Abstract
Rel/NF-κB transcription factors play critical roles in induction and regulation of innate immune response in organisms. In this study, the full length of a Relish homolog cDNA from Exopalaemon carinicauda named EcRelish was 2141 bp encoding a 660 amino-acid polypeptide. EcRelish cDNA contained a conserved Rel homology domain and two nucleus localization signals. Sequence analysis indicated that the deduced amino acid sequence of the EcRelish showed high similarities to that of other crustaceans. Real time RT-PCR analysis showed that EcRelish mRNA expressed with different levels in tested tissues, and the highest expression was observed in the hemocytes. With longer infection time, the cumulative mortality rates increased gradually followed by the proliferation of Vibrio anguillarum and WSSV. The expression profiles of EcRelish gene were analyzed after V. a nguillarum, white spot syndrome virus (WSSV) challenge, and ammonia-N stress. The results showed that the expression levels of EcRelish mRNA in the hemocytes were up-regulated at 1–24 h after V. a nguillarum challenge. Meanwhile, the expression levels of EcRelish mRNA were up-regulated at 3 h after WSSV challenge. The expression of EcRelish in hemocytes was down-regulated significantly under ammonia-N stress during the experimental time. The results indicated that EcRelish might be involved in immune defense against pathogens and ammonia-N stress in E. c arinicauda.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The ridgetail white prawn Exopalaemon carinicauda, which is distributed along the eastern coast of China, has contributed to one-third of the gross outcome of the polyculture ponds [1, 2]. Due to the merits of fast growth, high reproductive ability, and good environmental adaptability and trophic miscellaneous, the culture scale of E. c arinicauda has expanded in recent years [3]. However, with the development of intensive culture and the deterioration of ecological environment, some environmental stressors, highly virulent bacteria, or virus caused frequent outbreaks of diseases that resulted in huge economic losses [4–6]. Therefore, deep understanding of the immune mechanism of the ridgetail white prawn is necessary for healthy culture of crustaceans.
As an invertebrate, the ridgetail white prawn relies on the innate immunity to defense of the microbial invaders by activating various immune genes [7]. Antimicrobial peptides (AMPs) are one of the major components of the shrimp immune system [8]. In Drosophila melanogaster, AMP genes are regulated by the Toll and Imd signal pathways [9]. Toll and Imd pathways play different roles in resisting different pathogens by controlling different kinds of antimicrobial peptides [10, 11]. Previous studies have demonstrated that the Toll pathway was activated by gram-positive bacteria and fungi, while the Imd pathway responded to gram-negative bacteria infection [12–15]. When the organism is invaded by some pathogens, it will release one signal, which can be received by pattern-recognition receptors. Then, the signal will be transferred to activate the cell nuclear transcription factors (NF-κB) by a series of cascade reactions [9]. NF-κB family genes include Relish, Dorsal, and Dif. In the Imd pathway, Relish are activated to induce the gene expression of antibacterial peptides such as the anti-lipopolysaccharide factor (ALF) [16, 17], while Dorsal and Dif are activated in the successive signaling cascade of the Toll pathway for antifungal and antibacterial responses inducing product antimicrobial peptides such as penaeidins and crustin [8, 18–23].
The research of the immune signaling pathways and their regulatory mechanisms were initially reported in the fruit fly Drosophila [24]. Then some Relish genes were cloned from the mosquito Aedes aegypti [25] and Anopheles gambiae [26]. To date, some Rel/NF-κB family genes have been cloned in some crustacean species, including Penaeus monodon [27], Fenneropenaeus chinensis [28], Litopenaeus vannamei [29], and Eriocheir sinensis [30]. Normally, the full length Relish consists of a Rel homology domain (RHD), a nucleus localization signal, an IkB-like domain containing six ankyrin (ANK) repeats, and a death domain. The Relish is activated by cleaving away its C-terminal half, ANK repeats, and a death domain, with a caspase enzyme [23]. While the C-terminal fragment remains in the cytoplasma waiting for degradation, the N-terminal fragment containing the RHD translocates into the nucleus to regulate the synthesis of cognate antimicrobial proteins [63, 64]. The alternative splicing produced two types of REL gene: a full-length form (REL-F) includes the RHD, C-terminal ANK repeats and death domains, while a shorter form (REL-S) lacks ANK repeats or death domains [25, 46]. Two isoforms of Relish, both short and full length, have been reported in A. g ambiae [26], F. c hinensis [28, 46], and L. v annamei [29]. Neither REL-S nor REL-L has been reported in E. c arinicauda.
As an important environmental factor, toxic levels of ammonia can not only affect growth, molting, oxygen consumption, and ammonia excretion, but also affect the immune response of shrimp [31]. In crustaceans, there are lots of reports to support the hypothesis that ammonia nitrogen (ammonia-N) can decrease the value of immune parameters such as total haemocyte count (THC), prophenoloxidase (proPO), superoxide dismutase(SOD), peroxidase (POD), and expression of genes implicated in the defence system in L. v annamei [32], Marsupenaeus japonicus [33], and Macrobrachium rosenbergii [34]. Furthermore, NF-κB activation appears to contribute to the mechanism of ammonia-induced astrocyte swelling in fulminant hepatic failure (FHF) [35] and activate inducible nitric oxide synthase (iNOS) gene in Heteropneustes fossilis [36]. However, until recently, as a crucial element in the biological response to stress, little was known about the effect of ammonia on NF-κB in crustaceans. In this study, a full-length cDNA of Relish gene was cloned from E. c arinicauda (named EcRelish) and the gene structure was analyzed. The mortality and the proliferation of Vibrio anguillarum and white spot syndrome virus (WSSV) were recorded after infected with pathogens. In addition, the expression patterns of EcRelish in various tissues and the expression profiles of EcRelish in hemocytes after V. a nguillarum, WSSV challenge, and ammonia-N stress were investigated. These results will be essential to better understand the physiological function of Relish in the shrimp immune response to gram-negative bacteria, WSSV, and ammonia-N stress.
Materials and methods
Animals and rearing conditions
Healthy E. c arinicauda with body length of 4.37 ± 1.05 cm and body weight of 1.33 ± 0.76 g (n = 30) were collected from a commercial farm in Qingdao, China. They were reared in 200 l polyvinyl chloride polymer (PVC) tanks containing aerated sand-filtered seawater (salinity of 30 ppt, pH 7.8–8.4) at 22–25 °C for 7 days before processing. There were 40 shrimps in each tank. During the acclimation period, two thirds of the water in each tank was renewed twice daily and the shrimps were fed daily with a ration of 10 % of the body mass.
RNA extraction and cDNA synthesis in hemocytes
Hemolymph was extracted from the hearts of four individuals using 1 ml syringes containing 0.4 ml of cold anticoagulant (1.588 g sodium citrate, 3.92 g sodium chloride, 4.56 g glucose, 0.66 g EDTA-2Na, 200 ml ddH2O) [65] and placed into 1.5 ml plastic tubes. Then, the hemocytes were collected after the hemolymph was centrifuged at 4000 rpm (centrifuge instrument: Eppendorf/5415R, Germany; rotor: F-45-24-11), at 4 °C for 10 min. Total RNA was extracted from hemocytes using Trizol Reagent according to the manufacturer’s protocol (Invitrogen, USA), and contaminating genomic DNA was eliminated using RQ1 RNase-free DNase (Promega, USA) following the manufacturer’s instructions. RNA quantity, purity and integrity were verified spectrophotometrically (OD260/OD280) and by electrophoresis on 1.5 % agarose gels. The cDNA was synthesized from 2 μg of total RNA by M-MLV reverse transcriptase (Promega, USA) at 42 °C for 60 min with oligo (dT) 18 primer following the protocol of the manufacturer.
PCR and cloning of EcRelish cDNA
Full-length EcRelish cDNA of E. c arinicauda was obtained by the procedures of reverse transcription polymerase chain reaction (RT-PCR) and rapid amplification of the cDNA ends (RACE) method. The degenerate primers pairs F1 and R1 (Table 1) were designed based on the highly conserved nucleotide sequence of Relish of F. c hinensis (EU815055), L. v annamei (EF432734), P. m onodon (JQ728539), Carcinoscorpius rotundicauda (DQ345784), and E. s inensis (GQ871279) (http://www.ncbi.nlm.nih.gov/) applying DNAMAN v6.0.3.99 (Lynnon Biosoft) to multiple sequence alignment. The PCR reaction conditions were: 5 min initial denaturation at 94 °C for one cycle, then 30 cycles of 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 1 min, and 72 °C for 10 min. A partial EcRelish cDNA fragment of 587 bp (the annealing sites of the degenerate primers F1: 215–236 bp, R1: 781–801 bp) was obtained from the pair of degenerate primers. Based on the partial sequence data of EcRelish, its 3′ and 5′ ends were obtained using a SMART™ RACE cDNA Amplification Kit (Clontech, USA). The 3′ end RACE PCR reaction was performed with cDNA template from hemolymph RNA using the gene specific primer F2 and the anchor primer UPM (Table 1). The PCR reaction conditions were 1 cycle of 94 °C for 3 min, 5 cycles of 94 °C for 30 s, 72 °C for 3 min, 5 cycles of 94 °C for 30 s, 70 °C for 30 s, and 72 °C for 3 min, 25 cycles of 94 °C for 30 s, 68 °C for 30 s and 72 °C for 3 min, and 1 cycle of 72 °C for 10 min. For 5′ end RACE PCR, the gene specific primer R2 and the anchor primer UPM (Table 1) were used, and the PCR reaction systems and conditions were the same as those described above.
The PCR amplified products were analyzed on 1.8 % agarose gel and the target band was purified by a Sanprep PCR purification kit (Sangon, China) and cloned into a PMD18-T vector following the instructions provided by the manufacturer (TaKaRa, Japan). Recombinant bacteria were identified by blue/white screening and confirmed by PCR. Plasmids containing the insert were purified by Wizard Plus SV Minipreps DNA Purification System (Promega, USA) and sequenced by Sunny Biotechnology Company (Shanghai, China).
Sequence analysis
The EcRelish cDNA full sequence was spliced by DNASTAR.Lasergene.v7.1 (USA). The nucleotide sequence and deduced amino acid sequence of EcRelish was compared to non-redundant nucleotide and protein databases using the BLAST algorithm (http://www.ncbi.nlm.nih.gov/BLAST/). The nucleotide and deduced amino acid sequences of EcRelish cDNA were analyzed using the conserved domain database search programs (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and SMART 4.0 (http://smart.embl-heidelberg.de/). A multiple sequence alignment of Relish amino acid sequences was performed using the programs of DNAMAN v6.0.3.99 (Lynnon Biosoft) [66]. A phylogenetic tree of Relish was made by MEGA 4.0 using the Neighbor-Joining method [37].
Expression of EcRelish in different tissues
Hemocytes, gill, hepatopancreas, muscle, ovary, heart, mandibles, stomach, and intestine were dissected from unchallenged E. c arinicauda. The mRNA expression levels of EcRelish mRNA in different tissues were determined by real time RT-PCR. Total RNA was extracted as described above. The RNA samples were analyzed in 1.0 % agarose electrophoresis and quantitated at 260 nm, all OD260/OD280 were between 1.8 and 2.0. Total RNA (5 μg) was reverse transcribed using the PrimeScript™ Real time PCR Kit (TaKaRa, Japan) for real time RT-PCR analysis. The 18 s rRNA of E. C arinicauda (GenBank accession number: GQ369794) was used as an internal control for expression, because it was stably expressed among different tissues and varying experimental conditions in this study.
Experimental design of V. a nguillarum and WSSV challenge
This experiment was classified into three groups: V. a nguillarum challenged group (VA group), WSSV challenged group (WSSV group) and PBS group as control group, which were reared in the PVC tanks. Healthy shrimp were divided into nine PVC tanks evenly, each of which contained 40 individuals. Each group included three PVC tanks. V. anguillarum strains named V. a nguillarum vib72 were obtained from Germplasm Resources and Genetic Breeding Laboratory, Yellow Sea Fisheries Research Institute (Qingdao, China), and cultured on marine agar 2611E. WSSV crude extract was obtained from L. v annamei, which was provided by the Mariculture Disease Control and Pathogenic Molecular Biology Laboratory, Yellow Sea Fisheries Research Institute (Qingdao, China), according to the methods of absolute quantitative detection [38]. Briefly, for preparation of the WSSV crude extract for injection, 1 g gills of WSSV-infected of L. v annamei were homogenized separately in 10 ml 0.01 M phosphate-buffered saline (PBS, NaCl 137 mM, KCl 2.7 mM, Na2HPO4 10 mM, KH2PO4 2 mM, pH 7.4) on ice. The homogenized tissue was centrifuged at 1200 rpm at 4 °C for 20 min. The supernatant was filtered through a 0.45 μm filter. The filtrate was then stored at −80 °C for infectivity studies. Viral load was quantified by absolute quantitative detection. In the V. a nguillarum challenge experiment, individuals were injected with 20 μl live V. a nguillarum suspended in PBS solution (105 CFU/g) each as experimental shrimp. Bacterial strains were isolated from muscle of E. c arinicauda infected by V. a nguillarum, and then identified by PCR with bacterial universal primers of 16S rDNA (Table 1). DNA sequences of the PCR-amplified isolate from the infected E. c arinicauda showed high identity with the known V. a nguillarum sequence in GenBank; it showed that these bacterial strains belonged to V. a nguillarum isolates [67]. In the WSSV challenged group, individuals were injected with 20 μl WSSV crude extract (105 copies/g); control group individuals were injected with 20 μl PBS. After challenge, a WSSV-specific PCR reaction using the specific primers WS-F1 and WS-F2 was processed to check the infection results [38] (Table 1). Hemocytes of six individuals randomly collected from each group were extracted at 1, 3, 6, 12, 24, 48, 72 h post-injection (hpi), and preserved in −80 °C for RNA extraction. Muscles in each sampling time were collected to count the colony numbers of V. a nguillarum and detect the WSSV copy number. Subsequently, the number of dead shrimp in the VA, WSSV, and control group was recorded. Experiments were performed in triplicate. Cumulative mortality rate was calculated as follows: S (%) = [D 1 + D 2 + 549 +D t ]/N t × 100 (S indicates the cumulative mortality rate, D indicates the number of dead, t indicates the sampling time, N indicates the total number of shrimp in every tank).
Detection of the proliferation of V. a nguillarum and WSSV in muscle
The number of V. a nguillarum was determined by counting the number of colonies formed on an agar slant of 2216E medium (5.0 g tryptone, 1.0 g yeast extract, 20 g agar, 1 l synthetic seawater). Then 0.1 g muscle immersed in 1 ml 0.01 M PBS was homogenized with a grinding rod in a 1.5 ml centrifuge tube as the primary liquid. Using the gradient dilution ten times, the primary liquid was diluted to five concentrations. Every concentration was coated in three parallel marine agar slants of 2216E medium. All of these flats were put in 28 °C incubator for 16 h. Then the bacterial colonies with the same shape as the shape of a V. a nguillarum colony were recorded. Identification of bacterial strains was processed as described above.
Absolute quantitative real-time PCR was performed to determine the WSSV copy number in 0.1 g shrimp muscles collected at different sampling times by using Premix Ex Taq™ (Probe qPCR) Kit (Takara, RR390A). Briefly, the primers WS-F and WS-R were used to amplify a 68 bp fragment, which would be transferred into recombinant plasmid. The standard recombinant plasmids of WSSV were extracted by Mini Kit I of plasmids (Biomed, China). Viral genomic DNA was extracted using Column Marine Organization DNAout (TGreen, DP324). A standard curve was obtained using serial dilutions of recombinant plasmid, which were used to quantify the WSSV viral genomic copy number (Kwang, 2011). The absolute quantitative real-time PCR was carried out in a total volume of 20 μl, containing 10 μl of Premix Ex Taq™ (Probe qPCR) (2× conc.), 2 μl of viral DNA, 0.4 μl each of WS-F primer (10 μM) and WS-R primer (10 μM), 0.8 μl TaqMan Probe, 0.2 μl ROX Reference DyeII (50×), and sterile ddH2O was added to a total volume of 20 μl. Sterile ddH2O for the replacement of template was used as a negative control. The PCR program was 95 °C for 30 s, then 40 cycles of 95 °C for 5 s and 60 °C for 34 s. Each assay was carried out in triplicate.
Ammonia-N stresses experiment
The median lethal of total ammonia-N were found to be 140.28 mg/l for 72 h for adult E. carinicauda [39]. Basing on this research, a preliminary experiment was designed to confirm the suitable concentration of ammonia-N stress by four concentration gradients of 17.54, 35.07, 70.14, and 140.28 mg/l. Ultimately the concentration of ammonia-N stress was confirmed to be 35.07 mg/l for which the survival rate was 70 %, which was enough to take samples. Two groups of shrimp were placed on average in six PVC tanks serving as the ammonia-N stress group and the control group. In the ammonia-N stress test, the experimental shrimp (40 shrimps per tank) were kept in 50 l tanks containing 30 l of 31 ppt salinity seawater with 35.07 mg/l NH4Cl. Because of this experiment of ammonia-N challenge belonging to acute stress, the times for sampling were intensive. Therefore, the sampling times were 1, 3, 6, 12, 24, 48, and 72 h, respectively. The test water was renewed daily. The concentration of ammonia-N by Nessler’s reagent was tested every day during the experiment. The NH4Cl was added to keep the concentration of ammonia-N as 35.07 mg/l. The shrimp in the control group were kept in 31 ppt salinity seawater without ammonia-N stress. Subsequently, the number of dead shrimp in the ammonia-N group was recorded. Experiments were performed in triplicate. Cumulative mortality rate was calculated as described above.
Quantification of EcRelish mRNA expression by real time RT-PCR
Real time RT-PCR was performed on an ABI PRISM 7500 Sequence Detection System (Applied Biosystems, USA) to investigate the EcRelish expression pattern after the challenge of V. a nguillarum, WSSV, and ammonia-N stress in hemocytes of E. c arinicauda. The pair of specific primers F3 and R3 (Table 1) was used to amplify a PCR product of 158 bp. The primers 18 s-F and 18 s-R (Table 1) were used to amplify an 18S gene of 147 bp as an internal control for a quantitative real time. The real time RT-PCR was carried out in triplicate in a total volume of 20 µl containing 10 µl SYBRR Premix Ex Taq™ II (2×) (TaKaRa, Japan), 2 µl of the cDNA, 0.8 µl each of F3 and R3 primer (10 mM) (or 18 s-F and 18 s-R to amplify the 18 s), 0.4 µl ROX Reference Dye II (50×), and 6.0 µl DEPC-treated water. The PCR program was 95 °C for 30 s, then 40 cycles of 95 °C for 5 s and 60 °C for 34 s, followed by 1 cycle of 95 °C 15 s, 60 °C for 1 min and 95 °C for 15 s. DEPC-treated water for the replacement of template was used as a negative control. The standard curves of EcRelish and 18 s rRNA were made through a twofold dilution series of cDNA template of hemocytes. According to the formula: E = 10(−1/S)−1 (S was the slope of the standard curve), the amplification efficiencies (E) of EcRelish and 18 s rRNA were 104.8 % and 101.4 %, respectively, (the appropriate range of amplification efficiencies were 90–105 %). All analyzed were based on the C T values of the PCR products. The C T was defined as the PCR cycle at which the fluorescence signal crossed a threshold line that was placed in the exponential phase of the amplification curve. The amplification specificity for EcRelish and 18 s was determined by analyzing the dissociation curves. The EcRelish expression levels in response to V. a nguillarum, WSSV, and ammonia-N stress were calculated with \(2^{-\Delta\Delta\text{C}_T}\) methods [40]. Using this method, the data are presented as the fold change in gene expression normalized to an endogenous reference gene and relative to the control group. All data were presented as mean ± SE, and subjected to one-way analysis of variance followed by Duncan’s multiple range test. Significance was set at P < 0.05 [41].
Results
The cumulative mortality rates of E. c arinicauda after V. a nguillarum, WSSV challenge and ammonia-N stress
The variation of the cumulative mortality rates in different sampling times after V. a nguillarum, WSSV challenge, and ammonia-N stress are shown in Fig. 1. In the V. a nguillarum challenged group, the cumulative mortality rates increased gradually in the first 48 hpi, then no individuals died afterwards. Finally, the cumulative mortality rate of the VA group was 63.33 %. In the WSSV challenged group, there was no shrimp death in the first 6 hpi, and many shrimps died from 24 to 72 hpi. The cumulative mortality of the WSSV group was 70 % at the end of the experiment. In the ammonia-N stress group, the cumulative mortality rates increased gradually in the first 48 hpi after stress, and the cumulative mortality rate was 33.33 %. The cumulative mortality had no significant (P > 0.05) at the end of the experiment between the V. a nguillarum group and the WSSV group, however, they were much higher than the cumulative mortality of the control group (P < 0.05). There were no dead shrimp in the control group in the whole process of the experiment. The cumulative mortality had no significant difference (P > 0.05) at the end of the experiment between the V. a nguillarum group and the WSSV group, however, they were much higher than the cumulative mortality of the control group (P < 0.05).
The proliferation of V. a nguillarum and WSSV in survival shrimp muscle
The bacteria colony amount of V. a nguillarum was not detected in 0 h, and a small amount could be detected in the first 3 hpi. The numerical values of V. a nguillarum increased gradually from 3 hpi (4 × 104 CFU/g) and reached the peak value at 24 hpi (2.1 × 106 CFU/g), then a decrease of V. a nguillarum appeared at 48 hpi (6.2 × 105 CFU/g) to 72 hpi (9.1 × 104 CFU/g) (Fig. 2). The WSSV amount was 7.0 × 105 copies/g at 0 h without infection, and increased exponentially at every detection time. Then the viral load of WSSV reached the peak value (4.7 × 109 copies/g) at the end of this experiment.
Sequence analysis of the EcRelish gene
A full-length Relish cDNA of E. c arinicauda (EcRelish) was obtained by the procedures of reverse-transcription PCR and RACE, and the results are shown in Fig. 3. The EcRelish cDNA sequence has been submitted to the GenBank (GenBank accession number: JX867729). The full-length of EcRelish was 2141 bp, containing a 1983 bp open reading frame (ORF), which encoded 660 amino acids, a 31 bp of 5′-untranslated region (UTR), and a 127 bp of 3′-UTR. The predicted protein molecular mass was 75.27 kDa, and the estimated isoelectric point was 5.998. Analysis revealed that the EcRelish protein exhibits the typical architecture of Rel/NF-κB family members, including a conserved N-terminal RHD from 50 to 248 amino acids, two nucleus localization signals, an IPT domain of the transcription factor NF-κB, and the signature sequence FRYKSE of Relish from 62 to 67 amino acids, suggesting that it belongs to the REL/NF-κB family.
Nucleotide and deduced amino acid sequences of EcRelish. Start codon is boxed; stop codon is indicated with asterisk. The two nuclear localization signals are marked with shaded boxes. The RHD domain is shown with fine line. The IPT domain is shown with bold line. The signature sequence FRYKSE of Relish is circled by red oval
Homology analysis of EcRelish
The deduced amino acid sequence of EcRelish exhibited similarities with Relish of invertebrates. It displayed high similarities to Relishes of E. s inensis (73 %), L. v annamei (73 %), P. m onodon (73 %), and F. c hinensis (76 %).
Multiple sequence alignment of EcRelish with other crustaceans Relish showed high conservation in two conserved domains, IPT domain, and conserved N-terminal RHD while the differences are the absence of six anchored simple sequence repeats and a death domain (DD) (Fig. 4).
Multiple alignment of EcRelish with other crustacean Relish: Relish of Exopalaemon carinicauda (AGR39579.1), Fenneropenaeus chinensis (ACJ36224.1), Litopenaeus vannamei (ABR14713.1), Penaeus monodon (AFH66691.1), Eriocheir sinensis (ADM14334.1). The signature sequence FRYKSE of Relish is marked by gray frame, the IPT domain is underlined, the RHD is marked by a dotted line, six anchored simple sequence repeats are marked by black frames and the death domain (DD) is marked by black oval
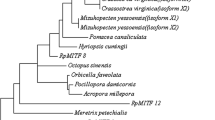
Based on the sequences of Relish, a NJ phylogenetic tree was constructed using MEGA 4.0 (Fig. 5). Insecta and crustacean Relishes were separated and formed two distinct branches in the tree. In the group of crustaceans, all the shrimps and crabs were clustered together and formed two branches. Relish of crustacean clustered with C. r otundicauda. Relishes of insecta were clustered together and formed a sister group to the branch of crustaceans and C. r otundicauda. The relationships displayed in the phylogenetic tree were in good agreement with the concept of traditional taxonomy. EcRelish belongs to the crustacean Rels subgroup and is closely related to NF-κB/Rel of crabs and shrimps.
The phylogenetic tree of the Rel/NF-κB family. The sequences used in the phylogenetic tree are as follows: Nasutitermes walkeri (GenBank accession no. AAZ08469), Nasutitermes comatus (GenBank accession no. AAZ08478), Nasutitermes pluvialis (GenBank accession no. AAZ08476), Glossina morsitans (GenBank accession no. AAZ91474), Drepanotermes rubriceps (GenBank accession no. AAZ08479), Drosophila melanogaster (GenBank accession no. AAF20133), Carcinoscorpius rotundicauda (GenBank accession no. ABC75034), Eriocheir sinensis (GenBank accession no. ADM14334), Exopalaemon carinicauda (GenBank accession no. JX867729), Fenneropenaeus chinensis (GenBank accession no. ACJ36224), Penaeus monodon (GenBank accession no. AFH66691), and Litopenaeus vannamei (GenBank accession no. ABR14713)
Expression of EcRelish in different tissues of E. c arinicauda
Real time RT-PCR was employed to quantify the EcRelish expression in different tissues. The mRNA transcripts of EcRelish could be detected in all the examined tissues with different expression levels including hemocytes, gill, hepatopancreas, muscle, stomach, intestine, heart, ovary, mandibles, and eyestalk (Fig. 6). The highest expression was observed in the hemocytes, and very low expression was detected in the ovary and mandibles.
EcRelish gene expression in hemocytes of E. c arinicauda after V. a nguillarum and WSSV challenges
Since the hemocytes showed the highest expression of EcRelish, the expression profiles of EcRelish mRNA were analyzed in hemocytes of E. c arinicauda after they were stimulated by V. a nguillarum and WSSV. Compared with that of blank (0 h), injection of V. a nguillarum caused the activation of EcRelish transcription at the first 24 hpi. Compared with that of the PBS group, V. a nguillarum injection significantly induced the up-regulated expression of EcRelish in hemocytes at 1–24 hpi (P < 0.01). In addition, the expression level of EcRelish had the highest expression at 1 hpi, then it decreased constantly and reached the control group level at 48 hpi until the experiment finished (P > 0.05) (Fig. 7).
The temporal expressions of EcRelish mRNA in hemocytes of E. c arinicauda after WSSV challenge are shown in Fig. 8. Compared with that of blank (0 h), injection of WSSV caused the activation of EcRelish transcription at 3 hpi, and caused suppression of EcRelish transcription at 6–72 hpi. Compared with that of the PBS group, the expression of EcRelish in the WSSV group was up-regulated significantly at 3 hpi (4.5-fold of the PBS group, P < 0.01), then decreased at 6 and 12 hpi. Afterwards, the expression was up-regulated again and reached to the second peak at 24 hpi (1.79-fold of the PBS group, P < 0.05), then gradually down-regulated at 72 hpi.
EcRelish gene expression in hemocytes of E. c arinicauda after Ammonia-N stress
The temporal expressions of EcRelish mRNA in hemocytes of E. c arinicauda after ammonia-N stress are shown in Fig. 9. Compared with that of the blank (0 h), exposure to ammonia-N caused suppression of EcRelish at 1–72 h. Compared with that of the control group, the expression levels of EcRelish mRNA in the group of ammonia-N stress showed down-regulation significantly during the whole experimental time (P < 0.01). Even so, the expression levels had a waving change and increased at 24 and 72 h after the shrimp were exposed to ammonia-N.
Discussion
Invertebrates rely entirely on a nonspecific immunity system as the first-line host defense to prevent or combat different microbial invaders by activating various genes [42–44]. The identification and characterization of genes involved in immune responses are essential for the elucidation of immune defence mechanisms and diseases control [45]. In this study, a new full-length 2141 bp of Relish cDNA (named EcRelish) was cloned from E. c arinicauda. In the deduced amino acid sequence of EcRelish, a N-terminal RHD domain, an IPT domain, the signature sequence FRYKSE of Relish, and two nuclear localization signals were found. The nuclear localization signals existed in the RHD domain of the cloned sequence. The results suggested that EcRelish was one of the nuclear transcription factors. Rel/NF-κB factors have a well-conserved RHD involved in DNA binding, dimerization, and interaction with the inhibitor kB (IkB). When the cell receives signals, Rel/NF-κB factors are released from the IkB and then rapidly enter the nucleus to activate expression of various down-stream genes [23]. The two different types of Relish were both found in F. c hinensis and L. v annamei, which could be searched in NCBI. In view of the absence of the six ANK repeats and a death domain in EcRelish shown in the multiple sequence alignment, the EcRelish may belong to the shorter isoform Relish, which was earlier reported in the F. c hinensis and L. v annamei (GenBank accession No. ACJ36224 and No. ACR24222, respectively) [29, 47]. Phylogenetic analysis showed that EcRelish was clustered together with Relish proteins of crustaceans and other arthropods.
Real time RT-PCR revealed that EcRelish was constitutively expressed in all the detected tissues of E. c arinicauda. Tissue distribution analysis of EcRelish showed that the highest expression of EcRelish was in hemocytes, intestines, and gills, which was consistent with that of P. m onodon [22]. It has been reported that hemocytes played a critical role in host-defense mechanisms against shrimp viral infection, including bacteria and WSSV [48, 49]. Hemolymph is one of the most important immune related organs, in which many immune related genes have high transcription levels [28, 50, 51]. Therefore, the high expression of EcRelish in the hemocytes indicated that EcRelish might be related to the immune reaction of the shrimp.
It was reported that the Toll and Imd pathways are the major regulators of the immune response in Drosophila [11]. Dif and Dorsal were involved in the Toll pathway, while Relish was involved in the Imd pathway [52]. The existence of a toll pathway has been verified in F. c hinensis [53]. EcRelish has the same characteristic structure of Rel-S like F. c hinensis and L. v annamei, in accordance with the resisting role in other shrimps, which indicated that EcRelish might also be involved in the immune response of E. c arinicauda to bacteria and virus infection. Therefore, the stress experiments of bacteria and virus were used to test our hypothesis. In the V. a nguillarum challenged group, the cumulative mortality rates showed an increasing tendency from 16.67 % to 60 % gradually in 1–24 hpi, whereas the increase in 48–72 hpi was less evident, which might be related to the variation of the quantity of V. a nguillarum in the survival shrimp's muscles. In the preinfection stage, the rapid proliferation of V. a nguillarum led to massive mortality of shrimp. It has been reported that the injected bacteria could be rapidly removed by the hemolymph of the shrimp [54], indicating that the V. a nguillarum in survival shrimp might be cleared gradually by hemolymph of E. c arinicauda. With the decrease of V. a nguillarum after 24 hpi, the number of V. a nguillarum was not enough to infect the shrimp, so the shrimp survived at the subsequent stage. To a large extent, the expression profile of EcRelish in hemocytes showed significant up-regulation during the first 24 hpi, which was consistent with proliferation of V. a nguillarum. In the first 24 hpi, the amount of gradually increased V. a nguillarum was of enough quantity to cause the shrimp death and stimulate the immune system, so EcRelish was transcribed on a large scale. These data suggested that EcRelish might be involved in the Imd pathway responsive to bacteria, which were already reported in Drosophila. However, there was no changes of EcRelish gene expression in the later stage of infection (48–72 hpi) compared to the PBS group, the reason for this may be that with the injection time and the number of V. a nguillarum was decreasing for the clearance of vibrio by hemocytes [54], therefore, V. a nguillarum was not enough to stimulate the transcripts of EcRelish, so that the expression of EcRelish recovered to a normal level after 48 hpi.
WSSV is one of the most devastating and virulent viral agents threatening the penaeid shrimp culture industry and has been responsible for serious economic losses in shrimp culture [4, 55, 56]. In the WSSV challenged group, the cumulative mortality increased linearly from 20 to 70 % with the infection times in 24–72 hpi, which showed the health of shrimp was seriously destroyed during WSSV-infection. The viral load increasing exponentially indicated that, before the copy number of WSSV increased to a certain quantity (108 copies/g), the immune system of shrimp was not strong enough to prevent the virus from invading, and a massive death of shrimp was caused. The expression of Relish is responsive to bacteria or WSSV, which can regulate the transcription of Penaeidins in L. v annamei and F. c hinensis [57]. In this study, the expression of EcRelish gene was induced in hemocytes after WSSV challenge. WSSV induced the rapid up-regulation of EcRelish transcription at 3 hpi compared to that of the blank (0 h) or PBS group, which showed that EcRelish might be involved in immune response to WSSV stimulation. However, the expression of EcRelish dropped to a very low level at 6 and 12 hpi of the WSSV challenged group compared with that of the PBS group, which might be the strategy of pathogens escaping from innate immunity of the host. WSSV infection intensively affected the transcription and the expression of EcRelish at an early time and decreased the transcriptional level of immune genes. The expression of EcRelish that increased at 24 hpi suggested that WSSV was promoted to express its packed genes by activating the NF-κB gene Relish of the host. As time progressed, the EcRelish mRNA expression levels decreased till the end of the experiment, which might be because as the virus infection progressed, the virus would unceasingly reproduce in hosts, then the health of shrimps was destroyed during WSSV-infection [58], so that the expression levels of immune related gene were reduced.
Ammonia is the major toxicant resulting from the excretion of cultured shrimp and mineralization of organic wastes such as feces and unconsumed feed in an intensive culture farm [59]. Elevated concentrations of ammonia can cause impairment in numerous organs and induce a modification of the immune system in aquatic animals [60]. In the present study, the cumulative mortality rates of shrimps increased gradually during the first 24 h after ammonia-N stress, which indicated that ammonia-N could cause a challenge to shrimp. The expression levels of EcRelish in the ammonia-N challenged group were down-regulation significantly in the whole experimental process compared with that of the control group. These observations suggested that ammonia-N might suppress the transcription of Relish gene and then prevent the factors involved in immune reactions, furthermore causing a reduction of immune capacity. Previous studies have demonstrated that the expression of the proPO and peroxinectin in Penaeus stylirostris ir decreased respectively by 60 % and 50 % in response to the ammonia stress [61]. The down-regulated expression of these immune related genes after ammonia stress indicated that ammonia can result in a depression in immune capacity of crustaceans [62]. The expression levels had a waving change and were increased at 24 h and 72 h after exposure to ammonia-N. It can be speculated that the immune related gene of the shrimp attempted to take part in the progress of resisting the ammonia-N stress at 24 h and 72 h after exposure to ammonia-N, but did not succeed.
In conclusion, a full-length Relish cDNA (EcRelish) was cloned from hemocytes of E. c arinicauda. The mRNA encoding EcRelish was synthesized in tested tissues. The expression of EcRelish in hemocytes changed rapidly in response to the infection of V. a nguillarum and WSSV, and ammonia-N stress. The expression pattern of EcRelish indicated that the EcRelish gene was inducible and might be involved in anti-bacterial and anti-viral action in E. c arinicauda. The expression profiles of EcRelish gene after ammonia-N stress showed EcRelish played a key role in environmental stresses.
References
Liu RY (1957) Palaemon and Macrobrachium. Bull Biol 6:14–23
Xu WJ, Xie JJ, Shi H (2010) Hematodinium infections in cultured ridgetail white prawns, Exopalaemon carinicauda, in eastern China. Aquaculture 300:25–31
Xia DQ (1999) Biological characteristics and aquaculture technique of Exopalaemon carinicauda. China Fish 3:42–43 (in Chinese)
Lightner DV, Revdman RM (1998) Shrimp diseases and current diagnostic methods. Aquaculture 164:201–220
Li GY (1995) Review of disease and immune mechanism in Penaeus chinensis. Marine Sciences 4:1–3 (in Chinese)
Le Moullac G, Taravao T, Française P (2000) Environmental factors affect immune response and resistance in Crustaceans. The Advocate 18−19
Aguirre-Guzman G, Sanchez-Martinez JG, Campa-Cordova AI (2009) Penaeid shrimp immune system. Thai J Vet Med 39:205–215
Amparyup P, Kondo H, Hirono I, Aoki T, Tassanakajon A (2008) Molecular cloning, genomic organization and recombinant expression of a crustin-like antimi-crobial peptide from black tiger shrimp Penaeus monodon. Mol Immunol 45:1085–1093
Hoffmann JA, Reichhart JM (2002) Drosophila innate immunity: an evolutionary perspective. Nat Immunol 3:121–126
Kush RS, Leulier F, Lemaitre B (2001) Drosophila immunity: two paths to NF-kB. Trends Immunol 22:260–264
De Gregorio E, Spellman PT, Tzou P (2002) The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J 21:2568–2579
Georgel P, Naitza S, Kappler C, Ferrandon D, Zachary D, Swimmer C et al (2001) Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev cell 1:503–514
Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, Georgel P et al (1995) A recessive mutation, immune deficiency (IMD), defines two distinct control pathways in the Drosophila host defense. Proc Natl Acad Sci USA 92:9465–9469
Naitza S, Rossé C, Kappler C, Georgel P, Belvin M, Gubb D et al (2002) The Drosophila immune defense against gram-negative infection requires the death protein dFADD. Immunity 17:575–581
Rutschmann S, Kilinc A, Ferrandon D (2002) Cutting edge: the Toll pathway is required for resistance to gram-positive bacterial infections in Drosophila. Immunology 168:1542–1546
Hedengren M, Asling B, Dushay MS (1999) Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol Cell 4:827–837
Wang DN, Liu JW, Yang GZ, Zhang WJ, Wu XF (2002) Cloning of anti-LPS factor cDNA from Tachypleus tridentatus, expression in Bombyx mori larvae and its biological activity in vitro. Mol Biotechnol 21:1–7
Belvin MP, Anderson KV (1996) A conserved signaling pathway: the Drosophila Toll-Dorsal pathway. Annu Rev Cell Dev Biol 12:393–416
Lemaitre B, Reichhart JM, Hoffmann JA (1997) Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. PNAS 94:14614–14619
Rutschmann S, Jung AC, Hetru C, Reichhart JM, Hoffmann JA, Ferrandon D (2000) The Rel protein DIF mediates the antifungal but not the antibacterial host defense in Drosophila. Immunity 12:569–580
Kang CJ, Wang JX, Zhao XF, Yang XM, Shao HL, Xiang JH (2004) Molecular cloning and expression analysis of the ch-penaedin, an antimicrobial peptide from Chinese shrimp, Fenneropenaeus chinensis. Fish Shellfish Immunol 16:513–525
Cuthbertson B, Bullesbach E, Bachere E, Fievet J, Gross P (2004) A new class (penaeidin class 4) of antimicrobial peptides from the Atlantic white shrimp (Litopenaeus setiferus) exhibits target specificity and an independent proline-rich-domain function. Biochem J 381:79–86
Visetnan S, Supungul P, Hirono I, Tassanakajon A, Rimphanitchayakit V (2015) Activation of Pm Relish from Penaeus monodon by yellow head virus. Fish Shellfish Immunol 42:335–344
Hoffmann JA, Reichhart JM, Hetru C (1995) Innate immunity in insects. Curr Opin Immunol 7:4–10
Shin SW, Kokoza V, Ahmed A, Raikhel AS (2002) Characterization of three alternatively spliced isoforms of the Rel/NF-κB transcription factor Relish from the mosquito Aedes aegypti. PNAS 99:9978–9983
Meister S, Kanzok SM, Zheng XL, Luna C, Li TR, Hoa NT (2005) Immune signaling pathways regulating bacterial and malaria parasite infection of the mosquito Anopheles gambiae. Proc Natl Acad Sci USA 102:11420–11425
Liu WJ, Yang LS, Li XL (2013) Cloning and mRNA expression analysis of Relish gene in Penaeus monodon following immune stimulation. J Fish Sci China 20:50–60 (in Chinese with English abstract)
Li FH, Yan H, Wang DD, Priya TAJ, Li SH, Wang B et al (2009) Identification of a novel relish homolog in Chinese shrimp Fenneropenaeus chinensis and its function in regulating the transcription of antimicrobial peptides. Dev Comp Immunol 33:1093–1101
Huang XD, Yin ZX, Liao JX (2009) Identification and functional study of a shrimp Relish homologue. Fish Shellfish Immunol 27:230–238
Li F, Wang L, Zhang H (2010) Molecular cloning and expression of a Relish gene in Chinese mitten crab Eriocheir sinensis. Int J Immunogenet 37:499–508
Frias-Espericueta MG, Harfush-Melendez M, Páez-Osuna F (2000) Effects of ammonia on mortality and feeding of postlarvae shrimp Litopenaeus vannamei. Bull Environ Contam Toxicol 65:98–103
Liu CH, Chen JC (2004) Effect of ammonia on the immune response of white shrimp Litopenaeus vannamei and its susceptibility to Vibrio alginolyticus. Fish Shellfish Immunol 16(3):321–334
Jiang GJ, Yu RC, Zhou MJ (2004) Modulatory effects of ammonia-N on the immune system of Penaeus japonicus to virulence of white spot syndrome virus. Aquaculture 241:61–75
Cheng WT, Chen JC (2002) The virulence of Enterococcus to freshwater prawn Macrobrachium rosenbergii and its immune resistance under ammonia stress. Fish Shellfish Immunol 12:97–109
D’Acquisto F, Luvone T, Rombola L (1997) Involvement of NF-KB in the regulation of cyclooxygenase-2 protein expression in LPS-stimulated J774 macrophages. FEBS Lett 418:175–178
Choudhury MG, Nirmalendu S (2012) Influence of environmental ammonia on the production of nitric oxide and expression of inducible nitric oxide synthase in the freshwater air-breathing catfish (Heteropneustes fossilis). Aquat Toxicol 116–117:43–53
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA 4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1609
Yan DC, Dong SL, Huang J, Zhang JS (2007) White spot syndrome virus (WSSV) transmission from rotifer inoculum to crayfish. J Invertebr Pathol 94:144–148
Liang JP, Li J, Li JT (2012) Acute toxicity of ridgetail white prawn Exopalaemon carinicauda. Fish Sci 31:526–529 (in Chinese with English abstract)
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 −ΔΔCT method. Methods 25:402–408
González-Rodríguez G, Colubi A, Gil MÁ (2012) Fuzzy data treated as functional data: a one-way ANOVA test approach. Comput Stat Data Anal 56:943–955
Duan YF, Liu P, Li JT (2013) Expression profiles of selenium dependent glutathione peroxidase and glutathione S-transferase from Exopalaemon carinicauda in response to Vibrio anguillarum and WSSV challenge. Fish Shellfish Immunol 35:661–670
Zhang QL, Li FH, Zhang XJ, Dong B, Zhang JQ, Xie YS et al (2008) cDNA cloning, characterization and expression analysis of the antioxidant enzyme gene, catalase, of Chinese shrimp Fenneropenaeus chinensis. Fish Shellfish Immunol 24:584–591
Iwanaga S, Lee BL (2005) Recent advances in the innate immunity of invertebrate animals. J Biochem Mol Biol 38:128–150
Zhao JM, Song LS, Li CH, Ni DJ, Wu LT, Zhu L et al (2007) Molecular cloning, expression of a big defensin gene from bay scallop Argopecten irradians and the antimicrobial activity of its recombinant protein. Mol Immunol 44:360–368
Wang DD (2012) Functional studies on the regulation of immunity in Chinese shrimp, Fenneropenaeus chinensis by the nuclear transcription factor, NF-κB family genes. PhD dissertation, Chinese Academy of Sciences, Beijing
Yan H (2009) Cloning and Expression of Nuclear Transcription Factors Rel/NF-κB family genes in Chinese shrimp Fenneropenaeus chinensis. Master’ Degree dissertation, Chinese Academy of Sciences, Beijing
Bachere E (2000) Shrimp immunity and disease control. Aquaculture 191:3–11
Holmblad T, Söderhäll K (1999) Cell adhesion molecules and antioxidative enzymes in a crustacean, possible role in immunity. Aquaculture 172:111–123
Wang B, Li FH, Luan W, Xie YS, Zhang CS, Luo Z et al (2008) Comparison of gene expression profiles of Fenneropenaeus chinensis challenged with WSSV and Vibrio. Mar Biotechnol 10:664–675
Mekata T, Kono T, Yoshida T, SakaiM Itami T (2008) Identification of cDNA encoding Toll receptor, MjToll gene from kuruma shrimp, Marsupenaeus japonicus. Fish Shellfish Immunol 24:122–133
Uvell H, Engström Y (2007) A multilayered defense against infection: combinatorial control of insect immune genes. Trends Genet 23:342–349
Chi YH (2012) Cloning and preliminary study on the function relationships of genes related to the Rac1/PI3K/Akt pathway in Chinese shrimp. Master’ Degree dissertation, Chinese Academy of Sciences, Beijing
Guo ZX, Feng J, Wang JY (2006) Clearance of Vibrio anguillarum by hemocytes in giant black tiger shrimp Penaeus monodonin vivo. J Fish Sci China. 13:28−32 (in Chinese with English abstract)
Shen H, Wan XH, Wang LB (2013) Study on experimental infection of Exopalaemon carinicauda Holehuis with white spot syndrome virus. Marine Sciences 37:55–60 (in Chinese with English abstract)
Lightner DV, Hasson KW, White BL, Redman RM (1998) Experimental infections of western hemisphere penaeid shrimp with Asian white spot syndrome virus and Asian yellow head virus. J Aquat Anim Health 10:271–281
Wang DD, Li SH, Li FH (2013) Screening of genes regulated by relish in Chinese shrimp Fenneropenaeus chinensis. Dev Comp Immunol 41:209–216
Liu YC, Li FH, Dong B, Wang B, Luan W, Zhang XJ, Xiang JH (2007) Molecular cloning, characterization and expression analysis of a putative C-type lectin (Fclectin) gene in Chinese shrimp Fenneropenaeus chinensis. Mol Immunol 44:598–607
Chen YY, Sim SS, Chiew SL (2012) Dietary administration of a Gracilaria tenuistipitata extract produces protective immunity of white shrimp Litopenaeus vannamei in response to ammonia stress. Aquaculture 370:26–31
Colt JE, Armstrong DA (1981) Nitrogen toxicity to crustaceans, fish and molluscs. In: Allenand LJ (ed) “Proceedings of the bio-engineering symposium for fish culture”, Kinney EC Section of the American Fisheries Society FCS publ.1, Bethesda, pp 34−47
Le Moullac G, Haffner P (2000) Environmental factors affecting immune responses in Crustacea. Aquaculture 191:121–131
Hong M, Chen L, Sun X (2007) Metabolic and immune responses in Chinese mitten-handed crab (Eriocheir sinensis) juveniles exposed to elevated ambient ammonia. Comp Biochem Phys C 145:363–369
Stöven S, Ando I, Kadalayil L, Engström Y, Hultmark D (2000) Activation of the Drosophila NF-κB factor Relish by rapid endoproteolytic cleavage. EMBO Rep 1:347–352
Kim M, Lee JH, Lee SY, Kim E, Chung J (2006) Caspar, a suppressor of antibacterial immunity in Drosophila. Proc Natl Acad Sci USA 103:16358–16363
Söderhäll K, Smith VJ (1983) Separation of the haemocyte populations of Carcinus maenas and other marine decapods. Dev Comp Immunol 7:229–239
Li X, Meng X, Kong J, Luo K, Luan S, Cao B, Liu N, Pang J, Shi X (2013) Molecular cloning an characterization of a cathepsin B gene from the Chinese shrimp Fenneropenaeus chinensis. Fish Shellfish Immunol 35:1604–1612
Moreno C, Romero J, Espejo RT (2002) Polymorphism in repeated 16S rRNA genes is a common property of type strains and environmental isolates of the genus Vibrio. Microbiology 148:1233–1239
Acknowledgments
The authors are grateful to all the laboratory members for experimental material preparation and technical assistance. This study was supported by the earmarked fund for National “863” Project of China (No. 2012AA10A409), Modern Agro-industry Technology Research System (No. CARS-47), and the Special Fund for Agro-scientific Research in the Public Interest (No. 201103034).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ge, Q., Liang, J., Li, J. et al. Molecular cloning and expression analysis of Relish gene from the ridgetail white prawn Exopalaemon carinicauda . Fish Sci 81, 699–711 (2015). https://doi.org/10.1007/s12562-015-0898-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12562-015-0898-z