in Sparus sarba after a Vibrio alginolyticus challenge [45], as was the case for Epinephelus coioides, where HSP70 mRNA expression peaked at 24 h post-infection with live V. alginolyticus [46]. After being challenged with A. hydrophila, hepatic HSP70 mRNA expression of Megalobrama amblycephala Yih was significantly increased at first, reaching a peak at 6 h post-infection and then decreasing [47]. Likewise, a clear time-dependent mRNA expression pattern of ScHSP70a in both head kidney and spleen of S. chuatsi was observed in this study when infected with A. hydrophila. On the other hand, ScHSP70b showed a different expression pattern after A. hydrophila infection. Although ScHSP70b was also up-regulated at 6 h post-infection in both immune tissues, its induction fluctuated from 6- to 48-h post-induction until it returned to the control levels. The difference in the expression profiles of ScHSP70a and ScHSP70b post-infection may be affected by complicated factors that are not clear. Still, their up-regulation after A. hydrophila infection indeed indicates that the two ScHSP70 isoforms participate in the fish immune response during bacterial infection. Further investigation might help the development of better disease management strategies for S. chuatsi.
In summary, two distinct HSP70 isoforms, ScHSP70a and ScHSP70b, were identified and characterized from the mandarin fish S. chuatsi for the first time. Their amino acid sequence characteristic features, DNA structures and mRNA expression patterns strongly suggest that the two ScHSP70 isoforms correspond to the inducible nuclear-cytosolic HSP70. Furthermore, mRNA expression patterns of ScHSP70a and ScHSP70b during embryonic development and after exposure to different environmental stresses were analyzed by real-time qRT-PCR. The results showed that both genes were expressed at low levels in various tissues under normal physiological conditions, but were strongly induced by stress stimulation in a time-dependent or tissue-specific manner. These observations indicate that they participate in different ways in embryonic development and in protection against damage from high temperature, hypoxia and bacterial infection. The identification and expression analysis of the two ScHSP70 genes have laid the foundation for study of the anti-adverse mechanism as well as possible improvement of stress tolerance and disease resistance of S. chuatsi.
References
Roberts RJ, Agius C, Saliba C, Bossier P, Sung YY (2010) Heat shock proteins (chaperones) in fish and shellfish and their potential role in relation to fish health: a review. J Fish Dis 33:789–801
Yamashita M, Yabu T, Ojima N (2010) Stress protein HSP70 in fish. Aquat BioSci Monogr 3:111–141
Mayer MP, Bukau B (2005) Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci 62:670–684
Li Z, Zhao X, Wei Y (2004) Regulation of apoptotic signal transduction pathways by the heat shock proteins. Sci China C Life Sci 47:107–114
Basu N, Todgham AE, Ackerman PA, Bibeau MR, Nakano K, Schulte PM, Iwama GK (2002) Heat shock protein genes and their functional significance in fish. Gene 295:173–183
Liu T, Daniels CK, Cao S (2012) Comprehensive review on the HSC70 functions, interactions with related molecules and involvement in clinical diseases and therapeutic potential. Pharmacol Ther 136:354–374
Brocchieri L, Conway DME, Macario AJ (2008) hsp70 genes in the human genome: conservation and differentiation patterns predict a wide array of overlapping and specialized functions. BMC Evol Biol 8:19. doi:10.1186/1471-2148-8-19
Kothary RK, Jones D, Candido EP (1984) 70-Kilodalton heat shock polypeptides from rainbow trout: characterization of cDNA sequences. Mol Cell Biol 4:1785–1791
Santacruz H, Vriz S, Angelier N (1995) Cloning and characterization of hsp70 cDNA during embryonic development of zebrafish (Danio rerio). Biol Cell 84:110
Molina A, Biemar F, Muller F, Iyengar A, Prunet P, Maclean N, Martial JA, Muller M (2000) Cloning and expression analysis of an inducible HSP70 gene from tilapia fish. FEBS Lett 474:5–10
Yamashita M, Hirayoshi K, Nagata K (2004) Characterization of multiple members of the HSP70 family in platyfish culture cells: molecular evolution of stress protein HSP70 in vertebrates. Gene 336:207–218
Zhang A, Zhou X, Wang X, Zhou H (2011) Characterization of two heat shock proteins (Hsp70/Hsc70) from grass carp (Ctenopharyngodon idella): evidence for their differential gene expression, protein synthesis and secretion in LPS-challenged peripheral blood lymphocytes. Comp Biochem Physiol B Biochem Mol Biol 159:109–114
Daugaard M, Rohde M, Jaattela M (2007) The heat shock protein 70 family: highly homologous proteins with overlapping and distinct functions. FEBS Lett 581:3702–3710
Lakhotia SC, Prasanth KV (2002) Tissue- and development-specific induction and turnover of hsp70 transcripts from loci 87A and 87C after heat shock and during recovery in Drosophila melanogaster. J Exp Biol 205:345–358
Ojima N, Yamashita M, Watabe S (2005) Comparative expression analysis of two paralogous Hsp70s in rainbow trout cells exposed to heat stress. Biochim Biophys Acta 1681:99–106
Fangue NA, Hofmeister M, Schulte PM (2006) Intraspecific variation in thermal tolerance and heat shock protein gene expression in common killifish, Fundulus heteroclitus. J Exp Biol 209:2859–2872
Lao HH, Sun YN, Yin ZX, Wang J, Chen C, Weng SP, He W, Guo CJ, Huang XD, Yu XQ, He JG (2008) Molecular cloning of two C1q-like cDNAs in mandarin fish Siniperca chuatsi. Vet Immunol Immunopathol 125:37–46
Chen CF, Li J (1996) Studies on virulence and isolation of pathogenic bacteria causing bacterial septicemia in mandarin fish (Siniperca chuatsi Basilewsky). J Huazhong Agric Univ 15:370–373 (in Chinese)
Xuan CW, Liang XF, Yu R, Fu Y, Ye W (2012) The plankton composition in mandarin fish Siniperca chuatsi ponds and bait fish ponds in hot season. Chin J Fish 25:25–29 (in Chinese)
Hall BG (2013) Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol 30:1229–1235
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622
Li MF (2010) Research progress on biology of mandarin fish. Mod Fish Inf 7:16–21 (in Chinese)
Boorstein WR, Ziegelhoffer T, Craig EA (1994) Molecular evolution of the HSP70 multigene family. J Mol Evol 38:1–17
Yost HJ, Lindquist S (1986) RNA splicing is interrupted by heat shock and is rescued by heat shock protein synthesis. Cell 45:185–193
Kay RJ, Russnak RH, Jones D, Mathias C, Candido EP (1987) Expression of intron-containing C. elegans heat shock genes in mouse cells demonstrates divergence of 3′ splice site recognition sequences between nematodes and vertebrates, and an inhibitory effect of heat shock on the mammalian splicing apparatus. Nucleic Acids Res 15:3723–3741
Huang Y, Wimler KM, Carmichael GG (1999) Intronless mRNA transport elements may affect multiple steps of pre-mRNA processing. EMBO J 18:1642–1652
Kane DA, Kimmel CB (1993) The zebrafish midblastula transition. Development 119:447–456
Blechinger SR, Evans TG, Tang PT, Kuwada JY, Warren JJ, Krone PH (2002) The heat-inducible zebrafish hsp70 gene is expressed during normal lens development under non-stress conditions. Mech Dev 112:213–215
Evans TG, Yamamoto Y, Jeffery WR, Krone PH (2005) Zebrafish Hsp70 is required for embryonic lens formation. Cell Stress Chaperones 10:66–78
Iwama GK, Thomas PT, Forsyth RB, Vijayan MM (1998) Heat shock protein expression in fish. Rev Fish Biol Fish 8:35–56
Murtha JM, Keller ET (2003) Characterization of the heat shock response in mature zebrafish (Danio rerio). Exp Gerontol 38:683–691
Tokyol C, Karaorman G, Bastug M (2005) Effects of acute and adaptive hypoxia on heat shock protein expression in hepatic tissue. High Alt Med Biol 6:247–255
Latchman DS (2001) Heat shock proteins and cardiac protection. Cardiovasc Res 637–646
Stenslokken KO, Ellefsen S, Larsen HK, Vaage J, Nilsson GE (2010) Expression of heat shock proteins in anoxic crucian carp (Carassius carassius): support for cold as a preparatory cue for anoxia. Am J Physiol Regul Integr Comp Physiol 298:R1499–R1508
Ton C, Stamatiou D, Dzau VJ, Liew CC (2002) Construction of a zebrafish cDNA microarray: gene expression profiling of the zebrafish during development. Biochem Biophys Res Commun 296:1134–1142
Mu W, Wen H, Li J, He F (2013) Cloning and expression analysis of a HSP70 gene from Korean rockfish (Sebastes schlegeli). Fish Shellfish Immunol 35:1111–1121
Delaney MA, Klesius PH (2004) Hypoxic conditions induce Hsp70 production in blood, brain and head kidney of juvenile Nile tilapia Oreochromis niloticus (L.). Aquaculture 236:633–644
Huang WJ, Xia LM, Zhu F, Huang B, Zhou C, Zhu HF, Wang B, Chen B, Lei P, Shen GX (2009) Transcriptional upregulation of HSP70-2 by HIF-1 in cancer cells in response to hypoxia. Int J Cancer 124:298–305
Sung YY, MacRae ATH (2011) Heat shock proteins and disease control in aquatic organisms. J Aquac Res Dev S 2:006. doi:10.4172/2155-9546.S2-006
Wang X, Zou Y, Wang Y, Li C, Chang Z (2001) Differential regulation of interleukin-12 and interleukin-10 by heat shock response in murine peritoneal macrophages. Biochem Biophys Res Commun 287:1041–1044
Shi Y, Tu Z, Tang D, Zhang H, Liu M, Wang K, Calderwood SK, Xiao X (2006) The inhibition of LPS-induced production of inflammatory cytokines by HSP70 involves inactivation of the NF-kappaB pathway but not the MAPK pathways. Shock 26:277–284
Wallin RPA, Lundqvist A, Moré SH, von Bonin A, Kiessling R, Ljunggren H (2002) Heat-shock proteins as activators of the innate immune system. Trends Immunol 23:130–135
Campisi J, Leem TH, Fleshner M (2003) Stress-induced extracellular Hsp72 is a functionally significant danger signal to the immune system. Cell Stress Chaperones 8:272–286
Zhang X, Pang H, Wu Z, Jian J (2011) Molecular characterization of heat shock protein 70 gene transcripts during Vibrio harveyi infection of humphead snapper, Lutjanus sanguineus. Fish Physiol Biochem 37:897–910
Deane EE, Li J, Woo NY (2004) Modulated heat shock protein expression during pathogenic Vibrio alginolyticus stress of sea bream. Dis Aquat Organ 62:205–215
Cui M, Zhang QZ, Yao ZJ, Zhang ZH (2011) Molecular cloning and expression analysis of heat-shock protein 70 in orange-spotted grouper Epinephelus coioides following heat shock and Vibrio alginolyticus challenge. J Fish Biol 79:486–501
Ming J, Xie J, Xu P, Liu W, Ge X, Liu B, He Y, Cheng Y, Zhou Q, Pan L (2010) Molecular cloning and expression of two HSP70 genes in the Wuchang bream (Megalobrama amblycephala Yih). Fish Shellfish Immunol 28:407–418
Acknowledgments
This research was funded by the National Key Technology R&D Program, China (No. 2012BAD25B04), the Educational Commission of Guangdong Province, China (No. cxzd1104), the Science and Technology Planning Projects of Guangdong Province, China (No. 2012A020800001, No. 2008A020100003, No. 2007A020300001-1). Funding was also provided by the Agriculture Science Technology Achievement Transformation project (No. 2012GB2E000338) and the Cooperative Project of Guangdong Province, China (No. 2011B090400179).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, P., Zeng, S., Xu, P. et al. Identification and expression analysis of two HSP70 isoforms in mandarin fish Siniperca chuatsi . Fish Sci 80, 803–817 (2014). https://doi.org/10.1007/s12562-014-0747-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12562-014-0747-5