Abstract
Chronic myeloid leukemia (CML) is a disease of bone marrow stem cells caused by excessive growth and accumulation of granulocytes in the blood. Aberrant expression of the BCR-ABL proteins in bone marrow stem cells have found out in 95 % cases of CML. Tyrosine Kinase domains (SH2 and SH3) of BCR-ABL proteins are the potent targets to inhibit the process. Initially, imatinib is preferred as an efficient inhibitor to control functional activity of disease. Recently, it has been reported that the advanced stage of CML developed resistance against imatinib. In continuation, dasatinib is the first drug to combat against this disease by targeting multiple receptors and proven better as compared to imatinib. Here, an attempt has been made to identify similar analogs of dasatinib. Virtual screening was performed against various natural compound databases to get some potent natural compounds which are able to inhibit more than one receptor. Binding affinity of screened natural compounds was compared with some of the well-known inhibitors like imatinib, dasatinib, nilotinib etc., by analyzing their docking score and binding efficiency with the receptor. Stability of the best ligand–receptor complex was checked by performing 10 ns molecular dynamics simulation. ADMET properties of the obtained screened compounds were analyzed to check drug like property. Based on the aforementioned analysis, it has been suggested that these screened potent compounds are capable to inhibit multiple receptor proteins like ABL and SRC and consequently combat against the deadly disease CML.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Chronic myeloid leukemia (CML) is a stem cell disorder caused by abnormal growth of granulocytes in the bone marrow and overruns in to the blood for accumulations [1, 2]. Three phases of disease development process of CML are (1) transformation from initial to chronic (2) chronic to accelerated and (3) accelerated to blast stage or fatal stage [3]. Occurrence of CML cases are increasing day by day, indeed number of people suffering from CML becomes doubled since 2001 [4]. At the molecular level investigation, CML is caused due to translocation of the Abelson (ABL) gene on chromosome 9 to the Breakpoint Cluster Region (BCR) of chromosome 22 [5]. In the most of CML cases, constitutive proliferation of BCR-ABL oncoproteins is found which leads to ABL kinase as a putative drug target [6]. Initially, CML was treated by chemotherapy, bone marrow and stem cell transplantation which is having many fatal side effects [7]. Nowadays, it is being treated by targeted therapy with the help of BCR-ABL tyrosine kinase inhibitors which led to the enhancement of survival rate of the patients. BCR-ABL protein contains SH2 and SH3 domains, constitutively activates the ABL tyrosine kinase protein, which participates in signal transduction mechanism. It uses the ATP as co-factor to phosphorylate the substrates present in the signal transduction mechanism and its aberrant functioning leads to the cancer. The discovery of imatinib mesylate in late 1990s as an inhibitor of BCR-ABL protein has changed the entire treatment arena of CML [6, 8]. It specifically kills the cancerous cell growth and minimally affects the normal cells [9]. imatinib mesylate had proven as first generation and most effective drug against this disease especially in initial stages of CML. Later on in some cases, CML cells developed resistant to the imatinib and reoccurrence of disease has been seen in newly treated patients of advanced stages [10]. Common causes of resistance are probably due to point mutation of the kinase domain of BCR-ABL protein or less bioavailability of the drugs which consequently leads to the insensitivity toward drugs [10]. To overcome these lacunas, various second generation inhibitors like boustinib, tozaseritib, nilotinib, dasatinib, and bafatinib had been discovered for the effective inhibition of BCR-ABL protein. Some of them are in clinical trials or some of the drugs are available in market [11]. Due to limitations and side effects of the above said inhibitors, extensive study of target-based therapy for CML is needed [12]. Effective inhibition of oncogenic proteins, identification of the multi-target inhibitory ligands are required to circumvent the mutated CML cases [13]. The first multi-targeted-based inhibitor, dasatinib had been recently approved for CML and it acts as dual inhibitor [14]. In this work, in-silico analysis of the natural inhibitors for tyrosine kinase protein has been done by targeting two important non receptor tyrosine kinase proteins like SRC and ABL [15, 16]. They are structurally different in C-terminal region but homologous in N-terminal region where SH2 and SH3 domains are present [17]. The SH2 and SH3 domains of the ABL protein play a significant regulatory role to activate it, and SH3 domain of ABL is homologous to SRC protein’s SH3 domain [18]. SH2 domain of the SRC or ABL is interacting with the inhibitor and mediated the inhibitory reactions. Small molecules/Inhibitors act as a competitive inhibitor for the ATP and bind to the SH2 domain of ABL or SRC kinase protein and causes hindrance to the phosphorylation process [19]. At the same time SH3 domain plays structurally important role during inhibition of SH2. Identification of common natural compounds which are effective against both of the oncoproteins becomes important for the treatment of CML.
This work is based on five steps approaches to find out the common natural inhibitory molecules for ABL as well as SRC protein. (a) Selected the SH2 domain of ABL and SRC protein as a target because its structure is homologous to each other, and investigated their active site. (b) Dataset of natural ligands was generated by considering Lipinski rule of five from ZINC natural database and collected some of the already available inhibitors like imatinib, dasatinib, boustinib, nilotinib etc as reference or control molecules. (c) Virtual screening was performed for both the protein against generated inhibitors datasets [20, 21]. (d) Molecular dynamic simulation study of the best protein–ligand complex was performed for 10 ns. (e) ADMET prediction was performed for the best screened inhibitors which are common for both receptors.
2 Material and Methods
The entire experiments were performed on Centos 6.5 of Linux operating systems with 12 GB RAM, NVIDIA graphics and 1 TB Hard disk computer system. Hardware of the system is as follows: Intel(R) Core(TM) i7-3770 CPU@3.40 GHz processor.
2.1 Target Preparation and Active Site Prediction
Inhibitors targeting to the SH2 domain could inhibit the phosphorylation process of ABL and SRC and consequently induce the arrest of cell cycle and apoptosis. The targeted SH2 domain of ABL and SRC proteins were selected on the basis of their structural similarity [22]. 3D structure of ABL and SRC were downloaded from Protein Data Bank [PDB-ID 2ABL and 2H8H, respectively] (Figs. 1a, b) [23, 24]. The PDB structure of both proteins was prepared by Protein Preparation Wizard of Maestro [25]. SiteMap, version 3.0, was used for active site prediction of both proteins which uses OPLS_2005 force field to detect top ranked potential receptor binding cavity. After preparation of protein and identification of active site, receptor grid was generated for both proteins by Grid Generation panel of GLIDE module, version 6.2, Maestro. Grid dimension for ABL protein was as follows: Inner box: X = 10, Y = 10, Z = 12, Outer box: X = 30.0, Y = 30.0, Z = 32.0, Grid contour X = 2.0, Y = 8.0, Z = 40.0. Likewise grid dimension for SRC protein is as follows: Inner box X = 10, Y = 10, Z = 10, Outer box: X = 28.82, Y = 28.82, Z = 28.82, Grid contour X = 21.106, Y = 20.08, Z = 58.56.
2.2 Dataset Generation and Virtual Screening
A dataset of natural compounds was generated by collecting compounds from ZINC natural product database. ZINC natural product database is a collection of 12 different natural product dataset which contains 142131 unique molecules and their individual distributions are described in Table 1. Datasets were converted into PHASE database format (.phdb) of the Schrödinger software. Virtual screening is a technique by which small molecules are identified from the large dataset which are most likely to bind the targeted protein [26]. Generated dataset was screened against the active site of SH2 domains of both proteins. On the basis of binding affinity of the interacting molecule and hydrogen bond formed between them, GLIDE assigns the glide scores to the each interacting complex. High scoring compounds are considered to be the highly interacting compound and placed into top position as compared to the other screened molecules. Screening process of GLIDE includes three steps like High Throughput Virtual Screening (HTVS), Standard Precision (SP) and Extra Precision (XP). Through this rigorous docking-based in-silico filtering, we obtained some compounds with significant glide and docking score [27]
2.3 Docking Studies
Some of the well-known inhibitors like imatinib, dasatinib, nilotinib, befatinib etc, were used as reference molecules. Molecular ligand docking was performed for ABL and SRC protein individually with these molecules for comparative analysis by using GLIDE module of Maestro.
2.4 Molecular Dynamics (MD) Simulation
Molecular Dynamics Simulation is a computer-based simulation of the atomic/molecular movements in the systems for a particular time. The best conformer of ABL and SRC proteins with ligands was subjected for Molecular Dynamics Simulations. Entire MD Simulation experiments for both the complex (ABL and SRC) were performed with the help of Desmond software [28, 29]. Neutral territory method was used to effectively explore the parallel computing in simulation process [30]. The OPLS 2005 force field was used to define the interactions of amino acids of system. Simple Point Charge (SPC) water model was embedded [31], with dimension 10 \(\times\) 10 \(\times\) 10 Å to cover the protein–ligand complex. System neutralization was done by adding the Cl\(^{-}\) ion to maintain the pH of the systems. Default steps and protocols of the Desmond were selected to optimize the systems equilibrium, which allows them for slow relaxations and without deviating from the initial co-ordinates. Finally, production run was carried out for 10 ns by maintaining the normal temperature and pressure. The structural and dynamic behavior of both the complexes was analyzed by calculating the root-mean-square deviations (RMSD) and potential energy [32].
2.5 Calculation of ADME and Toxicity
ADME properties like Molecular Weight, hydrophilicity, hydrophobicity, solvent accessible surface area, number of rotable bonds, donor-hydrogen bonds, acceptor-hydrogen bonds etc. of common four natural inhibitors were calculated by QuikProp v3.9, module of Maestro.
OSIRIS property explorer was used for toxicity prediction of common four inhibitors of ABL and SRC protein (http://www.organic-chemistry.org/prog/peo/). It is an online toxicity prediction tool for the analysis of drug relevant property like Mutagenic, Tumorigenic, Irritant, Reproductive effective, cLogP, Solubility, Molecular Weight etc.
3 Results and Discussion
3.1 Protein Preparation and Active Site Analysis
Preparation of ABL and SRC proteins included bond order refinement and their minimization. The cavity was selected and prepared by adding H-bonds, disulfide bonds and water molecules beyond 5 Å were removed. Finally, both the proteins were reviewed and modified by checking the metal binding states, chains balance and pH maintenance of the system. SiteMap has detected the largest volume of 527.8 Å3 as an active site of ABL protein. Likewise active site of SRC was also detected with volume 1434.4 Å3. Active site of the ABL protein was validated by analyzing the Hyun-Joo Nam et al. work which showed the presence of Tyr89, Trp118, Trp128, Asn133 and Tyr134 in the active site. Validation of the active site of SRC protein was done by analyzing the Laurent et al. work which showed donor H-bond with Tyr340 and acceptor H-bond with Met341.
3.2 Analysis of Virtual Screening and Molecular Docking
Screening used to perform sequential three layers of molecular docking during final selection of ligands namely HTVS, SP and XP. The LigPrep (module for preparation of ligands) treated compound libraries of 142131 compounds were flexibly docked into the active site of both the proteins using GLIDE and their screening at all the three stages have been summarized in Table 1. In first step the ligands were rapidly screened by HTVS (High Throughput Virtual Screening); the ligands passed out from HTVS were subsequently analyzed in Glide SP (Standard Preparation) in second step and Glide XP (Extra Precision) in the third step [33]. The scoring of the ligands were based on their g-score (Glide score) and Glide e-model score.
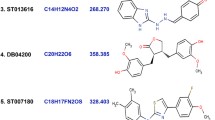
Detailed investigation of docking scores and binding affinity of natural screened inhibitors with both proteins was done. Out of which, four natural compounds have shown common inhibitory activity against both the receptors and these compounds had also good binding affinities with both receptors on the basis of their docking score and glide e-model scores (Table 2). 2D structures of screened molecules are shown in Fig. 2.
2D structure of screened natural inhibitors a ZINC14437962 = (2S,3S)-3,5,7-trihydroxy-6-methyl-2-(3,4,5-trihydroxyphenyl)chroman-4-one, b ZINC14645645 = (2S,3S)-2-(3,4-dihydroxyphenyl)-3,5,7-trihydroxy-8-[(4-hydroxyphenyl)methyl]chroman-4-one, c ZINC31169866 = (6R)-1,7-bis(3,4-dihydroxyphenyl)-6-hydroxy-heptan-3-one, d ZINC36470466 = 2-(3-hydroxybenzyl)-3-(3,4-dihydroxybenzyl) butane-1,4-diol
In Table 2, six reference molecules which are already known for their inhibitory effect against both these receptors were also listed and their scores were analyzed and compared with the screened inhibitors. Docking scores of ABL receptor with befatinib, boustinib, dasatinib, imatinib, nilotinib, and tozaseritib are \(-4.678\), \(-4.686\), \(-5.126\), \(-5.746\), \(-5.590\), and \(-5.673\) respectively. Whereas docking scores of the selected top scoring molecules C1, C2, C3, and C4 with ABL are \(-9.493\), \(-8.905\), \(-9.109\), and \(-7.475\), respectively. Docking scores of SRC receptor with befatinib, boustinib, dasatinib, imatinib, nilotinib, and tozaseritib are \(-7.085\), \(-5.719\), \(-8.794\), \(-7.831\), \(-8.129\), and \(-7.957\) respectively. Whereas docking scores of the selected top scoring molecules C1, C2, C3, and C4 with SRC are \(-9.513\), \(-11.304\), \(-10.598\), and \(-10.771\) respectively. Docking scores of all selected ligand are found better as compared to the reference molecules which indicate that these compounds can have more potent inhibitory action against both ABL and SRC receptors.
Various energies like solvation energy, van der Wall energy, and Electrostatic energy of the ABL and SRC with these screened four common ligands were also calculated by using MacroModel, version 10.3, Schrödinger, LLC, New York, NY, 2014 as depicted in Table 3.
The docking studies indicated that the screened top ranked compounds showed strong hydrogen bonding interactions with both the receptors. The docking score of these screened four compounds ranges from \(-7.457\) to \(-9.493\) for ABL protein and \(-9.513\) to \(-11.304\) for SRC receptor. The key interacting residues Gly178, Asp 83, Pro 83, 177, Glu 172, Leu 110, 141, P177, Ser 132 etc. were conserved within active site of the receptor and contributed to the strong binding affinity of these four top ranked compounds for ABL protein. Whereas in SRC receptor, key interacting residues Met 341, Asp 348, 391, Leu 273, Lys295, Iso 336, and Phe 405 were conserved within its active site which further contributed to the efficient and more potent binding affinity of these four top ranked compounds.
By analyzing docking complexes of ABL with natural compounds C1, C2, C3, and C4 number of hydrogen bond formed are 6, 5, 4, and 5, respectively, in its hinge region. Most common residues present in hydrogen bond interactions were Gly 178, Asp 83, Pro 82-83, Glu172, Leu 110, 141 Ser132, 140, Ile 135 and Thr 197. The binding modes of these four screened compounds were shown in Fig. 3a–d, respectively, and various strong interactions such as H-bond, Hydrophobic, Polar etc formed by these compounds with ABL receptor have been depicted in Table 4.
Likewise, by analyzing docking complexes of SRC with natural compounds C1, C2, C3, and C4 number of hydrogen bond formed are 4, 3, 4, and 5 respectively. Most common residues present in the hydrogen bond interactions were Met 341(2), Asp 348(2), 391, Leu 273, Lys295, Iso 336, and Phe 405 (2). The binding modes of these four screened compounds were shown in Fig. 4a–d, respectively, and various strong interactions such as H-bond, Hydrophobic, Polar etc formed by these compounds with SRC receptor have been shown in Table 4.
3.3 MD Simulation analysis
Molecular dynamics simulation was used to explore dynamic perturbations in the conformation of complex structure for both receptor with four compounds. Here, we have shown the detail analysis of MD simulation results of only one compound (C3) with receptor ABL and SRC (docking score = \(-9.109\) for ABL and docking score = \(-10.598\) for SRC), because C3 is showing better binding affinity for both the receptors. The Simulation process was run for 10 ns in order to check the stability of the protein-ligand complex. During the initial stage, there were more fluctuations in the potential energy and RMSD of the system. But after the equilibration phase till 5-6 ns, the system started showing consistent behavior during the production phase of the simulation process. The initial energy of the complex (ABL with compound 3) was \(-41,390\) kcal/mol. During the course of 10 ns simulation, the potential energy shows a decrease and after 7 ns, it was found that the graph shows not much significant differences which is indicative of stabilization of the system as shown in Fig. 5a.
Similarly for the complex (SRC with compound 3), the initial energy was found to be \(-17,8640\) kcal/mol and the potential energy shows a decrease during the course of 10 ns simulation (Fig. 5b). But here also the graph shows not much significant differences after 7 ns which is indicative of stabilization of the system. The trajectory files had been combined using standard commands/protocol of Desmond and were analyzed for flexibility of the backbone atoms of the docked protein complex. The RMSD graph plotted in Fig. 5c, d indicates the result of dynamic behavior of both ABL and SRC from the initial position to final position throughout the simulation time. First, we aligned all the frames and then the root-mean-square deviation (RMSD) values of the both docked protein complex’s backbone atoms were plotted against frames obtained during simulation of 10 ns using Visual Molecular Dynamics (VMD) software. Initially there were larger fluctuations (max = 3.484 Å) in the C\(\alpha\) atoms of the docked ABL protein backbone RMSD which in the production phase get reduced and was an average of 1.724 Å as depicted in Fig. 5c. Here, the overall standard deviation was 0.458 Å during 10 ns simulation time. In the same manner, large fluctuations (max = 2.820 Å) in the C\(\alpha\) atoms of the docked SRC protein backbone RMSD were also observed which in the production phase get reduced and was an average of 1.129 Å, and there also occurs a standard deviation of 0.319 Å during the course of 10 ns simulation as shown in Fig. 5d. Rest three natural compounds were also shown stability in course of 10 ns simulation run. Simulations were stopped after 10 ns because after 5-7 ns all graphs have attended the stability.
3.4 ADME Descriptors Analysis
Fifteen principal descriptors as shown in Table 5 [34] and fourteen predicted ADME properties as shown in Table 6 were calculated for selected natural inhibitors by QikProp module of the Schrödinger software [35, 36].
ADMET properties are significant measures for a molecule which can serve as a drug [37] as shown in Table 6. ADMET properties like percentage of human oral absorption of good drug should be more than 25 %; however, ligand C4 showed highest percentage as compared to others, i.e., 67.9 %. For aqueous solubility (logS) ligand C2 showed highest among all i.e \(-2.488\), apparent Caco-2 permeability in nm/s (PCaco) of ligand C4 showed highest among all i.e. 66.078, logHERG indicates the HERG K\(^{+}\) channel blockage: log IC50 which is higher in ligand C1, i.e., \(-4.678\). PMDCK descriptor is used for prediction of non-active transport which is highest in case of C4, i.e., 26.24. For polarizability (Polrz in angstrom) ligand C4 showed highest among all, i.e., 38.719, logP for hexadecane/gas (logPC16) is highest in ligand C3, i.e., 14.631, logP for octanol/gas (logPoct) is highest in ligand C3, i.e., 24.987, logP for octanol/water (logPo/w) is highest in C4, i.e., 1.443, Lipinski rule of 5 violations (rule of 5) is allowed till 4, whereas ligand C2 and C4 showed zero violation and ligand C1 and C3 showed single violation. Less violation of rule 5 indicates the higher drug likeness of the compound [38]. Jorgensen rule of 3 violations (rule of 3) is allowed maximum till 3, whereas ligand C2 and C4 showed 1 violation and ligand C1 and C3 showed 2 violations. Likewise less violations of the rule three shows higher oral bioavailability of the ligand [39].
3.5 Toxicity Analysis
Toxicity Risk Assessment panel of the Osiris Property Explorer (OPE) was used to predict toxic property of selected natural inhibitors as summarized in Table 7. OPE use to predict toxic property by analyzing available molecular toxic property in the Registry of Toxic Effects of Chemical Substances (RTECS) database. RTECS, a Accelrys database which is a repository of chemical structures and their toxic property. These properties are mutagenic, tumorigenic, irritating effect, reproductive effect etc. Some of other descriptors were also predicted like cLogP, Solubility, Drug likeness, Drug Score, Topological Polar Surface Area (TPSA) etc. Predicted results are color coded like red which shows high risk, whereas green confirms the drug likeness.
By analyzing the results of toxicity predictor, all four compounds were non mutagenic, non-tumorigenic, nonirritant, non-reproductive, etc. Other descriptor values of all drugs were also in acceptable range. cLogP is used to measure the hydrophilicity of the drugs which ensures the drugs absorbance probability and it should not be greater than 5.0. LogS is used to measure the solubility of drugs which affects the absorption and distribution property of the drugs and it should be greater than \(-4\). Drug likeness property of the drugs is calculated on the basis of numerous properties, and it should be in range of \(-4\) to +4. Drug score is calculated on the basis of cLogP, LogS, drug likeness values and predicts the overall potential of the compound to be a drug. Acceptable ranges are 0.60 –1.0, \(<\)0.60 indicates high risk, whereas toward 1 indicated no risk. The TPSA is associated with blood–brain barrier and oral bioavailability of the drugs and its acceptable range is about 140 Å.
4 Conclusion
To the best of our knowledge, none of the reported work explored the in-silico identification of natural inhibitors for Chronic myeloid leukemia. In this work, we have identified four multi-targeted natural compounds by virtual screening, molecular docking, and MD simulation studies. Bioavailability, drug likeness, and toxicity of the selected compounds were also calculated by ADMET analysis. The ADMET score of these screened ligands suggest an overall favorable pharmacokinetics to be accepted as lead molecule. Molecular docking studies showed that these four screened natural compounds are expected to bind the inactive form of ABL as imatinib and dasatinib do, although requiring a lower conformational stringency, with the ability of binding more intermediate conformations than known inhibitors. The improved binding affinity score and number of hydrogen bonds suggest better interactions in active site of candidate receptors. Taken together, these observations raised the possibility that the small-molecule inhibitors with dual activity could act against ABL and SRC, proving its effectiveness in CML patients resistant to known drugs. Thus, we can safely conclude that these four natural compounds individually can be used as potential lead to synthesize new drug molecules for multiple targets involved in chronic myeloid leukemia.
References
Besa EC, Buehler B, Markman M, Sacher RA, Krishnan K (2013) Chronic myelogenous leukemia. Medscape reference. WebMD, New York
Faderl S, Talpaz M, Estrov Z, Kantarjian HM (1999) Chronic myelogenous leukemia: biology and therapy. Ann Intern Med 131(3):207–219
Kantarjian HM, Talpaz M, Giles F, O’Brien S, Cortes J (2006) New insights into the pathophysiology of chronic myeloid leukemia and imatinib resistance. Ann Intern Med 145(12):913–923
An X, Tiwari A, Sun Y, Ding P, Ashby C Jr, Chen Z (2010) BCR-ABL tyrosine kinase inhibitors in the treatment of Philadelphia chromosome positive chronic myeloid leukemia: a review. Leuk Res 34(10):1255–1268
Rowley JD (1973) A new consistent chromosomal abnormality in chronic myelogenous leukemia identified by quinacrine fluorescence and Giemsa staining. Nature 243(5405):290–293
Pray L (2008) Gleevec: the breakthrough in cancer treatment. Nat Educ 1(1):37
Baccarani M, Saglio G, Goldman J (2006) Evolving concepts in the management of chronic myeloid leukemia: recommendations from an expert panel on behalf of the European LeukemiaNet. Blood 108(6):1809–1820
Savage DG, Antman KH (2002) Imatinib mesylate—a new oral targeted therapy. N Engl J Med 346:683–693
Druker BJ (2004) Imatinib as a paradigm of targeted therapies. Adv Cancer Res 91:1–30
Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, Sawyers CL (2001) Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science 293:876–880
Heinrich MC, Griffith DJ, Druker BJ, Wait CL, Ott KA, Zigler AJ (2000) Inhibition of c-kit receptor tyrosine kinase activity by STI 571, a selective tyrosine kinase inhibitor. Blood 96:925–932
Demetri GD, Van OT, Blackstein M, Garrett C, Shah M, Heinrich M, McArthur G, Judson I, Baum CM, Casali PG (2005). Phase 3, multicenter, randomized, double-blind, placebo-controlled trial of SU11248 in patients (pts) following failure of imatinib for metastatic GIST. American society of clinical oncology 41st annual meeting proceedings (Post-Meeting Edition). Orlando FL. J Clin Oncol 23:4000, pp 13–17
Hill Zachary B, Perera B Gayani K, Maly J (2011) Bivalent inhibitors of the tyrosine kinases ABL and SRC: determinants of potency and selectivity. Mol Biosyst 7(2):447–456
Tokarski JS, Newitt JA, Chang CYJ, Cheng JD, Susan MW, Kiefer E, Kiefer E, Francis Kevin K, Robert YFL, Louis B, Dianlin JL, Yaqun X, Herbert EKZ (2006) The structure of Dasatinib (BMS-354825) bound to activated ABL kinase domain Elucidates its inhibitory activity against imatinib-resistant ABL mutants. Cancer Res 66:5790–5797
Martinelli G, Soverini S, Rosti G, Baccarani M (2005) Dual tyrosine kinase inhibitors in chronic myeloid leukemia. Leukemia 19(11):1872–1879
Utkarsh R, Pritish KV (2015) Flavonoids as multi-target inhibitors for proteins associated with Ebola virus: in-silico discovery using virtual screening and molecular docking studies. Interdiscip Sci Comput Life Sci. doi:10.1007/s12539-015-0109-8
Sibo F, Chiyoshi K, Richard J Ricklest, Schreiber S (1995) Specific interactions outside the proline-rich core of two classes of SRC homology 3 ligands. Proc Natl Acad Sci USA 92:12408–12415
Zellefrow CD, Griffiths JS, Saha S, Hodges AM, Goodman JL, Paulk J, Kritzer JA, Schepartz A (2006) Encodable activators of SRC family kinases. J Am Chem Soc 128(51):16506–16507
Shao S, Yu R, Yu Y, Li Y (2014) Dual-inhibitors of STAT5 and STAT3: studies from molecular docking and molecular dynamics simulations. J Mol Model 20(8):2399
Kumar A, Rajendran V, Sethumadhavan RP (2013) Evidence of colorectal cancer-associated mutation in MCAK: a computational report. Cell Biochem Biophys 67(3):837–851
Yadav PK, Singh G, Gautam B, Singh S, Yadav M, Srivastava U, Singh B (2013) Molecular modeling, dynamics studies and virtual screening of fructose 1, 6 biphosphate aldolase-II in community acquired- methicillin resistant \(Staphylococcus aureus\) (CA-MRSA). Bioinformation 9(3):158–164
Hyun-Joo N, Wayne GH, Thomas MR, Christin AF (1996) Intramolecular interactions of the regulatory domains of the Bcr–Abl kinase reveal a novel control mechanism. Curr Biol Ltd 4:1105–1114
Hennequin LF, Allen J, Breed J, Curwen J, Fennell M, Green TP, Lambert-van der BC, Morgentin R, Norman RA, Olivier A, Otterbein L, Plé PA, Warin N, Costello G (2006) N-(5-chloro-1, 3-benzodioxol-4-yl)-7-[2-(4-methylpiperazin-1-yl) ethoxy]-5- (tetrahydro-2H-pyran-4-yloxy) quinazolin-4-amine, a novel, highly selective, orally available, dual-specific c-Src/Abl kinase inhibitor. J Med Chem 49(22):6465–6488
Nam HJ, Haser WG, Roberts TM, Frederick CA (1996) Intramolecular interactions of the regulatory domains of the Bcr–Abl kinase reveal a novel control mechanism. Structure 4:1105–1114
Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W (2013) Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des 27(3):221–234
Rajamani R, Good AC (2007) Ranking poses in structure-based lead discovery and optimization: current trends in scoring function development. Curr Opin Drug Discov Dev 10:308–315
Kamaraj B, Purohit R (2014) Computational screening of disease-associated mutations in OCA2 gene. Cell Biochem Biophys 68(1):97–109
Kevin JB, Edmond C, Huafeng X, Ron OD, Michael PE, Brent AG, John LK, Istvan K, Mark AM, Federico DS, John KS, Yibing S, David ES (2006) Scalable algorithms for molecular dynamics simulations on commodity clusters. In: Proceedings of the 2006 ACM/IEEE conference on supercomputing. ACM, Tampa
Purohit R (2014) Role of ELA region in auto-activation of mutant KIT receptor: a molecular dynamics simulation insight. J Biomol Struct Dyn 32(7):1033–1046
Bowers KJ, Dror RO, Shaw DE (2006) The midpoint method for parallelization of particle simulations. J Chem Phys 124:184109–184111
Jorgensen WL, Maxwell DS, Tirado-Rives J (1996) Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc 118:11225–11236
Kumar A, Purohit R (2014) Use of long term molecular dynamics simulation in predicting cancer associated SNPs. PLoS Comput Biol 10(4):e1003318
Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT (2006) Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem 49(21):6177–6196
Singh DB, Gupta MK, Kesharwani RK, Misra K (2013) Comparative docking and ADMET study of some curcumin derivatives and herbal congeners targeting \(\beta\)-amyloid. Netw Model Anal Health Inform Bioinform 2:13–27
Yadav DK, Meena A, Srivastava A, Chanda D, Khan F, Chattopadhyay SK (2010) Development of QSAR model for immunomodulatory activity of natural coumarino lignoids. Drug Des Dev Ther 4:173–186
Yadav DK, Khan F, Negi AS (2011) Pharmacophore modeling, molecular docking, QSAR, and in silico ADMET studies of gallic acid derivatives for immune modulatory activity. J Mol Model 18:2513–2525
Wang J (2009) Comprehensive assessment of ADMET risks in drug discovery. Curr Pharm 15(19):2195–2219
Lipinski CA, Lombardo F, Dominy BW, Fenney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46:3–26
Jorgensen WL (1998) BOSS—biochemical and organic simulation system. In: Schleyer PVR (ed) The encyclopedia of computational Chemistry. 5, pp 3281–3285
Acknowledgments
Authors would like to thank Department of Bioinformatics, Indian Institute of Information Technology Allahabad, for providing facilities required for this work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
Authors declare that there is no conflict of interest.
Rights and permissions
About this article
Cite this article
Kumar, H., Raj, U., Srivastava, S. et al. Identification of Dual Natural Inhibitors for Chronic Myeloid Leukemia by Virtual Screening, Molecular Dynamics Simulation and ADMET Analysis. Interdiscip Sci Comput Life Sci 8, 241–252 (2016). https://doi.org/10.1007/s12539-015-0118-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-015-0118-7