Abstract
DNA glycosylases are DNA repair enzymes capable of removing damaged nitrogenous bases, including those formed as a result of UV irradiation with sunlight (approximately 300–400 нм). DNA glycosylases are common not only among bacteria, archaea, and eukaryotes, but some groups of viruses can also encode them. The best-known viral glycosylase is endonuclease V (DenV, Pdg-T4) of Escherichia virus T4, the main substrate of which is cyclobutane pyrimidine dimers. DenV is isolated separately from other large families of glycosylases; it is quite common in nature and has homologs in a number of other viruses and even bacteria. However, the ways of its origin are poorly understood. The best-known DenV homolog is the glycosylase of Chlorella virus strain, PBCV-1 (Cv-pdg). This review contains the main known data on the structure and mechanism of operation of DenV and its homologs. The issues of biological importance and distribution of the enzyme and its homologs among viruses are considered and supplemented separately.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Point mutations are quite a common phenomenon resulting in the disruption of the processes of gene replication and expression, which can ultimately lead to cell death. One of the causes of such mutations is UV radiation. In regard to its biological effects, UV radiation is usually divided into three spectral regions: UV A (320–400 nm), UV B (295–320 nm), and UV C (100–295 nm). As an integral part of sunlight, UV radiation is a factor affecting most Earth ecosystems. Accordingly, living entities had to develop effective methods of protection from UV light in the course of their evolution. Living organisms have numerous protective mechanisms (repair systems) responsible for the restoration of DNA damaged by UV irradiation (Rastogi et al. 2010). One of such repair systems is the system of base excision repair (BER) (Sampath et al. 2012). BER involves a group of enzymes, the main part of which is DNA glycosylases. This type of repair makes it possible to remove damaged bases in small segments of the DNA chain. The UV-induced DNA abnormalities that are removed by BER are cyclobutane pyrimidine dimers (CPD), (6–4) photoproducts, and Dewar photoproducts (Yokoyama and Mizutani 2014; Ganesan and Hanawalt 2016; Chatterjee and Walker 2017).
The BER pathway and glycosylases seem to be an early evolutionary acquisition, since they are found in bacteria (Friedberg et al. 2006), archaea (Marshall and Santangelo 2020), eukaryotes, and viruses, including bacteriophages (Bernstein and Bernstein 2001). From the evolutionary point of view, the most interesting are glycosylases found in viruses. Most viruses have a rather compact genome: They carry the minimal set of genes, necessary to maintain their life cycle—genes, which are often absent in the host cell. DNA repair in case of its damage by various mutagenic factors, as a rule, occurs due to the repair systems of the host cell. Some viruses, however, carry genes encoding the homologs of prokaryotic repair systems. For example, bacteriophage T4 has several genes encoding homologs of DNA repair systems. There are genes for the recombinational repair system (uvsXWY), excision repair genes (in particular, denV), and a number of other genes involved in restoring DNA functions disturbed by the mutation process (Friedberg 1972; Bernstein 1981; Miller et al. 2003a). This raises questions about the biological significance of the presence of several repair systems for viruses, taking into account the characteristics of their life cycle. The recombinational repair system of bacteriophage T4, in addition to repairing damaged phage DNA, including after exposure to UV radiation, closely associated with the provision of T4 phage multiplication in the processes of replication and packaging of its DNA. It is not known exactly why phages need an additional system for the repair of DNA photoproducts, carried out with the participation of DenV. Moreover, DenV, unlike many other glycosylases, has the highest affinity for the substrate in the form of cyclobutane pyrimidine dimers, and not for purine and pyrimidine oxidation products (Bernstein 1981; Miller et al. 2003a). At the same time, DenV seems to be a fairly common glycosylase among a number of viruses. Researchers have shown that DenV is encoded not only by a number of bacteriophages—T4, RB69, RB70, 44RR2.8t, and KVP40—but also by viruses of unicellular algae Paramecium bursaria Chlorella Virus 1 (Zharov 2008; Miller et al. 2003b; McCullough et al. 1998). The DenV homolog of chlorella viruses was named Cv-pdg and is 41% identical to phage glycosylase (McCullough et al. 1998). In this review, we will discuss the current experimental studies of the specific structural and biochemical characteristics of the two most studied viral glycosylases, DenV T4 (T4-pdg) and Cv-pdg, and try to answer a number of questions related to why viruses need their own pyrimidine dimer glycosylases in the presence of similar enzymes in the host cell. Why is CPD repair and the enzymes responsible for it not found in all viruses? Does the presence of pyrimidine dimer glycosylases give an advantage to representatives of certain taxonomic groups of viruses? How common are DenV T4 glycosylase homologs among other viruses? Answers to these questions can both help to understand the role of viral pyrimidine dimer glycosylases in the biological world and help lay the foundations for using this enzyme or its homologs in the practice of protecting human and domestic animal health from the mutagenic effects of sunlight.
Mechanism of functioning of T4 glycosylase
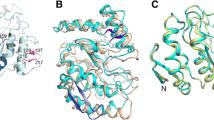
Glycosylase T4 (Pdg-T4), also known as endonuclease V (DenV), has been mostly investigated by R. S. Lloyd’s group (Lloyd 2005). Those studies resulted in the establishment of the mechanism and principles of operation of bifunctional glycosylases (Lloyd 2005). DenV is a small protein (138 aa). It is structured as a small single domain consisting of 3α-helices and connecting loops, with the whole macromolecule shaped as a comma (Morikawa et al. 1992, 1995; Lloyd 2005; Golan et al. 2006). The protein is characterized by an asymmetric arrangement of charged amino acid residues: basic residues on one side and acidic residues on the other. Such an arrangement ensures the correct orientation of the enzyme towards the substrate and affects the catalysis efficiency (Lloyd 2005).
The mechanism of DenV operation can be described as follows. The first step is binding of the enzyme to DNA, which is driven by a reaction of cationic substitution. The reaction is non-specific; after binding to DNA, the enzyme performs a one-dimensional scan of the DNA strand for pyrimidine dimers. The translocation of the enzyme along the strand is driven by Brownian motion. This is called processive search—because the enzyme is found to remain bound to DNA until most of pyrimidine dimers are removed from the strand (Lloyd et al. 1980; Gruskin and Lloyd 1986, 1988; Lloyd 2005). The degree of processivity was shown to be an important indicator of the efficiency of detection of target sites (Dowd and Lloyd 1990). The second step is specific binding of the enzyme to a pyrimidine dimer. The analysis of crystal structures of mutant DenV/DNA complexes revealed that, similar to some other glycosylases, DenV twists the DNA strand, flipping out the base into an extra-helical position, and trapping it in the enzyme “pocket” (Vassylyev et al. 1995; McCullough et al. 1997). What is unique to DenV, however, is that it is not the damaged base that is flipped out—it is the base complementary to the damaged one. This flipped out complementary base does not form hydrogen bonds with the enzyme: It is retained in the extra-helical position by van der Waals interactions. At that moment, the DNA chain bends sharply (up to 60°; Vassylyev et al. 1995; Lloyd 2005). Owing to these base exposure conformational changes, the amino acid residues of the active site of the enzyme get access to the substrate, making the target N-glycosidic bond more accessible for a nucleophilic attack. However, these conformational changes seem to be not obligatory for catalysis: Sometimes, base flip out does not occur, yet DenV can still recognize the target site. Rather than being essential for catalysis, base flip out may just facilitate the reaction (McCullough et al. 1997). The next step is catalytic: cleavage of the glycosidic bond of pyrimidine dimer. The active center of the enzyme is located at its N-terminus (Zharkov 2008). There are ten lysine residues at this end; probably, they ensure non-specific binding of the enzyme to DNA and its specific binding to CPD (Lloyd 2005). The α-amino group at the N-terminus of the enzyme (namely, at Thr2) performs a nucleophilic attack on the C1′ nucleoside of CPD (Schrock and Lloyd 1991, 1993; Dodson et al. 1993), which is accompanied by the dissociation of proton from Nα Thr2. The protonation of the O4′ ring of deoxyribose, which seems to be mediated by Glu23, stabilizes and opens the ribose ring, removing a second proton from Thr2 Nα (Doi et al. 1992). The outcome of the attack is the cleavage of the N-glycosidic bond with the formation of a Schiff base fragment (Dodson et al. 1993). As a result, an apyrimidine site (AP site) is formed. The subsequent reactions are determined by the AP-lyase activity of the enzyme (Kim and Linn 1988). First, there occurs β-elimination (through dissociation of proton from C2′, presumably by O4′), which is followed by the cleavage of phosphate at C3′ (Manoharan et al. 1988; McCullough et al. 2001). The regeneration of the enzyme occurs due to hydrolysis of the newly formed Schiff base. As a result of the reaction, a single-strand break is formed at the 3′ end. What remains in the end are 3′-open ribose ring, α,β-unsaturated aldehyde, and 5′ phosphate with 5′ pyrimidine dimer, still covalently bound to its 3′ pyrimidine (Lloyd 2005).
The subsequent reactions of the BER pathway are catalyzed by other proteins, which are encoded by both bacteriophage genes and genes of its host, E. coli—similar to the BER pathway mediated by bifunctional glycosylases (Bernstein 1981). A number of studies were carried out on mutant forms of DenV T4. They showed that a series of substitutions at certain sites of the enzyme amino acid sequence led to the disorder of its function. Apparently, Arg3 plays an important role in the binding of the enzyme to substrate, since its substitution for Lys3 was demonstrated to result in a complete loss of enzyme activity (Doi et al. 1992). The substitution of Glu23 for Asp23 or Gln23 completely inhibited glycosylase activity of the enzyme, yet did not affect its AP-lyase activity. This makes sense given that Glu23 is directly involved in the catalysis (Doi et al. 1992; Hori et al. 1992). Replacing Arg26 with Gln26 decreased glycosylase activity of the enzyme, confirming the fact that this site is involved in substrate binding (Doi et al. 1992). Arg22 is most likely involved in the DNA bending, which is responsible for the flip out of the base opposite to CPD (Doi et al. 1992). The substitution of Arg22 for Gln22 leads to the loss of glycosylase activity of the enzyme, indicating the importance of a positively charged amino acid residue at this position for the normal functioning of the enzyme. The substitutions of Arg17, Lys121, and Arg177 affect the efficiency of the enzyme binding to substrate. Lys121 is also crucial for glycosylase activity (Doi et al 1992). The substitution of His16 for other amino acids (Ala16, Cys16, Asp16, Glu16, Lys16, Gln16, or Ser16) decreased the enzyme activity to a varying degree. The authors of this study supposed that His16 might participate in the formation of an intermediate Schiff base (Meador et al. 2004).
Substrate specificity of DenV
According to the literature data, the main substrate of DenV is cyclobutane pyrimidine dimers. This type of abnormality consists in the formation of a cyclobutane ring between 5,6-bonds of two pyrimidine bases. Such cross-links can occur in the T^T, T^C, and C^C pairs; most often, however, they are found in the T^T pairs (Friedberg et al. 2006). It seems DenV can effectively cleave the glycosidic bond of the following photoproducts: T^T, C^T, C^C, and T^5-HMC. It was also noted that the efficiency of catalysis depended on the DNA sequence (Childs et al. 1983). A number of bacteriophages are known to contain non-canonical DNA bases (Nikulin and Zimin 2021), and some of these bases can form photoproducts, including CPD (Kim et al, 2013). In the DNA of wild-type T4 bacteriophage, cytosine is completely replaced with 5′-hydroxymethylcytosine (5-HMC) and its glucosylated variant (Miller et al. 2003a). These non-canonical bases are prone to the formation of photoproducts (Childs et al. 1983). Under UV C (254 nm), the probability of T^5-HMC dimerization is quite low, yet it increases significantly under sunlight (≥ 313 nm). Photoreactivation of photoproducts of non-canonical bases is inefficient: twice as low as that of canonical bases. Correspondingly, the lethality from 5-HMC^T dimerization in phages was shown to be substantially higher. Experiments with mutant phages, in which denV gene was knocked out, showed their decreased resistance to UV (Childs et al. 1978). These phages were 2–2.5 times more sensitive to UV C (254 nm) and 4–5 times more sensitive to UV B (302–305 nm) (Childs et al. 1978, 1983). The activity of DenV on substrates containing the oxidation products of purines and pyrimidines was not found to be high. Some studies demonstrated that DenV was also able to recognize other damaged bases: O-methylhydroxylamine and O-benzylhydroxylamine—although the efficiency of the recognition was rather low (Purmal et al. 1996). There were also studies indicating that DenV was able to recognize formadylpyrimidine products (FapyA). However, the efficiency of their elimination from DNA was less than 1% of the elimination rate of CPD (Dizdaroglu et al. 1996).
Glycosylase of chlorella virus (Cv-pdg)
One of the known homologs of DenV T4 among eukaryotic viruses is glycosylase of chlorella viruses (Cv-pdg). Cv-pdg has 41% homology with DenV T4 (McCullough et al. 1998) and a similar mechanism of action: It performs a nucleophilic N-terminal attack to cleave the glycosidic bond, with the protonation mediated by Glu23, as is case with DenV (Garvish and Lloyd 1999). This is followed by β-elimination, and the end product is, as in case of DenV, a 3′-α,β-unsaturated aldehyde formed after β-elimination at the abase site. If there is an excess of the enzyme, one can observe the formation of products characteristic of δ-elimination (Garvish and Lloyd 1999). The modeling of Cv-pdg structure upon interaction of the enzyme with DNA showed that the location of major regions of the active site is similar for both glycosylases (Zhu et al 1999). For DenV, these are Thr2, Arg3, and Gln15-Arg26; Lys33, His34, Gly55-Tyr61, Gln71, Gly82-Gln91, and Gln124-Lys130. For Cv-pdg, the catalytically important residues are located at the same sites, but there are also substitutions. Near the N-terminus, the main difference is Pro25-Arg26 in DenV versus Lys25-Met26 in Cv-pdg. Presumably, this substitution also increases the catalytic activity of Cv-pdg. On the basis of the results of modeling, the authors hypothesized that mutants of Cv-pdg by Met26Arg and Arg130 would have a strongly modified catalytic activity and binding of the enzyme to DNA, respectively (Zhu et al 1999).
A homolog of Cv-pdg was also found in the Acanthocystis turfacea chlorella virus (ATCV-1) (Fitzgerald et al 2007).
Substrate specificity of Cv-pdg
Despite the similarity between DenV and Cv-pdg, the latter has a broader substrate specificity. Cv-pdg can use various CPD photoisomers as substrates—in particular, trans-syn II dimers. The catalytic activity of Cv-pdg towards cis-syn dimers is higher than that of DenV (McCullough et al. 1998). Presumably, the broad substrate specificity of Cv-pdg, as compared to DenV, is related to a certain flexibility of its active site. In addition, Cv-pdg was shown to have a higher surface charge and a less dense packing of its polypeptide chain (Zhu et al. 1999). Cv-pdg is also characterized by a higher degree of processivity, which, in contrast to DenV, is not reduced at high salt concentrations. The Cv-pdg glycosylase was demonstrated to cleave not only cyclobutane dimers, but also products of the UV-induced free-radical oxidation—formamidopyrimidines (FapyAde and FapyGua) (Jaruga et al. 2002). An example of the enzyme that has catalytic activity towards such substrates is Fdg of E. coli.
Biological importance of DenV for viruses
The question of biological importance of DenV T4 and Cv-pdg for viruses has been repeatedly raised in the studies of these enzymes. As mentioned above, viruses (except giant ones) generally have a compact genome, which does not include genetic information about the systems whose analogs and homologs are available in the hosts of those viruses. At present, only a few groups of viruses are known to have homologs of DNA glycosylases, whose functions are also encoded in the host cells (Zharvov 2008; Dizdaroglu et al. 2017; Mechetin et al. 2020). For example, many herpesviruses are found to encode DNA glycosylases (UL2) which cut out uracil when it is included or formed in the phage DNA (Dogrammatzis et al. 2020). It was demonstrated that virus MHV68, mutant for UL2, had serious defects, appearing in the process of its replication and pathogenesis (Dong et al. 2018). Correspondingly, UL2 was suggested to be necessary to stabilize the genome of herpes viruses. It should be noted that Tevenvirinae bacteriophages, most of which seem to have DenV, are now united with herpes viruses in a single taxon, Heunggongvirae. At the same time, glycosylases of these viruses are different enough to be compared with each other. The main difference between them consists in their substrate specificity and the related functionality—in other words, in their biological importance for these viruses.
The main substrates for DenV T4 are pyrimidine dimers (Bernstein 1981). BER is not the only repair system known to deal with this type of photodamage in living organisms. Common in bacteria and eukaryotes are also NER (nucleotide excision repair) and the direct repair system (Friedberg et al. 2006; Chatterjee and Walker 2017). NER makes it possible to recognize and remove quite large damaged areas of DNA, including CPD-containing regions. The system of direct repair is based on the operation of DNA photolyases. Both systems are present in E. coli, the main host of Tevenvirinae bacteriophages. As found in the 1960s, the E. coli endonuclease UvrABC is not involved in the repair of the phage DNA: The experiments showed no difference in the UV sensitivity of phages cultivated in wild-type and UvrABC-deficient E. coli (Harm 1968). Moreover, it was demonstrated that most of the DNA damage caused by far UV light in phage T4, as well as other T-even bacteriophages, was repaired by photoreactivation (Harm 1968; Dulbecco 1950, 1952). At the same time, it was found that phage T4 was twice as resistant to UV radiation as phage T2 (Luria and Dulbecco 1949; Luria 1947). Presumably, this difference is related to DenV: It is absent in T2 (Bernstein and Wallace 1983). In this case, why does T4 need its own glycosylase specifically aimed at removing CPD? Evidently, the main function is additional protection of phage DNA from UV. UV is the strongest mutagenic factor of sunlight, which living organisms have had to cope with since the “dawn of time”—so they had to acquire effective methods of protecting their DNA. Moreover, cells often have multiple repair enzymes, the substrates of which are different photoproducts. Phage T4 also has several clusters of genes related to different repair systems, which can be implemented in the infected host cell (Bernstein and Wallace 1983; Miller et al. 2003a). For example, it carries genes of the UvsXWY system, a system of recombinational repair in which UvsX is a homolog of the E. coli protein RecA (Miller et al. 2003a, b). Proteins of the UvsXWY system are capable of repairing rather large DNA regions. UsvX was also demonstrated to remove different types of DNA abnormalities caused by various factors, including UV radiation (Bernstein and Wallace 1983). The comparison of the two repair systems showed that the DenV pathway mostly removed thymine dimers, whereas the UvsX pathway could deal with all kinds of lethal UV-induced DNA abnormalities (Meistrich 1972; Meistrich and Drake 1972). Another factor, which should be considered in the analysis of these two repair systems, is their role at different stages of the phage life cycle. DenV appears to work at the early stage: The enzyme was shown to remove up to 50% of all thymine dimers from phage DNA within the first 5 min of infection (Pawl et al 1976). It should also be noted that denV is located under the early promoter of the T4 genome (Friedberg and King 1971; Miller et al. 2003a). It seems one of the tasks of this repair system is to quickly restore damaged DNA before the start of replication. Put forward in some phylogenetic studies was also a theory that DenV might serve as a reserve for the NER system of the host cell, revitalizing defective repair systems in mutant host forms (Eisen and Hanawalt 1999). There are experimental data which support this assumption. It was shown, for example, that the infection of UvrABC-mutant E. coli strains with denV-containing T4 phages increased the survival rate of the bacterial cells after UV irradiation (Harm 1968). Furthermore, our earlier small-scale bioinformatic surveys showed that DenV and its homologs are quite common among living organisms of surface ocean waters (Karmanova and Zimin 2020). On the basis of those studies, we hypothesized that DenV might provide extra protection for the bacteria exposed to intense UV irradiation, maintaining vital functions of the bacterial cells during the phage infection.
Cv-pdg has a broader substrate specificity than DenV T4 and is able to interact with quite a large number of photoproducts, which makes this enzyme more versatile in repairing the UV-related damage. The researchers studying this glycosylase attributed its broad specificity to the supposition that CPD photoisomers were the most lethal threat for the virus, especially at the beginning of its replication (McCullough et al. 1998). Another study suggested that in the process of evolution, Cv-pdg underwent a strong selection, which particularly affected the active site of the enzyme (Jaruga et al. 2002). The main habitat of chlorella viruses is the surface waters of various water bodies, i.e., areas exposed to intense UV radiation. Thus, the broad substrate specificity of Cv-pdg was essential for chlorella viruses to survive.
How comes that two groups of viruses, which are quite distant from each other phylogenetically, have similar, in respect to their amino acid sequences, glycosylases? This is quite an interesting question, for which we did a little bioinformatics analysis, described in the next chapter.
Meta-analysis of viruses containing DenV homologs using bioinformatics tools
To look into this problem, we have performed a multifactorial analysis of literature data and genetic sequences from GenBank. The analysis of GenBank sequences was carried out using BLAST algorithms, the package GET_HOMOLOGUES and network analysis. Network analysis is an approach widely used in various fields of science, e.g., sociology or astronomy. In biology, it is applied, for example, to study gene expression or distribution of individual proteins on the tree of life. The objective of our study was to find an answer to the question posed above: Why DenV of phage T4 has homologous glycosylases found in the representatives of phylogenetically distant groups of viruses.
To see how widespread are homologs of DenV T4 glycosylase over viral taxa, we have reviewed literature data and analyzed all the viral sequences available in the NR databases using PSI-BLAST algorithms.
As a result, we have found 414 homologs, more than half of which (254) belongs to viruses of the phage T4 subfamily, Tevenvirinae (Table 1, Supplementary 1). It seems DenV is spread over most genera of this subfamily Tevenvirinae: Tequatrovirus, Dhakavirus, Gaprivervirus, Gelderlandvirus, Moonvirus, Mosigvirus, and Schizotequatrovirus. It should be noted that within this taxon, Schizotequatrovirus glycosylases are the most different from other homologs in terms of their amino acid sequence.
In addition, DenV homologs have been found in other Myoviridae taxa: Alcyoneusvirus, Asteriusvirus, Biquartavirus, Busanvirus, Eneladusvirus, Kafunavirus, Kanagawavirus, Marfavirus, Mimasvirus, Muldoonvirus, Tegunavirus, Shandongvirus, Tulanevirus, Winklervirus, and Vequintavirinae. A smaller number of homologs have also been revealed in the taxa Ackermanviridae, Demerecviridae, Podoviridae, and Siphoviridae. DenV homologs are also present in viruses of eukaryotic hosts: already discussed Cv-pdg in Chlorovirus and a homolog in Emiliania huxleyi virus 86, a virus of coccolithophores (Coccolithovirus). The latter consists of 128 amino acid residues; the authors who sequenced this protein suggested that this was a glycosylase of pyrimidine dimers, similar to DenV (Wilson et al. 2005). Some DenV homologs have also been found in unclassified Caudovirales viruses and in other, completely unclassified, viruses. Thus, DenV glycosylase seems to be widespread in the caudate bacteriophages Caudovirales—in particular, in Myoviridae and algal viruses (in the taxon Phycodnaviridae).
To compare DenV homologs with each other, we have built their phylogenetic tree, which is given in Fig. 1.
Phylogenetic tree of DenV homologs. The tree was constructed using the IQ-TREE software (Nguyen et al 2015; Minh et al 2013). The amino acid sequences were preliminarily aligned by MUSCLE algorithm using the MEGAX program (Kumar et al. 2018). The branches of DenV-containing viruses belonging to a common family (and, in case of Chlorovirus, to the family Phycodnaviridae) were collapsed. In some collapsed branches, a common downstream taxon was marked if it was common for all the DenV-containing viruses located in the branch. Ultrafast bootstrap support was used (Hoang et al. 2018). A full tree with non-collapsed branches is presented in Supplementary 2. A fragment of the amino acid sequence ROS1 by Arabidopsis thaliana (GenBank: AAP37178., region 884–1055, CDD: 419995), containing domain with bifunctional DNA glycosylase/lyase activity, was used to build the tree (Gong et al. 2002)
As one can see, the tree turned out to be quite heterogeneous. Three large clades can be distinguished. The first clade includes sequences of the majority of Myoviridae and unclassified phages of aeromonads and vibriobacteria. The second clade is formed by sequences of most of the Ackermanviridae taxa, vibriophages from the family Myoviridae, Roseobacter sp. phage of Siphoviridae, and some unclassified phages whose main habitat is water bodies. This clade also includes the DenV homolog of Emiliania huxleyi virus 86. The third clade is formed by sequences of Myoviridae phages, unclassified phages, and Cv-pdg of Chlorella viruses (Chlorovirus). Thus, each clade of the tree contains amino acid sequences from the Myoviridae family, with DenV of Emiliania huxleyi virus 86 occupying an intermediate position between the bacterial and algal domains.
To show relationships between the viruses that encode DenV homologs, a bipartite network was built (Fig. 2).
Bipartite network of viruses encoding DenV homologs. The bipartite network was built on the basis of a presence/absence matrix of homologous clusters in Tevenvirinae genomes obtained with GET_HOMOLOGUES (Contreras-Moreira and Vinuesa 2013). The network was visualized with Cytoscape v. 3.8.2., using the “Prefuse Force Directed Layout” algorithm (10,000 iterations) (Shannon et al. 2003). Nodes of partly or fully unclassified phages belonging to the same family, as well as nodes of viruses infecting eukaryotic hosts and belonging to the same genus, were collapsed
In the network, one can see two large and very distant nodes. The first is formed by genomes of Myoviridae viruses encoding DenV homologs. The second is formed by genomes of Chlorella sp. viruses (Chlorovirus) encoding DenV Cv-pdg homologs. The nodes are interconnected by the genome of another algovirus, Emiliania huxleyi virus 86, which belongs to the taxon of Coccolithovirus. The remaining viral genomes, encoding DenV homologs, are also grouped into nodes by their families; overall, they have more ties to Myoviridae genomes.
Both the genetic evolutionary network and phylogenetic tree indicate that, at some point, there was a horizontal transfer of DenV between the ancestral forms of Caudovirales and algaviruses. This supposition explains such a strange distribution of DenV over groups of viruses which are not strongly interconnected and whose hosts belong to different life domains. How the transfer happened is yet to be studied. One peculiar circumstance that should be noted, though, is that coccolithophore (the hosts of coccolithoviruses) can form around themselves a phycosphere, which includes many different types of bacteria, bacteriophages, and algal viruses. They all closely interact with each other, regulating each other’s vital activity (Nissimov et al. 2012; Kuhlisch et al. 2021). Perhaps, the transfer occurred in this unusual ecological niche.
On the basis of our multifactorial analysis, we also suppose that the biological importance of DenV for phages is related to the fact that, initially, this glycosylase was able to recognize and cleave CPD formed from non-canonical bases. A possible argument in favor of this hypothesis can be found in a recent theoretical paper about evolution of DNA repair systems. According to the paper, bifunctional glycosylases should have appeared very early; however, they were different from modern glycosylases. The authors also suggested that AP endonucleases and DNA glycosylases could have evolved in parallel with the emergence of DNA genomes, not only protecting them from damage, but also eliminating non-standard bases (including U, Hx, pseudouridine, N6-methyladenine, 5meC) (Prorok et al. 2021). Moreover, the BER pathway seems to have a functional advantage in this case, since photoreactivation of 5-HMC- and glucosylated 5-HMC-containing dimers is, as mentioned earlier, less efficient. This supposition is also supported by our data on the prevalence of DenV homologs among Tevenvirinae bacteriophages and other phage subfamilies that have non-canonical bases in their genomic DNA (Nikulin and Zimin 2021). Further studies are necessary, though, to either confirm or refute this hypothesis.
Conclusions
Thus, the main conclusion of this review is that the appearance of DenV, as a specific type of pyrimidine dimer glycosylase, is evolutionary linked to the emergence of non-canonical nitrogenous bases in DNA. Non-canonical bases, which are widespread in various groups of bacteriophages, are probably “remnants of the ancient experiments of nature” on the optimal composition of DNA, most resistant to certain environmental pressures. We suppose that the DenV-based repair pathway, which is aimed at the removal of abnormalities caused by shortwave sunlight, appeared and was later fixed in the process of evolution due to two factors: (1) high insolation during the times when the layer of Earth’s atmosphere was thin, and (2) a much wider spread of nucleotides of diverse chemical structure (which are currently called non-canonical). The DenV enzyme has been preserved—and now we can see its spread in the viruses that need to survive in high-insolation biotopes.
References
Bernstein C (1981) Deoxyribonucleic acid repair in bacteriophage. Microbiol Rev 45(1):72–98. https://doi.org/10.1128/mr.45.1.72-98.1981
Bernstein C, Bernstein H (2001) DNA repair in bacteriophage. In: Nickoloff JA, Hoekstra MF (eds) DNA damage and repair. Contemporary Cancer Research. Humana Press, Totowa. https://doi.org/10.1007/978-1-59259-095-7_1
Bernstein C, Wallace SS (1983) DNA repair. In: Mathews CK, Kutter E, Mosig G, Berget PB (eds) Bacteriophage T4. American Society for Microbiology, Washington, D.C., pp 138–151
Chatterjee N, Walker GC (2017) Mechanisms of DNA damage, repair, and mutagenesis. Environ Mol Mutagen 58(5):235–263. https://doi.org/10.1002/em.22087
Childs JD, Ellison MJ, Pilon R (1983) Formation of 5-hydroxymethylcytosine-containing pyrimidine dimers in UV-irradiated bacteriophage T4 DNA. Photochem Photobiol 37(5):513–519. https://doi.org/10.1111/j.1751-1097.1983.tb04510.x
Childs JD, Paterson MC, Smith BP, Gentner NE (1978) Evidence for a near UV-induced photoproduct of 5-hydroxymethylcytosine in bacteriophage T4 that can be recognized by endonuclease V. Mol Gen Genet MGG 167(1):105–112. https://doi.org/10.1007/BF00270326
Contreras-Moreira B, Vinuesa P (2013) GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol 79(24):7696–7701. https://doi.org/10.1128/AEM.02411-13
Dizdaroglu M, Zastawny TH, Carmical JR, Lloyd RS (1996) A novel DNA N-glycosylase activity of E. coli T4 endonuclease V that excises 4,6-diamino-5-formamidopyrimidine from DNA, a UV-radiation- and hydroxyl radical-induced product of adenine. Mutat Res 362(1):1–8. https://doi.org/10.1016/0921-8777(95)00025-9
Dizdaroglu M, Coskun E, Jaruga P (2017) Repair of oxidatively induced DNA damage by DNA glycosylases: mechanisms of action, substrate specificities and excision kinetics. Mutat Res Rev Mutat Res 771:99–127. https://doi.org/10.1016/j.mrrev.2017.02.001
Dodson ML, Schrock RD 3rd, Lloyd RS (1993) Evidence for an imino intermediate in the T4 endonuclease V reaction. Biochemistry 32(32):8284–8290. https://doi.org/10.1021/bi00083a032
Dogrammatzis C, Waisner H, Kalamvoki M (2020) “Non-essential” proteins of HSV-1 with essential roles in vivo: a comprehensive review. Viruses 13(1):17. https://doi.org/10.3390/v13010017
Doi T, Recktenwald A, Karaki Y, Kikuchi M, Morikawa K, Ikehara M, Inaoka T, Hori N, Ohtsuka E (1992) Role of the basic amino acid cluster and Glu-23 in pyrimidine dimer glycosylase activity of T4 endonuclease V. Proc Natl Acad Sci U S A 89(20):9420–9424. https://doi.org/10.1073/pnas.89.20.9420
Dong Q, Smith KR, Oldenburg DG, Shapiro M, Schutt WR, Malik L, Plummer JB, Mu Y, MacCarthy T, White DW, McBride KM, Krug LT (2018) Combinatorial loss of the enzymatic activities of viral uracil-DNA glycosylase and viral dUTPase impairs murine gammaherpesvirus pathogenesis and leads to increased recombination-based deletion in the viral genome. mBio 9(5):e01831-18. https://doi.org/10.1128/mBio.01831-18
Dowd DR, Lloyd RS (1990) Biological significance of facilitated diffusion in protein-DNA interactions Applications to T4 endonuclease V-initiated DNA repair. J Biol Chem 265(6):3424–3431
Dulbecco R (1950) Experiments on photoreactivation of bacteriophages inactivated with ultraviolet radiation. J Bacteriol 59(3):329–347. https://doi.org/10.1128/jb.59.3.329-347.1950
Dulbecco R (1952) Experiments on photoreactivation of inactive bacteriophages. J Cell Physiol Suppl 39(Suppl. 1):125–128. https://doi.org/10.1002/jcp.1030390413
Eisen JA, Hanawalt PC (1999) A phylogenomic study of DNA repair genes, proteins, and processes. Mutat Res 435(3):171–213. https://doi.org/10.1016/s0921-8777(99)00050-6
Fitzgerald LA, Graves MV, Li X, Hartigan J, Pfitzner AJ, Hoffart E, Van Etten JL (2007) Sequence and annotation of the 288-kb ATCV-1 virus that infects an endosymbiotic chlorella strain of the heliozoon Acanthocystis turfacea. Virology 362(2):350–361. https://doi.org/10.1016/j.virol.2006.12.028
Friedberg EC (1972) Studies on the substrate specificity of the T 4 excision repair endonuclease. Mutat Res 15(2):113–123. https://doi.org/10.1016/0027-5107(72)90024-3
Friedberg EC, King JJ (1971) Dark repair of ultraviolet-irradiated deoxyribonucleic acid by bacteriophage T4: purification and characterization of a dimer-specific phage-induced endonuclease. J Bacteriol 106(2):500–507. https://doi.org/10.1128/jb.106.2.500-507.1971
Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T (2006) DNA repair and mutagenesis. ASM Press, Washington DC. https://doi.org/10.1128/9781555816704
Ganesan A, Hanawalt P (2016) Photobiological Origins of the Field of Genomic Maintenance. Photochem Photobiol 92(1):52–60. https://doi.org/10.1111/php.12542
Garvish JF, Lloyd RS (1999) The catalytic mechanism of a pyrimidine dimer-specific glycosylase (pdg)/abasic lyase, Chlorella virus-pdg. J Biol Chem 274(14):9786–9794. https://doi.org/10.1074/jbc.274.14.9786
Golan G, Zharkov DO, Grollman AP, Dodson ML, McCullough AK, Lloyd RS, Shoham G (2006) Structure of T4 pyrimidine dimer glycosylase in a reduced imine covalent complex with abasic site-containing DNA. J Mol Biol 362(2):241–258. https://doi.org/10.1016/j.jmb.2006.06.059
Gong Z, Morales-Ruiz T, Ariza RR, Roldán-Arjona T, David L, Zhu JK (2002) ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111(6):803–814. https://doi.org/10.1016/s0092-8674(02)01133-9
Gruskin EA, Lloyd RS (1986) The DNA scanning mechanism of T4 endonuclease V. Effect of NaCl concentration on processive nicking activity. J Biol Chem 261(21):9607–9613
Gruskin EA, Lloyd RS (1988) Molecular analysis of plasmid DNA repair within ultraviolet-irradiated Escherichia coli. I. T4 endonuclease V-initiated excision repair. J Biol Chem 263(25):12728–12737
Harm W (1968) Recovery of UV-inactivated E. coli cells by the v-gene action of phage T4. Mutat Res 6(1):175–179. https://doi.org/10.1016/0027-5107(68)90115-2
Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS (2018) UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol 35:518–522. https://doi.org/10.1093/molbev/msx281
Hori N, Doi T, Karaki Y, Kikuchi M, Ikehara M, Ohtsuka E (1992) Participation of glutamic acid 23 of T4 endonuclease V in the beta-elimination reaction of an abasic site in a synthetic duplex DNA. Nucleic Acids Res 20(18):4761–4764. https://doi.org/10.1093/nar/20.18.4761
Jaruga P, Jabil R, McCullough AK, Rodriguez H, Dizdaroglu M, Lloyd RS (2002) Chlorella virus pyrimidine dimer glycosylase excises ultraviolet radiation- and hydroxyl radical-induced products 4,6-diamino-5-formamidopyrimidine and 2,6-diamino-4-hydroxy-5-formamidopyrimidine from DNA. Photochem Photobiol 75(2):85–91. https://doi.org/10.1562/0031-8655(2002)075%3c0085:cvpdge%3e2.0.co;2
Karmanova AN, Zimin AA (2020) Oceanic evolution of the enzyme repairing the UV-induced DNA lesions. J Phys Conf Ser 1701:012022. https://doi.org/10.1088/1742-6596/1701/1/012022
Kim J, Linn S (1988) The mechanisms of action of E. coli endonuclease III and T4 UV endonuclease (endonuclease V) at AP sites. Nucleic Acids Res 16(3):1135–1141. https://doi.org/10.1093/nar/16.3.1135
Kim SI, Jin SG, Pfeifer GP (2013) Formation of cyclobutane pyrimidine dimers at dipyrimidines containing 5-hydroxymethylcytosine. Photochem Photobiol Sci 12(8):1409–1415. https://doi.org/10.1039/c3pp50037c
Kuhlisch C, Schleyer G, Shahaf N, Vincent F, Schatz D, Vardi A (2021) Viral infection of algal blooms leaves a unique metabolic footprint on the dissolved organic matter in the ocean. Sci Adv 7(25):eabf4680. https://doi.org/10.1126/sciadv.abf4680
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549. https://doi.org/10.1093/molbev/msy096
Lloyd RS (2005) Investigations of pyrimidine dimer glycosylases–a paradigm for DNA base excision repair enzymology. Mutat Res 577(1–2):77–91. https://doi.org/10.1016/j.mrfmmm.2005.04.003
Lloyd RS, Hanawalt PC, Dodson ML (1980) Processive action of T4 endonuclease V on ultraviolet-irradiated DNA. Nucleic Acids Res 8(21):5113–5127. https://doi.org/10.1093/nar/8.21.5113
Luria SE (1947) Reactivation of irradiated bacteriophage by transfer of self-reproducing units. Proc Natl Acad Sci U S A 33(9):253–264. https://doi.org/10.1073/pnas.33.9.253
Luria SE, Dulbecco R (1949) Genetic recombinations leading to production of active bacteriophage from ultraviolet inactivated bacteriophage particles. Genetics 34(2):93–125. https://doi.org/10.1093/genetics/34.2.93
Marshall CJ, Santangelo TJ (2020) Archaeal DNA repair mechanisms. Biomolecules 10(11):1472. https://doi.org/10.3390/biom10111472
McCullough AK, Dodson ML, Schärer OD, Lloyd RS (1997) The role of base flipping in damage recognition and catalysis by T4 endonuclease V. J Biol Chem 272(43):27210–27217. https://doi.org/10.1074/jbc.272.43.27210
McCullough AK, Romberg MT, Nyaga S, Wei Y, Wood TG, Taylor JS, Van Etten JL, Dodson ML, Lloyd RS (1998) Characterization of a novel cis-syn and trans-syn-II pyrimidine dimer glycosylase/AP lyase from a eukaryotic algal virus, Paramecium bursaria chlorella virus-1. J Biol Chem 273(21):13136–13142. https://doi.org/10.1074/jbc.273.21.13136
McCullough AK, Sanchez A, Dodson ML, Marapaka P, Taylor JS, Lloyd RS (2001) The reaction mechanism of DNA glycosylase/AP lyases at abasic sites. Biochemistry 40(2):561–568. https://doi.org/10.1021/bi002404+
Meador MG, Rajagopalan L, Lloyd RS, Dodson ML (2004) Role of His-16 in turnover of T4 pyrimidine dimer glycosylase. J Biol Chem 279(5):3348–3353. https://doi.org/10.1074/jbc.M304714200
Mechetin GV, Endutkin AV, Diatlova EA, Zharkov DO (2020) Inhibitors of DNA glycosylases as prospective drugs. Int J Mol Sci 21(9):3118. https://doi.org/10.3390/ijms21093118
Meistrich ML (1972) Contribution of thymine dimers to the ultraviolet light inactivation of mutants of bacteriophage T4. J Mol Biol 66(1):97–106. https://doi.org/10.1016/s0022-2836(72)80008-1
Meistrich ML, Drake JW (1972) Mutagenic effects of thymine dimers in bacteriophage T4. J Mol Biol 66(1):107–114. https://doi.org/10.1016/s0022-2836(72)80009-3
Miller ES, Kutter E, Mosig G, Arisaka F, Kunisawa T, Rüger W (2003a) Bacteriophage T4 genome. Microbiol Mol Biol Rev 67(1):86–156. https://doi.org/10.1128/MMBR.67.1.86-156.2003
Miller ES, Heidelberg JF, Eisen JA, Nelson WC, Durkin AS, Ciecko A, Feldblyum TV, White O, Paulsen IT, Nierman WC, Lee J, Szczypinski B, Fraser CM (2003b) Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage. J Bacteriol 185(17):5220–5233. https://doi.org/10.1128/JB.185.17.5220-5233.2003
Minh BQ, Nguyen MA, von Haeseler A (2013) Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30(5):1188–1195. https://doi.org/10.1093/molbev/mst024
Morikawa K, Matsumoto O, Tsujimoto M, Katayanagi K, Ariyoshi M, Doi T, Ikehara M, Inaoka T, Ohtsuka E (1992) X-ray structure of T4 endonuclease V: an excision repair enzyme specific for a pyrimidine dimer. Science 256(5056):523–526. https://doi.org/10.1126/science.1575827
Morikawa K, Ariyoshi M, Vassylyev DG, Matsumoto O, Katayanagi K, Ohtsuka E (1995) Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants. J Mol Biol 249(2):360–375. https://doi.org/10.1006/jmbi.1995.0302
Manoharan M, Mazumder A, Ransom SC, Gerlt JA, Bolton PH (1988) Mechanism of UV endonuclease V cleavage of abasic sites in DNA determined by carbon-13 labeling. J Am Chem Soc 110(8):2690–2691. https://doi.org/10.1021/ja00216a074
Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32(1):268–274. https://doi.org/10.1093/molbev/msu300
Nikulin NA, Zimin AA (2021) Influence of non-canonical DNA bases on the genomic diversity of Tevenvirinae. Front Microbiol 12:632686. https://doi.org/10.3389/fmicb.2021.632686
Nissimov JI, Worthy CA, Rooks P, Napier JA, Kimmance SA, Henn MR, Ogata H, Allen MJ (2012) Draft genome sequence of the coccolithovirus Emiliania huxleyi virus 202. J Virol 86(4):2380–2381. https://doi.org/10.1128/JVI.06863-11
Pawl G, Taylor R, Minton K, Friedberg EC (1976) Enzymes involved in thymine dimer excision in bacteriophage T4-infected Escherichia coli. J Mol Biol 108(1):99–109. https://doi.org/10.1016/s0022-2836(76)80097-6
Prorok P, Grin IR, Matkarimov BT, Ishchenko AA, Laval J, Zharkov DO, Saparbaev M (2021) Evolutionary origins of DNA repair pathways: role of oxygen catastrophe in the emergence of DNA glycosylases. Cells 10(7):1591. https://doi.org/10.3390/cells10071591
Purmal AA, Rabow LE, Lampman GW, Cunningham RP, Kow YW (1996) A common mechanism of action for the N-glycosylase activity of DNA N-glycosylase/AP lyases from E. coli and T4. Mutat Res 364(3):193–207. https://doi.org/10.1016/s0921-8777(96)00032-8
Rastogi RP, Richa, Kumar A, Tyagi MB, Sinha RP (2010) Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair. J Nucl Acids 2010:592980. https://doi.org/10.4061/2010/592980
Sampath H, McCullough AK, Lloyd RS (2012) Regulation of DNA glycosylases and their role in limiting disease. Free Radic Res 46(4):460–478. https://doi.org/10.3109/10715762.2012.655730
Schrock RD 3rd, Lloyd RS (1991) Reductive methylation of the amino terminus of endonuclease V eradicates catalytic activities. Evidence for an essential role of the amino terminus in the chemical mechanisms of catalysis. J Biol Chem 266(26):17631–17639
Schrock RD 3rd, Lloyd RS (1993) Site-directed mutagenesis of the NH2 terminus of T4 endonuclease V The position of the alpha NH2 moiety affects catalytic activity. J Biol Chem 268(2):880–886
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504. https://doi.org/10.1101/gr.1239303
Vassylyev DG, Kashiwagi T, Mikami Y, Ariyoshi M, Iwai S, Ohtsuka E, Morikawa K (1995) Atomic model of a pyrimidine dimer excision repair enzyme complexed with a DNA substrate: structural basis for damaged DNA recognition. Cell 83(5):773–782. https://doi.org/10.1016/0092-8674(95)90190-6
Wilson WH, Schroeder DC, Allen MJ, Holden MT, Parkhill J, Barrell BG, Churcher C, Hamlin N, Mungall K, Norbertczak H, Quail MA, Price C, Rabbinowitsch E, Walker D, Craigon M, Roy D, Ghazal P (2005) Complete genome sequence and lytic phase transcription profile of a Coccolithovirus. Science 309(5737):1090–1092. https://doi.org/10.1126/science.1113109
Yokoyama H, Mizutani R (2014) Structural biology of DNA (6–4) photoproducts formed by ultraviolet radiation and interactions with their binding proteins. Int J Mol Sci 15(11):20321–20338. https://doi.org/10.3390/ijms151120321
Zharkov DO (2008) Base excision DNA repair. Cell Mol Life Sci 65(10):1544–1565. https://doi.org/10.1007/s00018-008-7543-2
Zhu H, Schein C, Braun W (1999) Homology modeling and molecular dynamics simulations of PBCV-1 glycosylase complexed with UV-damaged DNA. J Mol Model 5:302–316. https://doi.org/10.1007/s0089490050302
Funding
This work was supported by grant of the Russian Science Foundation №22–25-00669.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
About this article
Cite this article
Karmanova, A.N., Nikulin, N.A. & Zimin, A.A. Structural organization, evolution, and distribution of viral pyrimidine dimer-DNA glycosylases. Biophys Rev 14, 923–932 (2022). https://doi.org/10.1007/s12551-022-00972-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12551-022-00972-4