Abstract
Reactive oxygen species (ROS) play key roles in many physiological processes. In particular, the sterilization mechanism of bacteria using ROS in macrophages is a very important function for biological defense. Xanthine dehydrogenase (XDH) and aldehyde oxidase (AOX), members of the molybdo-flavoenzyme subfamily, are known to generate ROS. Although these enzymes occur in many vertebrates, some insects, and plants, little research has been conducted on XDHs and AOXs in crustaceans. Here, we cloned the entire cDNA sequences of XDH (MjXDH: 4328 bp) and AOX (MjAOX: 4425 bp) from Marsupenaeus japonicus (kuruma shrimp) using reverse transcriptase-polymerase chain reaction (RT-PCR) and random amplification of cDNA ends (RACE). Quantitative real-time RT-PCR transcriptional analysis revealed that MjXDH mRNA is highly expressed in heart and stomach tissues, whereas MjAOX mRNA is highly expressed in the lymphoid organ and intestinal tissues. Furthermore, expression of MjAOX was determined to be up-regulated in the lymphoid organ in response to Vibrio penaeicida at 48 and 72 h after injection; in contrast, hydrogen peroxide (H2O2) concentrations increased significantly at 6, 12, 48, and 72 h after injection with white spot syndrome virus (WSSV) and at 72 h after injection with V. penaeicida. To the best of our knowledge, this study is the first to have identified and cloned XDH and AOX from a crustacean species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Reactive oxygen species (ROS) that include free radicals are involved in many physiological processes, and can have both beneficial and harmful effects for organisms [1]. For instance, free radicals can damage both cells and DNA, and thus removal of free radicals is very important [2, 3]; in addition, hydrogen peroxide generated by ROS-producing enzymes is broken down by glutathione peroxidase (GPx), catalase (CAT), and peroxiredoxin (Prx) to water and oxygen [4,5,6]. At the same time, ROS such as superoxide (O2−) and superoxide-derived hydrogen peroxide (H2O2) are cytotoxic molecules associated with antibacterial and antiviral host defense mechanisms. Macrophages produce ROS against infection when an alien substance invades the system [7, 8]. Invertebrates, however, lack pro-acquired immunity, and thus it is generally believed that sterilization via ROS production is an important immune mechanism in these animals [9, 10].
In mammals, xanthine dehydrogenase (XDH) and aldehyde oxidase (AOX) are members of the molybdo-flavoenzyme subfamily that generate ROS [11, 12]. The molybdo-flavoenzyme subfamily comprises structurally related enzymes that require a molybdopterin cofactor for their catalytic activity. At present, five types of enzyme groups are known to occur in this subfamily, consisting of nitrate reductase (NR), aldehyde oxidase (AO), xanthine oxidoreductase (XOR), sulfite oxidase (SO), and mitochondrial amidoxime reducing component (mARC); these enzyme groups catalyze multiple biosynthetic pathways due to their broad substrate specificities [13, 14]. In the molybdo-flavoenzyme subfamily, XDH is known as a kind of XOR and catalyzes a reaction in which hypoxanthine is converted into xanthine and urea, releasing hydrogen peroxide [15]. In mammals, XOR is classified into XDH and XO (xanthine oxidase). Originally it was thought that these two enzymes derived from different genes and thus were categorized as separate enzymes, but it has since been determined that XDH is subsequently converted into XO and thus derive from the same gene. The conversion of XDH to XO is designated as the D/O conversion (dehydrogenase/oxidase conversion) and influences the increase in ROS production; as such, the D/O conversion is unique to XDH, and is an important process in ROS generation [16]. The XDH enzyme contains one molybdopterin domain (Mo-pt), one FAD domain, and two Fe/S domains. Mo-pt regulates the oxidation of xanthine, and the electrons thus introduced are rapidly transferred to the FAD domain via the Fe/S domains [17]. Various XDH enzymes have been identified in many species, including fishes and insects, the domain structure of which is highly conserved among these species [18, 19]. The AOX enzyme, a homodimer consisting of two subunits, catalyzes reactions that produce carboxylic acid and hydrogen peroxide from aldehyde, water, and oxygen, and composes a group of conserved proteins within the molybdo-flavoenzyme subfamily, along with the structurally related xanthine dehydrogenase enzyme [20]. Functional AOX genes (AOX1, AOX2, AOX3, and AOX4) are most abundant in rodents and marsupials, whereas only one type of functional AOX gene (hAOX1) and two non-functional pseudogenes—vestiges of the mouse AOX3 gene—have been identified in humans [13, 21]. Among invertebrates, four types of functional AOX (AOX1, AOX2, AOX3, and AOX4) genes, each one a distinct coding gene, have been identified in fruit flies (Drosophila melanogaster) [22].
In crustaceans, ROS play primary roles in the defense against microbial infection. Pathogenic intrusion into the hemolymph triggers the production of ROS, such as superoxide (O2−), hydroxyl radical (OH−), hydrogen peroxide (H2O2), and singlet oxygen (1O2), in response; these ROS have been shown to play important anti-microbial roles in the hemolymph of Pacific white shrimp (Litopenaeus vannamei), for example [23, 24]. In shrimp inoculated with white spot syndrome virus (WSSV), transcript levels of superoxide dismutase (SOD), an oxidation–reduction enzyme that converts O2− into more stable H2O2 and O2, were determined to increase 1 h post-infection but subsequently decreased 12 h post-infection. It has been suggested that hemolymph SOD induction could be part of an early ROS detoxification response, as the steady rate of decline in SOD expression as the viral infection progressed would be expected to generate higher ROS production in the hemolymph [23]. Thus, ROS may be key factors in the microbial defense systems of shrimp.
Shrimp farming is an especially profitable form of aquaculture, at the global scale second only to that of carp production. Among cultured shrimp species, production of Pacific white shrimp has especially undergone dramatic expansion over the past several decades; by 2014, this species accounted for US$18.46 billion of the total US$23.58 billion generated by shrimp farming worldwide [25]. Like Pacific white shrimp, kuruma shrimp (Marsupenaeus japonicus) is a penaeid species, and has traditionally been the most important shrimp culture in Japan [26]. In recent years, however, the spread of infectious diseases, such as WSSV and early mortality syndrome (EMS) among penaeid shrimp populations in culture systems has slowed the expansion of production [27], thereby increasing the urgency for better understanding the defense mechanisms of these shrimp. In light of this, our objective here was to identify and clone MjXDH and MjAOX in kuruma shrimp, which, to the best of our knowledge, represents the first time XDH and AOX enzymes have been analyzed in crustaceans.
Materials and methods
Kuruma shrimp
Adult specific pathogen free kuruma shrimp (average weight: 8 g) obtained from Matsumoto Fisheries, Miyazaki, Japan. Shrimps were acclimated in aerated seawater maintained at 23 °C and fed twice daily with a commercial diet at a rate equal to 1% of their body weight.
Hydrogen peroxide quantification
Concentrations of H2O2 were determined using phenol red as a substrate [28, 29], with modifications. Hemolymph (100 µl) was mixed with 100 µl anticoagulant (29.22 g l−1 of NaCl, 3.8 g l−1 of EGTA and 2.38 g l−1 of Hepes) in a centrifuge tube, to which 30 µl 0.1 M Tris–HCl (pH 8) and β-mercaptoethanol were subsequently added. The mixture was centrifuged at 12,000 g for 20 min at 4 °C, with the supernatant used for the assay. 6 µl assay sample and 196 µl phenol red mix (1 g l−1 of MES, 7 mg l−1 of phenol red and 50 mg l−1 of peroxidase) were placed in a 96 well microplate and incubated for 4 min at room temperature. The reaction was stopped by 4 µl 0.5 N NaOH, and the absorbance was measured at 550 nm in a microplate reader. A standard curve of 0–200 nM of H2O2 was used.
Degenerate primer design
Partial genes from XDH and AOX cDNA were initially obtained using RT-PCR with degenerate primers antagonistic to the conserved regions, which were identified from other organisms using sequence alignment with the ClustalW program [http://www.mbio.ncsu.ebu/BioEdit/bioedit.html]. The degenerate primers (Table 1) were designed to anneal DNA sequences from highly conserved regions.
RNA extraction and cDNA preparation
Total RNA was extracted from the lymphoid organs of the kuruma shrimp using RNAiso Plus (TaKaRa, Japan), in accordance with the manufacturer’s instructions, and quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific, USA). RNA purity was evaluated via determination of the OD260/OD280 ratio. The extracted RNA samples were converted to cDNA using ReverTra Ace qPCR RT Master Mix with gDNA Remover (TOYOBO, Japan), in accordance with the manufacturer’s instructions.
Cloning and sequencing
Degenerate primers for MjXDH and MjAOX (Table 1) were initially designed from the primers of the conserved regions of the XDH and the AOX genes, and PCR analyses were performed with cDNA prepared using these primers to amplify the initial predicted sequence. To further identify the gene sequences, RACE-PCR was performed using an SMART RACE cDNA Amplification Kit (TaKaRaBio, Inc., Japan), in accordance with the manufacturer’s instructions (TaKaRaBio, Inc., Japan). Following determination of partial sequences of MjXDH and MjAOX, the entire lengths were obtained using 5′- and 3′-RACE-PCRs with the gene-specific primers (Table 1). KAPA™ HiFiHotStart DNA Polymerase (Kapa Biosystem, USA), a high-fidelity PCR polymerase, was used for the RACE-PCR. The PCR products were cloned into the pGEM-T Easy vector (Promega, USA). Plasmid DNA was recovered from the three least independent clones via a QIAprep Spin Mini-prep Kit (QIAGEN, Japan), and sequencing analysis was performed using a CEQ 8000 Automated Sequencer (Beckman Coulter, Inc., USA). The structural domains of MjXDH and MjAOX were predicted using the simple molecular architecture research tool (SMART; Version 7.0) [30]. Sequences generated were analyzed for similarity with other known sequences using FASTA and the basic local alignment search tool (BLAST). Direct comparisons between cDNA sequences were performed using the gap program of Bioedit, and multiple sequence alignments were generated using ClustalW [http://www.mbio.ncsu.ebu/BioEdit/bioedit.html]. A phylogenetic chart based on the full-length amino acid sequences of previously published XDH and AOX proteins was constructed using the neighbor-joining (NJ) method with MEGA 6 [http://www.megasoftware.net].
Analysis of MjXDH and MjAOX expression in kuruma shrimp by qRT-PCR
Various tissues (gill, heart, lymphoid organ, muscle, hemolymph, stomach, and intestine) were removed from healthy shrimp (n = 3) to analyze expression levels of MjXDH and MjNOX. Total RNA was extracted and respective cDNAs were prepared (“Degenerate primer design” section). The gene-specific primers for MjXDH and MjAOX (Table 1) were used to amplify the conserved regions. The kuruma shrimp elongation factor 1α (MjEF1α) gene (Table 1) served as an internal control to confirm the quality and quantity of the cDNA used. Healthy shrimp (average weight of 7 g; n = 5) were inoculated with a PBS, WSSV, and V. penaeicida suspension, with bacterial numbers adjusted to 1 × 105 copies/shrimp (WSSV) and 1 × 105 CFU/shrimp (V. penaeicida). Gene expression analyses of the enzymes (MjXDH and MjAOX) were performed at specific times post-injection (0, 6, 12, 24, 48, and 72 h). V. penaeicida was obtained from the National Institute of Technology and Evaluation (NITE) Biological Resource Center (NBRC No. 15640) and cultured in marine broth (Becton, Dickinson, USA) liquid media. The concentration of the bacterial solution was adjusted by the McFarland No. 1 standard method. The relative expression ratio of MjXDH and MjAOX was calculated based on SYBR green quantitative real-time PCR (qRT-PCR). SYBR green qRT-PCR amplifications were carried out in triplicate with a total volume of 15 µl containing 7.5 µl THUNDERBIRD™SYBR qPCRMix (TOYOBO, Japan), 1.5 µl cDNA, 1.5 µl each of forward and reverse primers (5 ppm), and 3 µl distilled water. The q-PCR program consisted of 95 °C for 15 s, 60 °C for 45 s, followed by 40 cycles on a CFX connect™ (Bio-Rad). Melting curve analysis of amplified products was performed at the end of each cycle to confirm the specificity of amplification. The comparative Ct method was used to analyze the expression level of each gene, with the Ct for the target amplification of each gene and the Ct for the internal control determined for each individual sample. The average CT measurement for the three was used in the relative expression calculations, with MjEF1α set as the internal control. Data obtained from the qRT-PCR analysis were subjected to one-way analysis of variance (one-way ANOVA) followed by an unpaired two-tailed t-test.
Statistical analysis
Data obtained from the hydrogen peroxide assay and the qRT-PCR analyses were subjected to one-way analysis of variance (one-way ANOVA) followed by unpaired t-test. Two-tailed t-tests were used to identify significant differences in the hydrogen peroxide assay and the qRT-PCR analyses between controls and each inflectional section. Significance was set as P < 0.05.
Results
Cloning and sequencing
The entire cloned sequence of MjXDH cDNA was 4328 bp, and contained a 4086 bp open reading frame coding for 1362 amino acids with an estimated mass of 150 kDa (deposited in GenBank under the accession number LC208790). That of MjAOX cDNA was 4425 bp, and contained a 4017 bp open reading frame coding for 1339 amino acids with an estimated mass of 146 kDa (deposited in GenBank under the accession number LC208791).
Domain structure
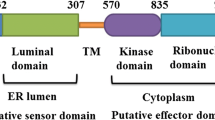
The complete domain structures of MjXDH and MjAOX were compared with those of other XDHs and AOXs to determine levels of similarity. MjXDH and MjAOX contained a molybdopterin domain (Mo-pt), an aldehyde/xanthine hammerhead domain, a CO dehydrogenase flavoprotein C-terminal domain, an FAD-binding domain, and two Fe/S redox center-containing domains. The length of each MjXDH domain was 538 aa (Mo-pt), 108 aa (aldehyde/xanthine hammerhead domain), 104 aa (CO dehydrogenase flavoprotein C-terminal domain), 181 aa (FAD-binding domain), 70 aa (Fe/S I domain), and 75 aa (Fe/S II domain), and the length of each MjAOX domain was 534 aa (Mo-pt), 109 aa (aldehyde/xanthine hammerhead domain), 107 aa (CO dehydrogenase flavoprotein C-terminal domain), 178 aa (FAD-binding domain), 72 aa (Fe/S I domain), and 75 aa (Fe/S II domain). These domains are typical of molybdo-flavoenzymes, to which class the XDH and AOX proteins belong (Fig. 1). Furthermore, induced protein alignment analyses of XDH and AOX sequences revealed that the location of each domain region and length were highly conserved among vertebrates and invertebrates, with the notable exception of the absence of four cysteine residues needed for D/O conversion from the XDHs of vertebrate (Figs. 2, 3).
Comparison of the deduced amino acid sequence of xanthine dehydrogenase from kuruma shrimp, fruit fly, human and rat. These sequences were aligned using the Crustal program and each domain regions were decided by SMART. GenBank accession numbers for using sequences is provided in supplemental material
Sequence homology
The amino acid sequences of MjXDH and MjAOX aligned well with the amino acid sequences of the various XDHs and AOXs in insects, fishes, and mammals (Table 2A and B). MjXDH had 52.4% sequence identity and 69.3% sequence similarity with fruit fly DmXDH; 52.4 and 69.7% with domestic silkworm BmXDH1; 55.4 and 70.7% with zebra fish DrXDH; and 43.9 and 62.8% with human HsXDH (Table 2A). MjAOX had 36.5% sequence identity and 53.5% sequence similarity with fruit fly, DmAOX1; 35.3 and 52.4% with domestic silkworm, BmAOX1; 27.3 and 44.1% with zebra fish, DrAOX; and 25.3 and 42.3% with human, HsAOX (Table 2B).
Phylogenetic analysis
A phylogenetic analysis was performed using XDH and AOX amino acid sequences from different species (Fig. 4). GenBank accession numbers of the sequences using phylogenetic analysis are listed (Supplemental material). The XDHs of insects and vertebrates and the AOXs formed different clusters. MjXDH was closely related to insects XDHs, whereas MjAOX was also closely related to insects AOXs, including southern house (Xiphophorus maculatus) and anopheles (Anopheles darling) mosquitoes, and the domestic silkworm (Bombyx mori). Mammalian AOX formed a cluster every kind of AOX, but, on the other hand, AOX of the invertebrate formed a cluster every species.
Gene expression analysis
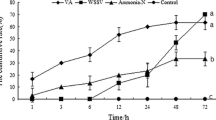
MjXDH and MjAOX expression levels were analyzed in various tissues, including the gill, heart, lymphoid organ, muscle, hemolymph, stomach, and intestine. Expression levels of MjXDH were higher in the heart and stomach tissues than in other organ tissues (Fig. 5a), whereas MjAOX expression was enhanced in the lymphoid organ and intestinal tissues compared to the other organ tissues, and expression was especially reduced in the hemolymph (Fig. 6 b). In a time-course experiment involving the PBS, WSSV, and V. penaeicida groups, no significant differences in MjXDH expression were observed in the heart and lymphoid organ tissues between the WSSV and V. penaeicida groups (Fig. 6a, c). On the other hand, although no significant changes were detected in heart tissue (Fig. 6d), MjAOX expression levels were highest in the lymphoid organ. Expression levels in the V. penaeicida group increased in the initial 24 h after injection, with levels reaching significance at 48–72 h after injection (P < 0.05). Between 48 and 72 h after injection, MjAOX expression was about 27-fold higher than in the PBS group at the same time points (Fig. 6b).
Kuruma shrimp MjXDH and MjAOX expressions in different organs of healthy kuruma shrimp. The expression of MjXDH and MjAOX were based on the ratio of these gene and MjEF-1α expression using quantitative real-time PCR. Bars represent mean ± standard errors (n = 3). a MjXDH expression analysis, b MjAOX expression analysis
Expression analyses of kuruma shrimp MjXDH and MjAOX in condition that injected PBS, WSSV and V. penaeicida and hydrogen peroxide concentration in hemolymph. PBS group served as a negative control. Heart and lymphoid organ were used for Expression analyses and hemolymph was used for. 0 h sample was collected from the group just before injection. Bars represent mean ± standard errors (gene expression analyses n = 5, hydrogen peroxide assay n = 4). Differences were considered at P < 0.05 and indicated by asterisks “*”. a MjXDH expression analysis in lymphoid organ. b MjAOX in lymphoid organ. c MjXDH in heart. d MjAOX in heart. e Hydrogen peroxide concentration in hemolymph
Hydrogen peroxide concentration
Concentrations of hydrogen peroxide in the hemolymph were measured in three groups, consisting of groups inoculated with PBS, WSSV, and V. penaeicida. Hydrogen peroxide levels were significantly higher in the WSSV group than in the PBS group at 6, 12, 48, and 72 h after injection, with concentrations reaching their highest points at 6 and 12 h, an approximately 1.7- to 2.8-fold increase over levels in the PBS group at the same time periods (P < 0.05). Hydrogen peroxide concentrations increased significantly in the V. penaeicida infected group at 72 h after injection, with levels roughly twofold higher than in the PBS group at the same time point (P < 0.05). An upward trend in the concentrations of hydrogen peroxide was observed in both the V. penaeicida and WSSV infected groups (Fig. 6 e).
Discussion
In this study, we identified the full-length MjXDH and MjAOX cDNA sequences of kuruma shrimp, which, to the best of our knowledge, is the first time that a crustacean XDH and AOX have been cloned and characterized following infection with WSSV and V. penaeicida.
The open reading frame (ORF) of MjXDH encodes a 1362 amino acid protein with an estimated mass of 150 kDa. The molecular mass of MjXDH was similar to those of XDH found in turnip sawfly, zebra fish, and chicken, which were ~ 157, ~ 148, and ~ 149 kDa, respectively. Furthermore, based on analysis of amino acid sequences, MjXDH sequence homology was determined to be highest with zebra fish (55.2% identity and 70.7% similarity). A molybdopterin (Mo-pt), aldehyde/xanthine hammerhead, CO dehydrogenase flavoprotein C-terminal, FAD-binding, and two Fe/S redox center-containing domains were predicted during domain structure analysis of MjXDH, domains that are highly conserved in the XDHs of other species. In mammals, a molybdopterin domain (Mo-pt) plays an important role in the catalysis of the hydroxylation reaction that produces uric acid from xanthine. The electron provided by the hydroxylation reaction in the Mo-pt domain moves to the FAD domain via the two Fe domains, which function as electron transports [31, 32]. Thus, these domains are fundamental for the oxidation–reduction reaction that produces uric acid, and the fact that these domains are conserved in MjXDH indicates that it is a functional protein. Conversely, four cysteine residues (Cys535, Cys992, Cys1316, and Cys1324) required for D/O conversion of XDH to XO in the Mo-pt domain were not conserved in MjXDH nor in insect XDHs, and therefore XDHs of invertebrates, including the kuruma shrimp, and XDHs of vertebrates formed separate phylogenetic clusters. In mammals, D/O conversion derives from linker cutting by proteases and disulfide-binding formation with these four cysteine residues, with hydrogen peroxide produced as a substrate by XO [33, 34].
The ORF of MjAOX encodes a 1,339 amino acid protein with an estimated mass of 146 kDa. The molecular mass of kuruma shrimp MjAOX was similar to that of AOXs in rats and domestic silkworms, at ~ 148 and ~ 143 kDa, respectively. All domains characteristic of the molybdo-flavoenzyme subfamily were present in MjAOX. In contrast to MjHDX, MjAOX sequence homology was highest with that of the fruit fly AOX1 (36.5% identity and 53.5% similarity), and moreover MjAOX sequence homology with other species was generally lower than that for MjXDH, suggesting the development of a variety of AOXs over the course of evolutionary history. Four types of AOX (AOX1–AOX4) are known to have originated from a common ancestral XDH and gradually differentiated into diverse enzymatic functions [21]. In our phylogenetic analysis, different clusters were formed among mammalian AOX1, AOX2, AOX3, AOX4, and XDH, whereas MjAOX was grouped with insect AOXs.
In expression analyses targeting mRNA, MjXDH was expressed in all sampled tissues, including the gill, heart, lymphoid organ, muscle, hemolymph, stomach, and intestine, with the highest expression levels observed in heart tissues. In humans, XDH is expressed in a wide variety of organs, an indication that XDH participates in the pathogens and plays an important role in cellular metabolic processes [35, 36]. However, no significant changes in MjXDH expression were detected following viral and bacterial injections, implying that MjXDH expression is not induced by viral or bacterial infection and thus MjXDH does not play a direct role in ROS generation. Alignment analyses revealed that four cysteine residues essential for D/O conversion were not conserved in MjXDH, and consequently kuruma shrimp are less likely to have a functioning hydrogen peroxide production mechanism regulated by XO; in an experiment involving fruit flies, for example, E. coli abundance was higher in a DmXDH mutant that lacked the four cysteine residues than in the wild type [37]. In addition, the increased XOR expression by cycloheximide previously observed in mice epithelial cells (HC11 cells) and the high sequence homology between MjXDH and the XDHs of other species detected in our study suggests that although MjXDH does not produce ROS directly, it may act as a secondary messenger in the immune system [38]. Moreover, the variation in MjAOX expression in each type of body tissue was more pronounced than for MjXDH, and the highest expression levels occurred in the lymphoid organ. Because the AOX enzyme is evolutionarily derived from an ancestral XDH and fulfills a wider variety of roles than XDH, which has a more or less stable and universal role, we believe MjAOX to be far more likely to have site-specific functions than MjXDH. Mammalian AOXs oxidize not only aromatic and aliphatic aldehydes into corresponding carboxylic acids, but also hydroxylate a series of heteroaromatic rings, the broad substrate specificity of which is depend on the diversity of isoforms [20]. It has been shown in fruit flies that each of the four identified DmAOXs have different substrate specificities and gene expression patterns [22]; moreover, in the rice leaf roller (Cnaphalocrocis medinalis), it is generally thought that CmAOX functions as an odorant-degrading enzyme (ODE) that has an essential role in the degradation of volatile odorants and maintenance of olfactory sensitivity [39]. Thus, it is possible that kuruma shrimp also have several AOX isoforms in addition to MjAOX, and that these isoforms have different substrate specificities, as is conjectured for other insects. In a time-course experiment of MjAOX with PBS, WSSV, and V. penaeicida injected groups, MjAOX expression in the lymphoid organ started to increasing at 24 h after V. penaeicida injection and showed significant increases at 48–72 h after injection. The highest expression level of MjAOX was confirmed at 48–72 h and was increased about 26–27 fold compared to the PBS group at the same time point. The lymphoid organ of penaeid shrimp plays important roles in the innate immune system [40], and thus, in light of its high level of expression in the lymphoid organ and its up-regulation following inoculation with pathogens, we hypothesize that MjAOX is therefore essential to the immune system of kuruma shrimp. In mammals, hydrogen peroxide concentrations are dependent on AOX activity and it reports show AOX is one of the fatal hydrogen peroxide generating enzymes [41, 42]. In N. benthamiana plants infected with tomato bushy stunt virus (TBSV), non-denaturing PAGE analysis indicated that AOX expression increased, as did levels of hydrogen peroxide [43]. Furthermore, it was previously shown that knockdown dual oxidase (MjDuox), a type of hydrogen peroxide synthetase similar to MjAOX, increased mortality rates of kuruma shrimp infected with V. penaeicida infection [44]. The upswing in MjAOX expression is linked to a rise in the concentration of hydrogen peroxide in the vibrio ward, and MjAOX is thought to be one of the ROS synthetases at the time of infection by V. penaeicida.
In the hydrogen peroxide assay, higher hydrogen peroxide concentrations in the hemolymph were observed in shrimp infected with WSSV and V. penaeicida; in particular, hydrogen peroxide concentrations at 6, 12, 48, and 72 h in the WSSV-injected group, and at 72 h in the V. penaeicida-injected group, were significantly higher than in the PBS group at the same time points. Although reaction activity and sterilization rates are higher for free radicals than for hydrogen peroxide, free radicals are less stable than hydrogen peroxide. Moreover, excessive production of superoxide can result in severe cell and DNA damage, but because of its high reactivity, superoxide is rapidly converted into hydrogen peroxide by superoxide dismutase (SOD), and hydrogen peroxide is able to pass through cellular membranes, whereas superoxide cannot [45]. In addition, it has been reported that hydrogen peroxide serves as a secondary messenger in cell signaling pathways and can up-regulate phagocytic activity via SOD [46]. As such, hydrogen peroxide plays a key role in the sterilization of bacteria by ROS. That concentrations of hydrogen peroxide were observed to increase in the hemolymphs of shrimp in response to inoculation with WSSV and V. penaeicida underscored the importance of hydrogen peroxide in kuruma shrimp biological defense mechanisms.
In conclusion, we determined that hydrogen peroxide concentrations in the hemolymph of kuruma shrimp fluctuated in response to pathogen exposure, and analyzed the gene-expression response of MjXDH and MjAOX to pathogens. Characteristic domains of molybdo-flavoenzymes are highly conserved in MjXDH and MjAOX, and both enzymes formed a cluster with the XDHs and AOXs of insects. In regard to tissue-specific distribution, MjXDH expression was highest in heart tissues, whereas that of MjAOX was highest in the lymphoid organ, leading us to hypothesize that MjAOX is an ROS-producing enzyme, in light of the fact that expression levels in the lymphoid organ increased 48–72 h after vibrio injection and hydrogen peroxide concentrations also increased. In order to thoroughly understand the ROS-producing mechanism via molybdo-flavoenzyme response to infection, further research on XDH and AOX is necessary. The results of our study represent an important step toward understanding the innate immune system of crustaceans.
References
Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, Telser J (2007) Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol 39(1):44–84. https://doi.org/10.1016/j.biocel.2006.07.001
Cohn CA (2005) Quantifying hydrogen peroxide in iron-containing solutions using leuco crystal violet. Geochem Trans 6(3):47. https://doi.org/10.1063/1.1935449
Graves JA, Metukuri M, Scott D, Rothermund K, Prochownik EV (2008) Regulation of reactive oxygen species homeostasis by peroxiredoxins and c-Myc. J Biol Chem 284(10):6520–6529. https://doi.org/10.1074/jbc.m807564200
Chelikani P, Fita I, Loewen PC (2004) Diversity of structures and properties among catalases. Cell Mol Life Sci 61(2):192–208. https://doi.org/10.1007/s00018-003-3206-5
Nordberg J, Arnér ESJ (2001) Reactive oxygen species, antioxidants, and the mammalian thioredoxin system. Free Radic Biol Med 31:1287–1312
Paglia DE, Valentine WN (1967) Studies on the quantitative and qualitative characterization of erythrocyte glutathione peroxidase. J Lab Clin Med 70:158–169
Juven BJ, Pierson MD (1996) Antibacterial effects of hydrogen peroxide and methods for its detection and quantitation. J Food Prot 59(11):1233–1241. https://doi.org/10.4315/0362-028x-59.11.1233
Skulachev VP (1998) Possible role of reactive oxygen species in antiviral defense. Biochem 63:1438–1440
Iwanaga S, Lee B (2005) Recent advances in the innate immunity of invertebrate animals. BMB Rep 38(2):128–150. https://doi.org/10.5483/bmbrep.2005.38.2.128
Lavine M, Strand M (2002) Insect hemocytes and their role in immunity. Insect Biochem Mol Biol 32(10):1295–1309. https://doi.org/10.1016/s0965-1748(02)00092-9
Manevski N, Balavenkatraman KK, Bertschi B, Swart P, Walles M, Camenisch G, Litherland K (2014) Aldehyde oxidase activity in fresh human skin. Drug Metab Dispos 42(12):2049–2057. https://doi.org/10.1124/dmd.114.060368
Pope SD, Chen L, Stewart V (2008) Purine utilization by Klebsiella oxytoca M5al: genes for ring-oxidizing and -opening enzymes. J Bacteriol 191(3):1006–1017. https://doi.org/10.1128/jb.01281-08
Garattini E, Mendel R, Romão MJ, Wright R, Terao M (2003) Mammalian molybdo-flavoenzymes, an expanding family of proteins: structure, genetics, regulation, function and pathophysiology. Biochem J 372(1):15–32. https://doi.org/10.1042/bj20030121
Llamas A, Chamizo-Ampudia A, Tejada-Jimenez M, Galvan A, Fernandez E (2017) The molybdenum cofactor enzyme mARC: moonlighting or promiscuous enzyme? BioFactors 43(4):486–494. https://doi.org/10.1002/biof.1362
Bray R (1975) 6 Molybdenum iron-sulfur flauin hydroxylases and related enzymes. Enzymes. https://doi.org/10.1016/s1874-6047(08)60229-2
Berry CE, Hare JM (2004) Xanthine oxidoreductase and cardiovascular disease: molecular mechanisms and pathophysiological implications. J Physiol 555(3):589–606. https://doi.org/10.1113/jphysiol.2003.055913
Nishino T, Okamoto K, Eger BT, Pai EF, Nishino T (2008) Mammalian xanthine oxidoreductase—mechanism of transition from xanthine dehydrogenase to xanthine oxidase. FEBS J 275(13):3278–3289. https://doi.org/10.1111/j.1742-4658.2008.06489.x
Ng’oma E, Groth M, Ripa R, Platzer M, Cellerino A (2014) Transcriptome profiling of natural dichromatism in the annual fishes Nothobranchius furzeri and Nothobranchius kadleci. BMC Genomics 15(1):754. https://doi.org/10.1186/1471-2164-15-754
Zhou X, Riddiford LM (2008) rosy function is required for juvenile hormone effects in Drosophila melanogaster. Genetics 178(1):273–281. https://doi.org/10.1534/genetics.107.080754
Terao M, Romão MJ, Leimkühler S, Bolis M, Fratelli M, Coelho C, Garattini E (2016) Structure and function of mammalian aldehyde oxidases. Arch Toxicol 90(4):753–780. https://doi.org/10.1007/s00204-016-1683-1
Kurosaki M, Bolis M, Fratelli M, Barzago MM, Pattini L, Perretta G, Garattini E (2012) Structure and evolution of vertebrate aldehyde oxidases: from gene duplication to gene suppression. Cell Mol Life Sci 70(10):1807–1830. https://doi.org/10.1007/s00018-012-1229-5
Marelja Z, Dambowsky M, Bolis M, Georgiou ML, Garattini E, Missirlis F, Leimkuhler S (2014) The four aldehyde oxidases of Drosophila melanogaster have different gene expression patterns and enzyme substrate specificities. J Exp Biol 217(12):2201–2211. https://doi.org/10.1242/jeb.102129
Gomezanduro G, Barillasmury C, Peregrinouriarte A, Gupta L, Gollasgalvan T, Hernandezlopez J, Yepizplascencia G (2006) The cytosolic manganese superoxide dismutase from the shrimp Litopenaeus vannamei: molecular cloning and expression. Dev Comp Immunol 30(10):893–900. https://doi.org/10.1016/j.dci.2006.01.002
Muñoz M, Cedeño R, Rodríguez J, Van der Knaap WP, Mialhe E, Bachère E (2000) Measurement of reactive oxygen intermediate production in haemocytes of the penaeid shrimp, Penaeus vannamei. Aquaculture 191(1–3):89–107. https://doi.org/10.1016/s0044-8486(00)00420-8
Tacon AG (2017) Biosecure shrimp feeds and feeding practices: guidelines for future development. J World Aquac Soc 48(3):381–392. https://doi.org/10.1111/jwas.12406
Bulbul MG, Kader MA, Asaduzzaman M, Ambak MA, Chowdhury AJ, Hossain MS, Koshio S (2016) Can canola meal and soybean meal be used as major dietary protein sources for kuruma shrimp Marsupenaeus japonicus? Aquaculture 452:194–199. https://doi.org/10.1016/j.aquaculture.2015.10.036
Thitamadee S, Prachumwat A, Srisala J, Jaroenlak P, Salachan PV, Sritunyalucksana K, Itsathitphaisarn O (2016) Review of current disease threats for cultivated penaeid shrimp in Asia. Aquaculture 452:69–87. https://doi.org/10.1016/j.aquaculture.2015.10.028
García-Triana A, Peregrino-Uriarte AB, Yepiz-Plascencia G (2016) Selenoprotein M gene expression, peroxidases activity and hydrogen peroxide concentration are differentially regulated in gill and hepatopancreas of the white shrimp Litopenaeus vannamei during hypoxia and reoxygenation. Comp Biochem Physiol Part A Mol Integr Physiol 199:14–20. https://doi.org/10.1016/j.cbpa.2016.04.019
Messner B, Boll M (1994) Cell suspension cultures of spruce (Picea abies): inactivation of extracellular enzymes by fungal elicitor-induced transient release of hydrogen peroxide (oxidative burst). Plant Cell Tissue Organ Cult 39(1):69–78. https://doi.org/10.1007/bf00037594
Letunic I, Doerks T, Bork P (2011) SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. https://doi.org/10.1093/nar/gkr931
Kobayashi K, Miki M, Okamoto K, Nishino T (1993) Electron transfer process in milk xanthine dehydrogenase as studied by pulse radiolysis. J Biol Chem 268:24642–24646
Olson JS, Ballou DP, Palmer G, Massey V (1974) The mechanism of action of xanthine oxidase. J Biol Chem 249:4363–4382
Kanda M, Brady FO, Rajagopalan KV, Handler P (1972) Studies on the dissociation of flavin adenine dinucleotide from metalloflavoproteins. J Biol Chem 247:765– 70
Komai H, Massey V, Palmer G (1969) The preparation and properties of deflavo xanthine oxidase. J Biol Chem 244:1692–1700
Harrison R (2002) Structure and function of xanthine oxidoreductase: where are we now? Free Radic Biol Med 33(6):774–797. https://doi.org/10.1016/s0891-5849(02)00956-5
Wright RM, Vaitaitis GM, Wilson CM, Repine TB, Terada LS, Repine JE (1993) cDNA cloning, characterization, and tissue-specific expression of human xanthine dehydrogenase/xanthine oxidase. Proc Natl Acad Sci USA 90(22):10690–10694. https://doi.org/10.1073/pnas.90.22.10690
Kim YS, Nam HJ, Chung HY, Kim ND, Ryu JH, Lee WJ, Yoo MA (2001) Role of xanthine dehydrogenase and aging on the innate immune response of Drosophila. J Am Aging Assoc 24(4):187–193. https://doi.org/10.1007/s11357-001-0020-6
Seymour KJ, Roberts LE, Fini MA, Parmley LA, Oustitch TL, Wright RM (2006) Stress activation of mammary epithelial cell xanthine oxidoreductase is mediated by p38 MAPK and CCAAT/enhancer-binding protein-β. J Biol Chem 281(13):8545–8558. https://doi.org/10.1074/jbc.m507349200
Zhang Y, Wang W, Li M, Li S, Liu S (2017) Identification of putative carboxylesterase and aldehyde oxidase genes from the antennae of the rice leaffolder, Cnaphalocrocis medinalis (Lepidoptera: Pyralidae). J Asia Pac Entomol 20(3):907–913. https://doi.org/10.1016/j.aspen.2017.06.001
Pongsomboon S, Wongpanya R, Tang S, Chalorsrikul A, Tassanakajon A (2008) Abundantly expressed transcripts in the lymphoid organ of the black tiger shrimp, Penaeus monodon, and their implication in immune function. Fish Shellfish Immunol 25(5):485–493. https://doi.org/10.1016/j.fsi.2008.07.010
Kamli MR, Kim J, Pokharel S, Jan AT, Lee EJ, Choi I (2014) Expressional studies of the aldehyde oxidase (AOX1) gene during myogenic differentiation in C2C12 cells. Biochem Biophys Res Commun 450(4):1291–1296. https://doi.org/10.1016/j.bbrc.2014.06.126
Kundu TK, Hille R, Velayutham M, Zweier JL (2007) Characterization of superoxide production from aldehyde oxidase: an important source of oxidants in biological tissues. Arch Biochem Biophys 460(1):113–121. https://doi.org/10.1016/j.abb.2006.12.032
Yergaliyev TM, Nurbekova Z, Mukiyanova G, Akbassova A, Sutula M, Zhangazin S, Omarov RT (2016) The involvement of ROS producing aldehyde oxidase in plant response to Tombusvirus infection. Plant Physiol Biochem 109:36–44. https://doi.org/10.1016/j.plaphy.2016.09.001
Inada M, Kihara K, Kono T, Sudhakaran R, Mekata T, Sakai M, Itami T (2013) Deciphering of the dual oxidase (Nox family) gene from kuruma shrimp, Marsupenaeus japonicus: full-length cDNA cloning and characterization. Fish Shellfish Immunol 34(2):471–485. https://doi.org/10.1016/j.fsi.2012.11.026
Blokhina O (2003) Antioxidants, oxidative damage and oxygen deprivation stress: a review. Ann Bot 91(2):179–194. https://doi.org/10.1093/aob/mcf118
Arbi M, Pouliliou S, Lampropoulou M, Marmaras VJ, Tsakas S (2011) Hydrogen peroxide is produced by E. coli challenged haemocytes and regulates phagocytosis, in the medfly Ceratitis capitata. The active role of superoxide dismutase. Dev Comp Immunol 35(8):865–871. https://doi.org/10.1016/j.dci.2011.03.020
Acknowledgements
This work was supported by JSPS-KAKENHI a Grant-in-Aid for Scientific Research (C) 15K07555 and Grant- in-Aid for Young Scientists (B) 16K18747. We would like to thank Editage (http://www.editage.jp) for English language editing.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No potential conflict of interest are disclosed.
Research involving human participants and/or animals
The kuruma shrimp that our target species in this study is edible shrimp in Japan. So, this study is not applicable study in this item. However, before dissecting, the kuruma shrimp was immersed into the iced water for around 5 mm into fall into suspended animation.
Informed consent
We agree that the information in this study is disclosed.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Okamura, Y., Inada, M., Elshopakey, G.E. et al. Characterization of xanthine dehydrogenase and aldehyde oxidase of Marsupenaeus japonicus and their response to microbial pathogen. Mol Biol Rep 45, 419–432 (2018). https://doi.org/10.1007/s11033-018-4177-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-018-4177-9