Abstract
Environmental DNA (eDNA) metabarcoding is revolutionizing biodiversity monitoring, but comparisons against traditional data rarely include long-term historical inventories. We targeted eukaryotes by amplifying a fragment of the 18S gene from eDNA isolated from seawater samples at 20 sites in the Gulf of California (GC) and contrasted regional taxonomic diversity against 316 simultaneous visual surveys and a historical database with over 5k species. From 61k Amplified Sequence Variants, we identified 850 eukaryotic families, of which half represent new compiled records, including 174 families of planktonic, benthic, and parasitic invertebrates. The 18S eDNA metabarcoding analysis revealed many overseen taxa, highlighting higher taxonomic ranks within micro-invertebrates, microscopic fungi, and other micro-eukaryotes from the supergroups Stramenopiles, Alveolata, and Rhizaria. The database combining all methods has doubled the number of distinct phyla, classes, and orders compared to the historical baseline, indicating biodiversity levels in the GC are much higher than previously assumed. The estimated proportion of historical taxa included in public reference databases was only 18% for species, partially explaining the small portion of 18S eDNA reads that were taxonomically assigned to species level (13%). Each method showed different taxonomic biases, with 18S eDNA metabarcoding detecting few vertebrates, visual surveys targeting only seven metazoan phyla, and the historical records focusing on macroinvertebrates, fish, and algae. Although all methods recovered the main known biogeographic regionalization, the 18S eDNA metabarcoding data did not support the historical pattern of higher diversity in the Central than Northern GC. While combining methods provides a novel view of biodiversity that is much more comprehensive than any individual approach, our study highlights many challenges in synthesizing biodiversity data from traditional and novel sources.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
All levels of marine biodiversity play key roles in supporting human life, including food provision, nutrient cycling and climate regulation, among others (Blasiak et al. 2020). Biodiversity can be described within three main nested components, including genetic (within species), species, and ecosystems (Convention on Biological Diversity 1992). Recent evidence shows species and habitats are shifting in response to ocean warming and other biogeochemical changes in the environment driven by anthropogenic activities (Lenoir et al. 2020; Pinsky et al. 2020). Changes include geographic range extensions and the development of new ecological interactions that could cascade into altered ecosystem structure and function (Pörtner et al. 2019). Thus, there is a growing need to improve the estimations and monitoring of biodiversity to track the current rate of environmental change (Hoban et al. 2020; Pollock et al. 2020). A key step in understanding and predicting tipping points in marine ecosystem health is a detailed and comprehensive description of the taxonomic diversity present across the tree of life, as biodiversity could be linked to ecological interactions related to nutrients, biomass, and energy fluxes (D’Alelio et al. 2016; Pawlowski et al. 2018; Seymour et al. 2020).
To address current needs, environmental DNA (eDNA) metabarcoding has emerged as an innovative technique that aims to discriminate multiple species simultaneously by capture, amplification, and high-throughput sequencing of conserved genomic regions (i.e., metabarcodes) from traces of eDNA occurring in water, air, and sediment samples (Taberlet et al. 2012). The resultant DNA variants are compared against a genetic reference database (e.g., BOLD, maintained by the International Barcode of Life) (Ratnasingham and Hebert 2007) to match them to a species, or lowest common ancestor known (Jo et al. 2021). Public sequence repositories like GenBank (Pruitt et al. 2005) also provide a diverse array of DNA sequences from which metabarcodes associated with described species could be extracted and used as references (Deiner et al. 2017a; Rodríguez-Ezpeleta et al. 2021; Jo et al. 2021). The method is considered a cost-effective and replicable tool for complementing biodiversity assessments of particular taxa (Fediajevaite et al. 2021) and across the tree of life (Keck et al. 2022).
Studies conducting eDNA metabarcoding in aquatic environments have made comparisons against traditional approaches, yet many have focused on freshwater systems, with fewer marine studies, frequently focused on fish (Fediajevaite et al. 2021; Takahashi et al. 2023), revealing higher diversity than previously assumed and considerable complementarity (Yamamoto et al. 2017; Gold et al. 2021; Valdivia-Carrillo et al. 2021). Other ground-truthing studies of marine eDNA metabarcoding have compared local taxonomic inventories resulting from simultaneous visual surveys or species collection methods targeting specific taxonomic groups, including stony corals (Nichols and Marko 2019; Dugal et al. 2022; Nishitsuji et al. 2023), sessile benthic fauna (West et al. 2022), or multiple invertebrate phyla (Leduc et al. 2019; Robinson et al. 2023). Compared to traditional methods, DNA and eDNA metabarcoding of benthic and planktonic marine communities typically uncovers higher local but similar regional diversity, and different sets of taxa, particularly for macroinvertebrates, plankton, and microphytobenthos (Keck et al. 2022). Traditional marine biodiversity monitoring in coastal oceanic areas has focused on the diversity and abundance of a limited set of conspicuous benthic macroinvertebrates, limiting our ability to describe ecosystem-level structure and function (Mora et al. 2011; Baird and Hajibabaei 2012). Field validation of species presence in the oceans is logistically challenging and time-consuming, and very few marine studies have compared eDNA metabarcoding data across the tree of life against regional long-term observational data (Deiner et al. 2017a; Fediajevaite et al. 2021).
Despite its perceived potential, eDNA metabarcoding monitoring is still developing worldwide, and its performance and effectiveness considering traditional surveys needs to be systematically assessed (Calderón-Sanou et al. 2019; Fediajevaite et al. 2021; Jo et al. 2021). In contrast to time-specific local surveys, historical biodiversity databases can represent a more comprehensive description of local and regional taxa, often built over a longer period and accumulating distinct monitoring campaigns, including multiple techniques by numerous people. Although marine biodiversity databases are available for some hotspots worldwide (e.g., OBIS; https://obis.org), taxonomic representation varies by region and lineage (Mugnai et al. 2021). Curated databases of regional marine biodiversity across the tree of life are scarce or non-existent in most cases (Appeltans et al. 2012). Thus, regional eDNA metabarcoding studies monitoring entire marine ecosystem biodiversity (by targeting all animal and plant phyla) usually cannot put their contributions in perspective using long-term baselines (Stat et al. 2017; Bakker et al. 2019).
Some important challenges in the application of eDNA metabarcoding include the taxonomic resolution of the selected metabarcode and the incompleteness of public reference databases that generally have strong taxonomic and geographical biases (Ruppert et al. 2019; Cordier et al. 2021; Mugnai et al. 2021). The fast-evolving mitochondrial COI gene, which is commonly used as a barcode for metazoans, shows high variability and excellent resolution at the species level, but poses challenges for designing conserved primers that usually have multiple degenerate bases and varying amplification efficiencies across broad taxonomic groups (Geller et al. 2013; Collins et al. 2019). The slower-evolving nuclear 18S gene shows conserved sections that allows anchoring primers for a broader array of taxa (e.g., all eukaryotes), while displaying comparatively lower variability that compromises species-level resolution (Leray and Knowlton 2016; Wangensteen et al. 2018). Estimates of macrofaunal coverage in public genetic reference databases have been reported for the global north in sites where marine biodiversity has been characterized with multiple techniques over decades (Lacoursière-Roussel et al. 2018; Hestetun et al. 2020). However, reports on proportional taxonomic coverage in public reference databases are less common in tropical and subtropical regions of the ocean around developing countries, which are both more biologically diverse (Rogers et al. 2022), and significantly less studied (de Santana et al. 2021; Keck et al. 2022; Takahashi et al. 2023).
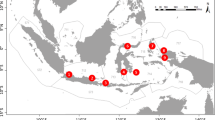
The Gulf of California, Mexico (GC; Fig. 1) is a 1130 km long and 80–200 km wide semi-enclosed subtropical sea in the Eastern Pacific. It is a marine biodiversity hotspot (Roberts et al. 2002) that provides about half of Mexico’s national fisheries catch (Lluch-Cota et al. 2007; Munguia-Vega et al. 2018). The GC has a history of biodiversity monitoring that goes back more than 150 years (Brusca et al. 2005) and has continued into modern times. Monitoring has mainly focused on reef fish (Hastings et al. 2010; Galland et al. 2017; Olivier et al. 2018; Valdivia-Carrillo et al. 2021), benthic macro-invertebrates (Brusca and Hendrickx 2010; Munguía-Vega et al. 2015; Ulate et al. 2016), and planktonic fish eggs and larvae (Peguero-Icaza et al. 2008; Sánchez-Velasco et al. 2017; Ahern et al. 2018). Recently, Morzaria-Luna et al. (2018) compiled a comprehensive, curated, and georeferenced historical database for the biodiversity of the GC that combines monitoring information available in regional and international public repositories, as well as subtidal reef monitoring campaigns. This historical compilation includes 12,105 species of terrestrial and marine prokaryotes and eukaryotes, providing a unique opportunity to contrast the performance of 18S eDNA metabarcoding against a regional historical baseline.
Most studies recognize three distinct biogeographic regions within the GC: Northern, Central, and Southern (Thomson et al. 2000; Brusca et al. 2005). These biogeographic regions possess different oceanographic and environmental conditions, and distinguishable species assemblages (Brusca et al. 2005; Munguia-Vega et al. 2018). The central GC has been recognized for having the highest species diversity (Morzaria-Luna et al. 2018; Munguia-Vega et al. 2018), gradually decreasing towards neighboring sub-regions (Ulate et al. 2016; Galland et al. 2017; Fernández-Rivera Melo et al. 2018). For example, out of 5228 named species of invertebrates in the Gulf of California (16% of which are endemic), 3467 occur in the Central Gulf, 2348 in the Northern Gulf, and 3464 in the Southern Gulf (Brusca and Hendrickx 2023).
Our goal was to assess the challenges and benefits of combining 18S eDNA metabarcoding data to long-term historical baselines and simultaneous visual surveys. Specifically, we (1) compared the number and identity of taxa recovered across techniques at species to phylum levels for different ad hoc groups of eukaryotes: invertebrates, vertebrates, algae, fungi, and other eukaryotes. We then (2) mapped and compared family richness and community structure per method across the Northern and Central GC (Fig. 1).
Material and methods
eDNA collection and underwater visual surveys
Environmental DNA collection and underwater visual surveys were carried out simultaneously at 20 rocky reefs within the GC onboard a research vessel during fall 2016. The focus was set on comparing global patterns of biodiversity within the GC, and between the North (n = 7) and Central (n = 13) regions of the GC (Fig. 1, Table 1), rather than individual per-site comparisons. Likewise, our experimental design was focused on comparing taxonomic inventories per method based on presence/absence, without attempting to thoroughly identify rare or abundant taxa. At each location, one replicate consisting of 1 L of seawater was collected simultaneously to visual surveys at depths of 3–21 m (Table 1 and S1). A group of six experienced monitoring divers conducted underwater visual surveys (25 m long x 4 m wide x 2 m high) targeting conspicuous bony fish at a minimum of eight transects per site. The same transects were used to record all visible benthic macroinvertebrates within the phyla Annelida, Arthropoda, Cnidaria, Echinodermata, Mollusca, and Platyhelminthes. All species surveyed were at least 3 cm long and identified underwater to the lowest taxonomic level possible, and in few cases, underwater pictures were taken and used to confirm identifications. Seawater samples were taken simultaneously at approximately 1–3 m from the bottom using sterile Nalgene bottles (Thermo Scientific) by 2–3 divers close (~10 m) to the group of divers conducting the surveys and at an intermediate location from where the fourth transect was placed. On board the vessel, eDNA filtration was done using 0.45-μm hydrophilic nitrocellulose Millipore filters placed in a Millipore Sterifil filtration system connected to a vacuum pump. A decontaminated area was set for filtration using sterile material, carefully cleaning the space and materials with a 1% sodium hypochlorite solution before and after each sample was processed. No filtration negative controls were processed in the field, but negative controls were included in the laboratory (see below). Further details of field methods have been previously described for these eDNA samples (Valdivia-Carrillo et al. 2021).
eDNA laboratory work and data processing
Environmental DNA extraction was performed using the DNeasy Blood and Tissue kit (Qiagen) in a dedicated eDNA-only laboratory space. We used a two-step PCR protocol for library construction as previously described (Valdivia-Carrillo et al. 2021). The first PCR round amplified the v7 region of the 18S rDNA gene using 18s_allshorts_F (5´ TTTGTCTGSTTAATTSCG 3´) and 18s_allshorts_R (5’ TCACAGACCTGTTATTGC 3’) primers (Guardiola et al. 2015), with a documented average amplicon length of 123 bp (min: 36, max: 892) (Taberlet et al. 2018). The parameters for the thermocycling were the following: 98 ° C for 3 min, 35 cycles of 98 ° C for 30 s, 45 ° C for 30 s, 72 ° C for 30 s, and a final extension of 72 ° C for 5 min. A second PCR step added multiplexing adapters (8 bp long with three mismatches each) and Illumina sequencing adapters (Table S2). Three PCR1 replicates were done for each sample, and 1 ul of each was used as a template in three independent PCR2 replicates per sample that were pooled and purified with AmpureXP beads (Beckman and Coulter). Negative controls were included during each PCR to monitor for external contamination. A mock community made of equimolar DNA concentrations from 25 taxa within six metazoan phyla (Table S3) was added to the sequencing library to assess the efficiency of taxonomic assignments, primer bias, and the relationship between DNA abundance versus read abundance. Twenty eDNA samples, the mock community, and a pool of four negative controls from the PCR steps were sequenced in a single Illumina NextSeq 500 MID (35 Gb) v2 chemistry flow cell (2 × 150 bp pair-end).
Sequences were processed using the Anacapa toolkit (Curd et al. 2019) which incorporates three analysis modules. Their first module, Creating Reference libraries Using eXisting tools (CRUX), was implemented to obtain a 18S custom reference database using default parameters. The NCBI annotated nucleotide repository (5th version, downloaded June 7th 2021; Pruitt et al. 2005) and the European annotated nucleotide (ENA) repository of plants, fungi, invertebrates, and vertebrates (143rd version, downloaded June 7th 2021; Kanz et al. 2005) were screened for available 18S references.
Anacapa’s second module, Sequence Quality Control and ASV Parsing (QC-ASV), was used to get amplicon sequence variants (ASVs). ASVs are sequence clusters obtained by modelling study-specific error rates to discriminate expected errors from rare diversity represented by sequence variants with low read counts (Callahan et al. 2016). We chose ASVs over Operational Taxonomic Units (OTUs) because using a fixed similarity threshold to determine taxonomic identity has been shown to inflate diversity estimates (Callahan et al. 2017), and it is likely that lineages within Eukaryota have independent evolutionary identity rates (De Santiago et al. 2021). The main parameters used in ASV parsing were 30% mismatch allowed for adapter and primer removal, average minimum quality of 30 (Phred 33), minimum sequence length before the alignment of 50 bp, and minimum sequence overlap of 40 bp with up to two mismatches (for complete parametrization see Table S4).
The third module, Anacapa Classifier, aligned ASVs to the 18S custom reference database built in the first module, using a modified version of the BCLA algorithm executed in Bowtie 2 v2.3.5 (Langmead and Salzberg 2012) with 100 bootstrap replicates added to test assignment robustness (see Table S4 for details). Anacapa executes ASV parsing and taxonomic assignment for single forward and reverse reads, aligned reads (merged), and paired reads (unmerged, with no overlap region), taking the most advantage of good quality reads recovered. A raw annotated ASV table was created containing ASV source (forward, reverse, merged, or unmerged ASVs), ASV count per sample, sequence identity, and length, whether it was a single/multiple and full/partial alignments to the 18S custom reference database, along with taxonomic annotations (from phylum to species level) and bootstrap score for each ASV recovered. Singleton ASVs (with one global read count) and all ASVs found in the negative controls were removed as likely contamination. Uncertain taxonomic annotations with bootstrap test scores below 70 and wrong species-level annotations (i.e., higher taxonomic rank annotations accompanied by “sp.,” “nan,” “uncultured,” or “incertae sedis”) were also removed. A 18S eDNA contingency table with phylum to species level annotated ASVs per site was generated.
Visual surveys and historical data processing
Eight visual transects per site for fish and macroinvertebrates were randomly chosen when needed and evaluated at each site, except for two sites from Angel de la Guarda Island (04.AGU1 and 05.AGU2), where only six invertebrate transects were available. For each species detected, family to phylum level taxonomic annotations were manually retrieved using the NCBI taxonomy browser, generating a contingency table of species occurrences per site.
A published database of historical species records of the GC (Morzaria-Luna et al. 2018) was used as a third biodiversity source. The database has georeferenced historical occurrences with taxonomic annotations from species to class rank according to the World Record of Marine Species (WoRMS; https://www.marinespecies.org), and local expertise.
While visual surveys and eDNA were monitored simultaneously at each location, historical records encompass hundreds of thousands of geographically referenced occurrences, posing a challenge when deciding how to match spatial occurrences with 18S eDNA metabarcoding detections. Environmental DNA in the ocean could be transported by strong asymmetric currents in the GC (Munguia-Vega et al. 2018), as suggested by a previous study targeting the 12S teleo metabarcode for the same eDNA samples used here, that showed a high level of fish taxonomic discrimination (119 Actinopterygii OTUs) over a broad geographic and bathymetric scale (Valdivia-Carrillo et al. 2021). To address this issue, we tested the degree of overlap between the 18S eDNA metabarcoding detections and historical records using two spatial scales (wide and narrow) representing distance thresholds to assign historical records around our sampling sites and establish the potential spatial fingerprint of the 18S eDNA detections before comparing methods. Using QGIS v3.14.1 (QGIS Development Team 2009), a wide merge was set using a 0.5 latitudinal degree radius (~55 km) from each location, which allowed occurrences to be included in up to three nearby sites. A narrow merge was set to a radius of 0.1 latitudinal degrees (~11 km) allowing occurrences to be included only in the closest sampling location. These distance criteria were established according to the potential eDNA transport and persistence. The wide merge was assumed to have an eDNA transport and persistence of 48 h (Wood et al. 2020), considering an average particle flow in the GC of 0.3 m s -1 (Santiago-García et al. 2014). In contrast, for the narrow merge, eDNA would be transported at an equal current speed for approximately 10 hours before sampling. The wide and narrow merge of historical occurrences were used to construct two contingency tables of species counts with taxonomic annotations (up to phylum level) per sampling location.
Homogenization of taxonomic annotations across datasets
Taxonomic inconsistencies due to synonymies and unresolved phylogenetic relationships within Eukaryota for all species-level entries of each biodiversity dataset were homogenized using the biodiversity data cleaning package bdc v1.0.0 (Ribeiro et al. 2022) in R (R Core team 2018). We used the full NCBI annotated repository (https://www.ncbi.nlm.nih.gov/taxonomy, accessed March 3rd, 2022) as a reference for taxonomic homogenization, allowing a 20% mismatch between species query and reference. Each biodiversity dataset was compared using Venn diagrams built with input from TaxonTableTools (Macher et al. 2021) to identify, correct, and/or complete missing or inconsistent annotations at all taxonomic levels. We manually removed terrestrial species from the historical and 18S eDNA metabarcoding datasets to focus on marine eukaryotes based on information from the WoRMS database. When missing taxonomy at phylum level occurred due to unresolved phylogenetic relationships, “<group name> NA” was added (e.g., Cryptophyceae were grouped as “Algae NA” at phylum level). Likewise, for many taxa, not all higher taxonomic ranks were assigned, i.e., taxa containing unresolved taxonomy at class, order, or family level were annotated as HLNA (Higher-Level taxonomy Not Available). Since at least 15 major lineages of micro-eukaryotes uncovered by the 18S eDNA metabarcoding data (and not previously compiled in the historical baseline) did not have phylum assignment in the NCBI taxonomy browser, we used the classification of major lineages and supergroups following de Vargas et al. (2015) to include them in our analyses, and collected information about their main trophic/symbiotic modes and habitats, to illustrate the potential ecological role of these cryptic taxa within the ecosystem (Table S5).
Biodiversity estimates across methods
Above phylum level, we created five eukaryotic ad hoc groups to highlight each method’s taxonomic composition: invertebrates, vertebrates, algae (containing mostly photosynthetic entities), fungi, and all “other eukaryotes.” We compared taxonomic γ-richness (i.e., the total number of taxa for each taxonomic rank in our sampled sites) using stacked barplots for each method and calculated a merged list of taxa combining information from all methods for each taxonomic rank. This was used to estimate the contribution of each source of biodiversity information to the total. We also compared overall eukaryotic taxonomic composition between methods from species to phylum rank with Venn diagrams calculated using TaxonTableTools (Macher et al. 2021), and plots were edited using Inkscape (Inkscape Project 2020). We included a small sample of those comparisons in the results to illustrate some of the patterns observed. For the analyses and comparisons across methods, we used richness estimates at the family level, since this intermediate rank provided a balanced comparison (see Results) and the best quantifiable resolution across sampled sites (i.e., higher ranks were highly similar among sites, while genus and species were poorly represented in the 18S eDNA metabarcoding data, see Discussion). Examples included three well represented phyla in the historical database and in visual surveys (Arthropoda, Mollusca, Echinodermata), and Rhodophyta, for which no visual survey data was available.
Contrasting spatial patterns of biodiversity
To evaluate the spatial patterns obtained with the three biodiversity sources, we visualized family level richness per location in QGIS v3.14.1 (QGIS Development Team 2009), plotted boxplots of family richness per region (following biogeographic regions shown in Fig. 1; Table 1), and accumulation curves across methods and regions (Fig. S1). We tested for statistical differences between gamma family richness between methods, and between regions (Northern GC n = 7; Central GC n = 13) within each method. We used the Kruskal-Wallis rank test and post hoc Dunn test for multiple comparisons using the stats and FSA packages in R (R Core team 2018; Ogle et al. 2023).
We then analyzed community structure at family rank for each method and all methods combined, to test for spatial concordance with a recognized biogeographic break between Northern and Central GC (Fig. 1), including comparisons for each ad hoc group independently and for all eukaryotes together. Family community structure was analyzed with non-metric multidimensional scaling (nMDS) based on Jaccard’s pairwise index with 5000 maximum initiations. We tested for statistical differences between regions using an analysis of similarity (ANOSIM) with 1000 permutations. Accumulation curves, nMDS (metaMDS), and ANOSIM were executed using the package vegan v2.5-7 (Oksanen et al. 2018) in R, as previously described (Valdivia-Carrillo et al. 2021).
Results
18S eDNA metabarcoding database
We obtained 20.6 million raw sequences, from which 83% were retained after quality control and ASV parsing (Table S6). Raw sequences are available in the Short Read Archive under Bioproject PRJNA919092. Cleaned reads comprised 16,085,704 (94.3%) paired merged reads, 433,868 (2.5%) paired unmerged reads, 512,184 (3.0%) single forward reads, and 33,604 (0.2%) single reverse reads. Read counts per sampled site ranged from 594k to 944k, with an average of 811k reads. The custom reference database contained 170,737 unique sequences from 119 phyla, 267 classes, 1220 Orders, 6438 families, 27,414 genus and 76,128 species of eukaryotes.
ASV parsing resulted in a total of 222,331 raw ASVs, of which 93% aligned multiple times to the reference database, 2% had a single alignment, and 5% did not match any reference. As for alignment type, 51% of raw ASVs had complete sequence length overlap, and the remaining 49% had partial alignment. The average percent identity between sequences in the custom reference database and the ASV query sequences was 98.7 ± 2.2. From the total raw ASVs, 61,346 ASVs (28%) remained after removal of singletons (160,600 ASVs) and negative control sample (343 ASVs, mainly assigned to fungus associated with human skin, human DNA and few common edible plants, Table S7). From these, 42,740 ASVs (69.8%) only appeared in a single sampled site.
Homogenization of 18S eDNA metabarcoding taxonomic annotations and removal of non-target taxa (mostly land plants) resulted in a final database of 16,077,043 amplicon reads from 61,197 ASVs, of which ~13K (22%) could not be annotated at any level within Eukaryota with the criteria used in our pipeline. Taxonomic annotations varied between ranks, with most ASVs ~47K (77%) annotated to phylum and class rank, ~31K (51%) identified to order, ~21K (34%) to family, ~17K (28%) to genus, and only ~8K (13%) to species-level, representing 1467 different species (Fig. 2a). ASV distribution between groups of eukaryotes showed a majority belonging to algae (~63%), followed by other eukaryotes (~18%) and invertebrates (16%), while fungi (2.1%) and vertebrate (~0.5%) ASVs showed smaller contributions (Fig. 2b–f).
18S eDNA metabarcoding rank taxonomic resolution. a Number of ASVs annotated to each taxonomic rank (from phylum to species) for all eukaryotes, and each ad hoc group; b algae; c other eukaryotes; d invertebrates; e fungi; f vertebrates. Note that ASVs in lower categories are a subset of those found in higher categories
All 25 expected taxa were recovered from the sequenced mock community to varying taxonomic resolutions (Table S3), accounting for 91% of reads in the mock sample. We did not recover any species level annotation. However, only five species had a genetic reference in our custom reference database. Multiple observed ASVs matched various single expected taxa, and taxa showed highly uneven read representations ranging from 0.001 to 56.3%.
Visual surveys and historical databases
Visual surveys registered 28,269 occurrences within 316 transects conducted at all monitored sites, containing 79% vertebrate (fish) and 21% invertebrate occurrences from 167 taxa, of which most (83%) were annotated to species level. Diversity here was composed of 96 vertebrate and 71 invertebrate taxa.
The wide merge of historical records retained 131,980 occurrences, representing 60% of all eukaryotic records in the original database. Kept occurrences were mostly of vertebrates (58.19%), followed by invertebrates (25.89%), algae (13.09%), other eukaryotes (2.77%), and very few fungi (0.05%). Diversity within this database included 5054 species, of which 61% were invertebrates, 19% vertebrates, 28% algae, 5% other eukaryotes, and 0.6% fungi. The narrow historical merge retained 5892 occurrences (8% from total) from 2103 species. Common taxa between historical records and 18S eDNA metabarcoding were higher for the wide merge (Fig. S2), particularly at lower taxonomic levels, showing a 41% wide-to-narrow increase of shared species, 43% increase of shared genera, and 33% increase of shared families. Shared orders, classes, and phyla also increased by 21%, 19%, and 12%, respectively with the wide merge, which was employed for all subsequent analyses.
Biodiversity across methods
The global list of eukaryotes combining 18S eDNA metabarcoding, visual surveys, and historical data from the monitored sites in the GC included 52 phyla (considering three groups of lineages labeled as algae NA, dino NA for dinoflagellates, and other NA), 160 classes, 577 orders, 1599 families, 3525 genera, and 6375 species (Fig. 3; complete taxonomic list in Table S8). The main contribution of each of the three sources of biodiversity information focused on different levels of the taxonomic hierarchy. At higher taxonomic ranks (from phylum to order), 18S eDNA metabarcoding showed the main contribution to the total diversity detected, representing 98% of all phyla, 93% of all classes, and 73% of all orders. In contrast, historical data included 48% of all phyla, 41% of all classes, and 57% of all orders. At lower taxonomic ranks (from family to species), historical data showed the greatest taxonomic contribution, ranging from 79% of all species to 67% of all the families recovered, whereas 18S eDNA metabarcoding had 23% of all species and 52% of all families recovered.
Eukaryotic taxonomic richness of the Gulf of California. Stacked bar charts showing the number of taxa from phylum to species level, per method (18S eDNA metabarcoding, historical and visual surveys), and for each ad hoc eukaryotic group (invertebrates, vertebrates, algae, fungi, and other eukaryotes)
We found a pattern of decreasing overlap in shared taxa between 18S eDNA metabarcoding and historical data from higher to lower ranks. Both methods shared 46% of phyla and 33% of classes, while only 9% of genus and 2% of species were shared (Fig. 4). The 18S eDNA metabarcoding database increased taxonomic diversity by 164% at phylum level (Fig. 5, Table S9), while increases at lower taxonomic levels were 146% for class, 76% for order, 51% for family, 38% for genus, and 26% for species (Fig. 5; Table S9). The 18S eDNA metabarcoding contributed at least 27 new phyla, including ten micro-eukaryotes, nine invertebrates, seven fungi, and two algae phyla (Fig. 4, Table S9). Additionally, at least 15 other major lineages of micro-eukaryotes with unresolved phylum level taxonomy were compiled within the supergroups Holozoa, Stramenopiles, Alveolata, Rhizaria, Viridiplantae and CRuMs (Table S5). The main contributions of 18S eDNA metabarcoding in terms of new phyla and major lineages were members of the plankton, with fewer benthic and parasitic groups. Most of them are microscopic eukaryotes, including fungi, algae, and invertebrates (Table S5). In contrast, visual surveys covered 13% of phyla and 8% of classes and orders, while recovering 2% of all species and 5% of all families (Fig. 3). Visual surveys added five species and one genus to the combined database (Fig. 4; Table S10).
Venn diagrams of eukaryotic taxonomic richness from phylum to species level found by each method. Phyla recovered per method are shown (for details on label “Other NA” refer to Table S5)
Taxonomic contribution of 18S eDNA metabarcoding and historical data. Bar plot showing the number of taxa (main y-axis, left side) added by the 18S eDNA metabarcoding to the historical database, from phylum (including major lineages) to species rank for each ad hoc eukaryotic group. The solid line depicts percentage of historical taxa possessing a genetic reference for the segment of the 18S gene analyzed in the custom reference database, dashed line shows percentage of ASVs that were taxonomically annotated, and point line shows the percentage increase in richness derived from 18S eDNA metabarcoding data compared to the historical baseline (secondary y-axis, right side)
Matching the custom-made reference database against the historical records allowed us to estimate the number of historic taxa possessing a reference sequence in the NCBI and ENA public repositories. For the fragment of the 18S gene analyzed, historical taxa representation in public reference databases was 100% at phylum rank, 98% at class, 87% at order, 80% at family rank, and decreased to 51% and 18% for genus and species ranks, respectively (Fig. 5; Table S9).
Comparing the contribution of each method at family level for four selected phyla present in the historical database illustrated some of the different patterns observed. Rhodophyta, a phylum for which data is absent from visual surveys due to challenges in identification in the field, showed a similar number of taxa at family rank that were new and exclusive to 18S eDNA metabarcoding (7) compared to taxa exclusive to historical records (11), with most families shared between these two methods (25; Fig. 6a). In contrast, the contribution of 18S eDNA metabarcoding was proportionately smaller for well-studied invertebrate phyla compared to historical records. For example, for highly diverse phyla like Arthropoda (201 families in the combined dataset including all three methods) and Mollusca (225 families in the combined dataset), 18S eDNA metabarcoding contributed 40 and 12 new and exclusive families (respectively). For Arthropoda and Mollusca, visual surveys contributed four and 21 families (respectively) but all of them were already present within the historical records. For Echinodermata, a phylum that is conspicuous and readily identifiable in the field (66 families total), 18S eDNA metabarcoding added only one new family, while visual surveys recorded 13 families already present in the historical database (Fig. 6, Table S10 and S11).
Spatial patterns of biodiversity
Taxonomic beta diversity at the family level showed a similar pattern in the nMDS plots separating Northern and Central GC regions for the combined database and each method individually, with samples showing relatively weak ties (stress range = 0.10-0.16; Fig. 7). ANOSIM tests for differences between regions were significant for all methods, with historical records showing slightly better agreement than the rest. Results for specific method and ad hoc eukaryotic group combinations presented more nuanced results, although most comparisons between Northern and Central GC were significant except within invertebrates and algae (Fig. S3). For invertebrates, only the historical records showed significant differences between Northern and Central region (ANOSIM R = 0.597, p = 0.001), whereas 18S eDNA metabarcoding (ANOSIM R = 0.018, p = 0.335), visual surveys (ANOSIM R = 0.171, p = 0.063), and all methods combined (ANOSIM R = 0.032, p = 0.337) were not significant. For algae, results were significant for 18S eDNA metabarcoding (ANOSIM R = 0.417, p = 0.002) but not for historical data (ANOSIM R = -0.003, p = 0.447)
Family level beta diversity of eukaryotes for the Northern and Central Gulf of California. a All methods; b 18S eDNA metabarcoding; c visual surveys; and d historical records. nMDS plots and ANOSIM test statistics based on Jaccard-distance of family composition between North (blue) and Central (red) regions. Ellipses represent standard deviation of points (site numbers follow Fig. 1)
We found significant differences in gamma family richness estimated by each method and all methods combined (Kruskal-Wallis chi-squared = 72.655, df = 3, p-value = 1.153e-15; Table S12). The family richness map combining all methods for all eukaryotic taxa ranged from 503 to 743 families per site in the GC (Fig. 8a, Table S1). The highest richness was found in the Central GC, with significantly lower average richness in the Northern GC (Kruskal-Wallis chi-squared = 5.465, df = 1, p-value = 0.019; Fig. 8e). The 18S eDNA metabarcoding data recovered from 186 to 348 families per site (Fig. 8b) with non-significant differences between Central and Northern GC family richness (Kruskal-Wallis chi-squared = 0.631, df = 1, p-value = 0.427; Fig. 8f). Visual surveys (Fig. 8c) had between 22 and 41 families, and differences between regions were also non-significant (Kruskal-Wallis chi-squared = 2.665, df = 1, p-value = 0.103; Fig. 8g). Historical records (Fig. 8d) had the largest contribution to family richness with 292-576 taxa per site, showed high resemble to the combined methods map, and family richness was likewise significantly higher at the Central than Northern GC (Kruskal-Wallis chi-squared = 4.749, df = 1, p-value = 0.029; Fig. 8h). The Northern GC (n = 7) accumulated 34% of 18S eDNA metabarcoding reads, 25% of visual surveys sightings and 21% of historical occurrences, whereas 66% of eDNA reads, 75% of visual surveys detections, and 79% of historical occurrences were found in the Central GC (n = 13). Accumulation curves for each method considering all sites and each region independently did not plateau under any scenario (Fig S1).
Geographic distribution of family taxonomic diversity for the North (blue) and Central (red) Gulf of California. Number of families per site size-coded into five quantiles, and boxplot distribution (min-Q1-median-Q3-max) per region (see Fig. 1) for: a, e all combined methods, and individually for: b, f 18S eDNA metabarcoding; c, g visual surveys; d, h historical records. Letters within boxplot note the Kruskal-Wallis statistical test results (see text) for each method (for site names and full sampling metadata see Table 1 and S1)
Discussion
Environmental DNA metabarcoding has revolutionized our ability to produce taxonomic inventories at an unprecedented scale, speed, and resolution, providing the opportunity to revisit our understanding of marine biodiversity within historically recognized hotspots. By combining a historical biodiversity database with traditional survey methods and 18S eDNA metabarcoding, our study showed that marine eukaryotic biodiversity in the GC is much higher than previously recognized. Given the limitations in our study related to a single replicate per site, in a single season and restricted to < 21 m deep, the extent of biodiversity present in the Gulf of California is likely much larger than presented here. While combining methods provides a novel view of biodiversity that is much more comprehensive than any approach alone, our study also highlighted many challenges faced in synthesizing biodiversity data from traditional and novel sources.
Environmental DNA’s contribution of eukaryotic taxa to the compiled historical baseline peaked at higher taxonomic ranks and included organisms that are only a few millimeters long (or much smaller), highlighting a historical bias of traditional monitoring techniques towards macro fauna (Table S5). Previously unrecorded taxa included micro-invertebrate phyla (Kinorhyncha, Entoprocta, Phoronida, Tardigrada, Hemichordata, Gastrotricha, Rotifera) and microscopic fungi (Basidiomycota, Oomycota, Blastocladiomycota, Zoopagomycota, Chytridiomycota, Mucoromycota, Cryptomycota). Protists (algae and other micro-eukaryotes) accounted for 80% of all taxonomically assigned sequencing reads in the library, akin to other studies of marine eukaryotic diversity using similar metabarcoding methods (de Vargas et al. 2015; Bakker et al. 2019). Environmental DNA also showed a large fraction of overseen diversity within micro-eukaryotes, including many recognized phyla (Tubulinea, Discosea, Euglenozoa, Hemimastigophora, Fornicata, Picozoa) along with a large list of major lineages with unresolved high-level phylogenetic relationships on NCBI. Particularly diverse were the supergroups Alveolata (Apicomplexa, Ciliophora, Perkinsozoa), Rhizaria (Endomyxa, Imbricata, Acantharea), Stramenopiles (Bigyra, Developea, Pirsoniales, Sticholonchida, Nassellaria, Spumellaria, Collodaria) and Holozoa (Aphelidae, Choanoflagellatea, Filasterea, Ichthyosporea; Table S5). These marine planktonic protists are phylogenetically and ecologically diverse and have been described previously in global metabarcoding campaigns (de Vargas et al. 2015; Cordier et al. 2022). They represent a large but poorly understood fraction of marine biodiversity, that could nonetheless play critical roles in nutrient and energy cycles in the ocean. For example, some mixotrophic taxa within Stramenopiles can flexibly use both phototrophic and heterotrophic metabolism, they can grow in limiting nutrient conditions, play a key role in increasing carbon transfer to higher trophic levels, and in the sinking of organic material (Cohen 2022). Large rhizarians (>0.6 mm) have been estimated to contribute to over 5% of the global carbon stock in the oceans (Biard et al. 2016). Including these previously overseen micro-eukaryotic groups to baseline inventories could significantly improve our understanding of marine food webs.
Overall, 174 new metazoan families of macrofauna were added by 18S eDNA metabarcoding, mainly from Arthropoda, Cnidaria, Bryozoa, Foraminifera, Mollusca, Platyhelminthes, Annelida, Porifera, and Nemertea (Table S11). These are phyla known to include large proportions of undescribed species globally (Appeltans et al. 2012). Most of the newly compiled taxa are planktonic (copepods, ostracods, hydrozoans, sea anemones, and ctenophores), parasitic (mites, trematodes, and nematodes), infaunal, or living in cryptic or inaccessible habitats like the deep sea (including anemones Actinostolidae and bivalves Xylophagaidae), making them difficult to monitor using classic monitoring techniques.
The total eukaryotic diversity estimated via our eDNA metabarcoding study (850 families) that targeted the V7 region of the 18S gene in the GC was larger than similar studies from coastal regions. Differences in family richness between studies can reflect real biological differences among places, but results are also highly dependent on the choice of the metabarcode leading to differential primer biases, and are also affected by the spatial, temporal and bathymetric scale of the experimental design, level of replication, sequencing depth, among other factors (Kelly et al. 2016; Collins et al. 2019; DiBattista et al. 2019). Even within the same 18S gene, the V9 region is known to have higher taxonomic resolution than V7 (Taberlet et al. 2018). A study using the 18S-V9 metabarcode in water samples from the Black Sea reported 259 families at a similar spatial but wider bathymetric scale than our study, also having higher replication (Zhang et al. 2020). Another study conducted within a spatial scale of ~100 km detected 498 families using the V1 region of the 18S metabarcode from water and sediment samples in Okinawa, Japan (DiBattista et al. 2020). At a smaller spatial scale, the 18S-V4 metabarcode was the main contributor (close to half) to a total of 287 families recorded using diverse metabarcodes (including 18S, COI and 16S genes) at a single bay in Ningaloo Reef, Australia (Stat et al. 2017). Even though such comparisons should be treated with caution given technical differences between studies and variation in habitats and environmental gradients among regions, our results reinforce the view that tropical and subtropical regions of the ocean like the GC are highly diverse (Rogers et al. 2022).
We estimated 22.5% of all ASVs were not assigned taxonomically, similarly to other marine studies reporting a proportion of taxonomically unassigned ASVs or OTUs between 14 to 50 % (de Vargas et al. 2015; Leray and Knowlton 2015; Bakker et al. 2019; Cordier et al. 2022). These results indicate that a significant global fraction of marine diversity remains to be included in reference databases even at higher taxonomic ranks, and thus, their sequences found in our 18S eDNA metabarcoding data cannot be placed yet with confidence within Eukaryota. Metagenome analyses of field samples aided by models to predict taxonomy and ecological function could help fill some of these gaps (Manu and Umapathy 2021; Cordier et al. 2021).
We found that no single method provided complete taxonomic coverage within every ad hoc eukaryotic lineage (Table S1). Visual surveys depend on knowledge of diagnostic traits of many species and the ability of trained scuba divers, allowing detection of taxa with specific characteristics (i.e., conspicuous, living in shallow reefs, commercially important, endangered). Visual surveys included a particular subset of species from only seven out of 25 phyla included in the historical database (Annelida, Arthropoda, Cnidaria, Chordata, Echinodermata, Mollusca, and Platyhelminthes). While 18S eDNA metabarcoding targeted a much larger set of phyla and major lineages, it had a poor representation of some groups, including vertebrates that represented only 0.4% of all ASVs but that are major components of the visual surveys and historical datasets. As mentioned earlier, a 12S fish-specific metabarcode applied to the same eDNA samples successfully recovered the diversity of bony fish (Valdivia-Carrillo et al. 2021), indicating eDNA from fish was present but overall represents a small fraction of the total eukaryotic eDNA detected with the 18S metabarcode used. The historical database contains mainly fish, macroinvertebrates, and algae records from multiple sources over several decades, with a marginal representation of fungi and other microeukaryotes. In contrast, visual surveys and eDNA metabarcoding measure biodiversity at a particular time point. These snapshots have the advantage of capturing biodiversity dynamics in response to particular protection efforts (e.g., the establishment of a no-take marine reserve) or anthropogenic stressors (e.g., overfishing or climate change).
The differences in the array of taxa targeted by each method could help explain some of the dissimilar patterns observed. For example, differences in spatial richness at the family level between the two GC regions. Historical data showed higher average family richness in the Central compared to the Northern GC, reproducing a known pattern (Morzaria-Luna et al. 2018; Munguia-Vega et al. 2018). The fact that 79% of occurrences in the historical database are in the Central GC could imply a bias related to a larger sampling effort over time, due to better accessibility of field work, leading to more documented species per site in the Central than the Northern GC. However, 18S eDNA metabarcoding data (and visual surveys) suggested similar richness values for sites between the two regions while also most reads were allocated to the Central GC. Despite the mentioned differences, accumulation curves for the compared sub-regions appear far from reaching saturation, especially for eDNA data (Fig S1). This indicates that the number of samples (20 in total for 18S eDNA metabarcoding and visual surveys) and the sequencing effort (average 800k reads/site for 18S eDNA metabarcoding, and eight transects/site for visual surveys) was insufficient to describe the full diversity present, therefore similar richness between regions would be likely related to under-sampling, than a true biological pattern. Another clue supporting the need to increase effort was that only 30% of all the taxonomically assigned ASVs were observed in more than one site. Overall, 18S eDNA metabarcoding taxonomic diversity from the Midriff Islands might be indicative of higher eukaryotic diversity in the Northern GC than previously recognized for macro-fauna (Brusca et al. 2005; Ulate et al. 2016; Munguia-Vega et al. 2018).
The nMDS plots for family level beta diversity showed consistent differences between Northern and Central regions for every method, in agreement with previous studies, and despite targeting different taxa (Fig. 7) (Brusca et al. 2005; Ulate et al. 2016; Galland et al. 2017; Olivier et al. 2018). However, 18S eDNA metabarcoding suggested that invertebrate communities were more similar between regions than suggested by historical records. Previous studies have found distinct levels of agreement in eDNA community structure depending on taxa and region. On the coast of South Africa, animals, protists, and bacteria shared a similar community structure pattern following three previously recognized biogeographic regions (Holman et al. 2021). In turn, DiBattista et al. (2022) found a more nuanced result on the coasts of the Arabian Sea and Sea of Oman, recovering the expected geographic breaks with varying degrees of concordance between corals, sponges, and other eukaryotes, while vertebrates showed mixed results.
We showed a close relationship between diminishing proportions in the availability of genetic references and the success of the 18S eDNA metabarcoding taxonomic assignment from higher to lower taxonomic ranks. This pattern could be explained both by the lack of reference sequences and the taxonomic resolution of the 18S metabarcode. At higher taxonomic ranks, we showed that many ASVs could not be taxonomically assigned (33% at phylum and 66% at family levels), likely due to a lack of reference sequences matching higher branches of the tree of life within an established similarity threshold. This problem could be alleviated in the future by complementing reference databases of metabarcodes by strategically sequencing the missing branches. Although the 18S metabarcode proved useful to survey very diverse phyla of marine eukaryotes using conserved primers, its slower evolutionary rate also means that low levels of variation, or complete lack of it, might be present at lower taxonomic ranks (e.g., family, genus and species), particularly among closely related or recently diverged taxa. Thus, a low taxonomic resolution for the 18S metabarcode might persist at these lower taxonomic levels, even if the proportion of reference sequences increases within these taxa, due to observed ASVs matching multiple species in the reference database. For example, the taxonomic resolutions reported for the 18S gene segment used in our study are 67–68% (Order and Family), 59% (Genus) and 47% (Species) (Taberlet et al. 2018). The issue of taxonomic resolution could be improved by simultaneously analyzing multiple metabarcodes with different evolutionary rates (e.g., Questel et al. 2021). The availability of genetic references for species from the GC (18% based on the historical dataset) seems low compared to other regions. For example, the North Sea showed a coverage of 36% for the 18S metabarcode and 50% for the COI metabarcode from 1800 species, when considering macro-fauna (Hestetun et al. 2020). Similarly, there was 50% coverage from 900 metazoans for the COI metabarcode in the Canadian Artic (Lacoursière-Roussel et al. 2018). In comparison, coverage for COI varied between 37 and 62% at the species level for metazoans among five marine biodiversity hotspots (Mugnai et al. 2021).
The limitations of 18S eDNA metabarcoding at lower taxonomic ranks forced us to focus our analyses at the family level to match the historical and visual survey datasets, an alternative used previously by other authors (DiBattista et al. 2020). This provided a balance for maximizing several parameters, including resolution (1599 total families present in the combined dataset), the amount of 18S eDNA metabarcoding data used (dictated by the proportion of ASVs that were taxonomically assigned to that level, 43% from all assigned reads), and representation of the historical biota in reference databases (estimated at 80%). Another challenge to merging datasets was related to the spatial scale at which biodiversity is captured by each method. While historical records and visual surveys are based on direct observations, the indirect inference of the presence of a taxon based on its DNA in the environment makes the spatial fingerprint of eDNA records much more dynamic and dependent on several factors, including eDNA release and degradation rates, and transport by ocean currents (Harrison et al. 2019). Our analyses indicated that this fingerprint tends to be a few dozen kilometers wide around the point of sampling for the strong currents found in the GC, although this parameter is expected to be specific for each region. Further studies that explicitly consider eDNA transport based on ocean currents at the time of sampling could improve our estimates about the spatial signature of eDNA detections.
Our study has some other limitations. Although the lack of field and DNA extraction negative controls did not allow us to rule out the amount of eDNA contamination between field sites or in the DNA extraction step, the fact that 70% of ASVs were found exclusively within single sites suggests that cross-contamination in the field does not seem to be a major concern. Also, the sequencing of the PCR negative controls indicated the presence of a limited set of human fungus, human DNA, and terrestrial species associated with contamination in the laboratory steps that were excluded from the analyses. While the historical database represents a vast effort and has been manually curated as thoroughly as possible, some taxa mentioned here as new or eDNA exclusive could have been reported previously in taxon-specific individual studies or gray literature that was overseen in our analyses, thus overestimating the contribution of taxa by eDNA. Additionally, even with the help of dedicated software to harmonize taxonomy between different datasets, some uncertainty remains due to human errors and the constant state of flow in marine taxonomy (Miya and Nishida 2014; Ruggiero et al. 2015), particularly in the process of merging the historical and eDNA datasets with the NCBI taxonomy for thousands of different entries.
Future studies could take advantage of developments in the fast-paced evolving field of eDNA metabarcoding, such as incorporating multiple metabarcodes (Questel et al. 2021), sequencing with long-read technologies (Patin and Goodwin 2022), sequencing complete mitogenomes amplified via long-range PCR (Deiner et al. 2017b), the use of taxonomy-free methods (Apothéloz-Perret-Gentil et al. 2017; Marques et al. 2020), and custom-built genetic reference databases (Valdivia-Carrillo et al. 2021; Dugal et al. 2022). We showed 18S eDNA metabarcoding is an untapped source to improve biodiversity inventories in marine ecosystems, showing many eukaryotic lineages that are difficult to observe and underrepresented in historical data. Although we showed that ASVs abundance was unrelated to eDNA abundance, recent research lines show promising results in estimating relative abundance using allometric scaling in taxon-specific studies with fish (Yates et al. 2020), or spiking eDNA with different standards (Harrison et al. 2020; Tsuji et al. 2022). If the frequent use and concomitant decrease in cost persist, molecular screenings of biodiversity could become routinely used for decision-making and for continued updating of eDNA biodiversity baselines.
References
Ahern ALM, Gómez-Gutiérrez J, Aburto-Oropeza O et al (2018) DNA sequencing of fish eggs and larvae reveals high species diversity and seasonal changes in spawning activity in the southeastern Gulf of California. Mar Ecol Prog Ser 592:159–179. https://doi.org/10.3354/meps12446
Apothéloz-Perret-Gentil L, Cordonier A, Straub F et al (2017) Taxonomy-free molecular diatom index for high-throughput eDNA biomonitoring. Mol Ecol Resour 17:1231–1242. https://doi.org/10.1111/1755-0998.12668
Appeltans W, Ahyong ST, Anderson G et al (2012) The magnitude of global marine species diversity. Curr Biol 22:2189–2202. https://doi.org/10.1016/j.cub.2012.09.036
Baird DJ, Hajibabaei M (2012) Biomonitoring 2.0: a new paradigm in ecosystem assessment made possible by next-generation DNA sequencing. Mol Ecol 21:2039–2044. https://doi.org/10.1111/j.1365-294x.2012.05519.x
Bakker J, Wangensteen OS, Baillie C et al (2019) Biodiversity assessment of tropical shelf eukaryotic communities via pelagic eDNA metabarcoding. Ecol Evol 9:14341–14355. https://doi.org/10.1002/ece3.5871
Biard T, Stemmann L, Picheral M et al (2016) In situ imaging reveals the biomass of giant protists in the global ocean. Nature 532:504–507. https://doi.org/10.1038/nature17652
Blasiak R, Wynberg R, Grorud-Colvert K et al (2020) The ocean genome: conservation and the fair, equitable and sustainable use of marine genetic resources. High Level Panel for a sustainable ocean economy. World Resources Institute, Washington, DC., pp 62. Available online at: https://oceanpanel.org/publications/
Brusca RC, Findley LT, Hastings PA et al (2005) Macrofaunal diversity in the Gulf of California. In: Cartron J, Ceballos G, Felger RS (eds) Biodiversity, ecosystems and conservation in Northern Mexico. Oxford University Press, New York, pp 72–95
Brusca RC, Hendrickx ME (2010) Invertebrate biodiversity and conservation in the Gulf of California. In: Brusca RC (ed) The Gulf of California. Biodiversity and conservation. The University of Arizona Press, Tucson, pp 72-95
Brusca RC, Hendrickx ME (2023) An online database of Sea of Cortez invertebrates, maintained by the Arizona-Sonora Desert Museum and managed by R. C. Brusca. https://www.desertmuseum.org/center/seaofcortez/database.php. Accessed 5 Jan 2023
Calderón-Sanou I, Münkemüller T, Boyer F et al (2019) From environmental DNA sequences to ecological conclusions: how strong is the influence of methodological choices? J Biogeogr 47:193–206. https://doi.org/10.1111/jbi.13681
Callahan BJ, McMurdie PJ, Holmes SP (2017) Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J 11:2639–2643. https://doi.org/10.1038/ismej.2017.119
Callahan BJ, McMurdie PJ, Rosen MJ et al (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
Cohen NR (2022) Mixotrophic plankton foraging behaviour linked to carbon export. Nat Commun 13:1302. https://doi.org/10.1038/s41467-022-28868-7
Collins RA, Bakker J, Wangensteen OS et al (2019) Non-specific amplification compromises environmental DNA metabarcoding with COI. Methods Ecol Evol 10:1985–2001. https://doi.org/10.1111/2041-210X.13276
Convention on Biological Diversity (1992) Biodiversity Outlook - United Nations Treaty Series. United Nations, Rio de Janeiro
Cordier T, Alonso-Sáez L, Apothéloz-Perret-Gentil L et al (2021) Ecosystems monitoring powered by environmental genomics: a review of current strategies with an implementation roadmap. Mol Ecol 30:2937–2958. https://doi.org/10.1111/mec.15472
Cordier T, Angeles IB, Henry N et al (2022) Patterns of eukaryotic diversity from the surface to the deep-ocean sediment. Sci Adv 8:eabj9309. https://doi.org/10.1126/sciadv.abj9309
Curd EE, Gold Z, Kandlikar GS et al (2019) Anacapa Toolkit: an environmental DNA toolkit for processing multilocus metabarcode datasets. Methods Ecol Evol 10:1469–1475. https://doi.org/10.1111/2041-210X.13214
D’Alelio D, Libralato S, Wyatt T, Ribera d’Alcalà M (2016) Ecological-network models link diversity, structure and function in the plankton food-web. Sci Rep 6:21806. https://doi.org/10.1038/srep21806
de Santana CD, Parenti LR, Dillman CB et al (2021) The critical role of natural history museums in advancing eDNA for biodiversity studies: a case study with Amazonian fishes. Sci Rep 11:18159. https://doi.org/10.1038/s41598-021-97128-3
De Santiago A, Pereira TJ, Mincks SL, Bik HM (2021) Dataset complexity impacts both MOTU delimitation and biodiversity estimates in eukaryotic 18S rRNA metabarcoding studies. Environ DNA 4:363–384. https://doi.org/10.1002/edn3.255
de Vargas C, Audic S, Henry N et al (2015) Eukaryotic plankton diversity in the sunlit ocean. Science 348:1261605. https://doi.org/10.1126/science.1261605
Deiner K, Bik HM, Mächler E et al (2017a) Environmental DNA metabarcoding: transforming how we survey animal and plant communities. Mol Ecol 26:5872–5895. https://doi.org/10.1111/mec.14350
Deiner K, Renshaw MA, Li Y et al (2017b) Long-range PCR allows sequencing of mitochondrial genomes from environmental DNA. Methods Ecol Evol 8:1888–1898. https://doi.org/10.1111/2041-210X.12836
DiBattista JD, Reimer JD, Stat M et al (2019) Digging for DNA at depth: rapid universal metabarcoding surveys (RUMS) as a tool to detect coral reef biodiversity across a depth gradient. PeerJ 7:e6379. https://doi.org/10.7717/peerj.6379
DiBattista J, Reimer JD, Stat M et al (2020) Environmental DNA can act as a biodiversity barometer of anthropogenic pressures in coastal ecosystems. Sci Rep 10:8365. https://doi.org/10.1038/s41598-020-64858-9
DiBattista JD, Berumen ML, Priest MA et al (2022) Environmental DNA reveals a multi-taxa biogeographic break across the Arabian Sea and Sea of Oman. Environ DNA 4:206–221. https://doi.org/10.1002/edn3.252
Dugal L, Thomas L, Wilkinson SP et al (2022) Coral monitoring in northwest Australia with environmental DNA metabarcoding using a curated reference database for optimized detection. Environ DNA 4:63–76. https://doi.org/10.1002/edn3.199
Fediajevaite J, Priestley V, Arnold R, Savolainen V (2021) Meta-analysis shows that environmental DNA outperforms traditional surveys, but warrants better reporting standards. Ecol Evol 11:4803–4815. https://doi.org/10.1002/ece3.7382
Fernández-Rivera Melo FJ, Reyes-Bonilla H, Ramírez-Ortiz G, Alvarez-Filip L (2018) Latitudinal variation in structure and function of conspicuous reef fish assemblages along the western Gulf of California. Rev Mex Biodivers 89:1154–1166
Galland GR, Erisman BE, Aburto-Oropeza O, Hastings PA (2017) Contribution of cryptobenthic fishes to estimating community dynamics of sub-tropical reefs. Mar Ecol Prog Ser 584:175–184. https://doi.org/10.3354/meps12364
Geller J, Meyer C, Parker M, Hawk H (2013) Redesign of PCR primers for mitochondrial cytochrome c oxidase subunit i for marine invertebrates and application in all-taxa biotic surveys. Mol Ecol Res 13:851–861. https://doi.org/10.1111/1755-0998.12138
Gold Z, Sprague J, Kushner DJ et al (2021) eDNA metabarcoding as a biomonitoring tool for marine protected areas. PLoS One 16:1–19. https://doi.org/10.1371/journal.pone.0238557
Guardiola M, Uriz MJ, Taberlet P et al (2015) Deep-sea, deep-sequencing: metabarcoding extracellular DNA from sediments of marine canyons. PLoS One 10:e0139633. https://doi.org/10.5061/dryad.520gq
Harrison JB, Sunday JM, Rogers SM (2019) Predicting the fate of eDNA in the environment and implications for studying biodiversity. Proc R Soc B: Biol Sci 286:20191409. https://doi.org/10.1098/rspb.2019.1409
Harrison JG, Calder J, Shuman B et al (2020) The quest for absolute abundance: the use of internal standards for DNA-based community ecology. Mol Ecol Resour 21:30–43. https://doi.org/10.1111/1755-0998.13247
Hastings P, Findley L, van der Heiden A (2010) Fishes of the Gulf of California. In: Brusca RC (ed) The Gulf of California. Biodiversity and conservation. The University of Arizona Press, Tucson, pp 96–118
Hestetun JT, Bye-Ingebrigtsen E, Nilsson RH et al (2020) Significant taxon sampling gaps in DNA databases limit the operational use of marine macrofauna metabarcoding. Mar Biodivers 50:70. https://doi.org/10.1007/s12526-020-01093-5
Hoban S, Bruford M, Urban JD et al (2020) Genetic diversity targets and indicators in the CBD post-2020 Global Biodiversity Framework must be improved. Biol Conserv 248:108654. https://doi.org/10.1016/j.biocon.2020.108654
Holman LE, de Bruyn M, Creer S et al (2021) Animals, protists and bacteria share marine biogeographic patterns. Nature Ecol Evol 5:738–746. https://doi.org/10.1038/s41559-021-01439-7
Inkscape Project (2020) Inkscape. Version 0.92.5. https://inkscape.org
Jo T, Takao K, Minamoto T (2021) Linking the state of environmental DNA to its application for biomonitoring and stock assessment: targeting mitochondrial/nuclear genes, and different DNA fragment lengths and particle sizes. Environ DNA 4:271–283. https://doi.org/10.1002/edn3.253
Kanz C, Aldebert P, Althorpe N et al (2005) The EMBL nucleotide sequence database. Nucleic Acids Res 33:21–26. https://doi.org/10.1093/nar/gki098
Keck F, Blackman RC, Bossart R et al (2022) Meta-analysis shows both congruence and complementarity of DNA and eDNA metabarcoding to traditional methods for biological community assessment. Mol Ecol 31:1820–1835. https://doi.org/10.1111/mec.16364
Kelly RP, O’Donnell JL, Lowell NC et al (2016) Genetic signatures of ecological diversity along an urbanization gradient. PeerJ 4:e2444. https://doi.org/10.7717/peerj.2444
Lacoursière-Roussel A, Howland K, Normandeau E et al (2018) eDNA metabarcoding as a new surveillance approach for coastal Arctic biodiversity. Ecol Evol 16:7763–7777. https://doi.org/10.1002/ece3.4213
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. https://doi.org/10.1038/nmeth.1923
Leduc N, Lacoursière-Roussel A, Howland KL et al (2019) Comparing eDNA metabarcoding and species collection for documenting Arctic metazoan biodiversity. Environ DNA 1:342–358. https://doi.org/10.1002/edn3.35
Lenoir J, Bertrand R, Comte L et al (2020) Species better track climate warming in the oceans than on land. Nat Ecol Evol 4:1044–1059. https://doi.org/10.1038/s41559-020-1198-2
Leray M, Knowlton N (2015) DNA barcoding and metabarcoding of standardized samples reveal patterns of marine benthic diversity. Proc Nat Acad Sci 112:2076–2081. https://doi.org/10.1073/pnas.1424997112
Leray M, Knowlton N (2016) Censusing marine eukaryotic diversity in the twenty-first century. Philos Trans R Soc Lond B Biol Sci 371:20150331. https://doi.org/10.1098/rstb.2015.0331
Lluch-Cota SE, Aragón-Noriega EA, Arreguín-Sánchez F et al (2007) The Gulf of California: review of ecosystem status and sustainability challenges. Prog Oceanogr 73:1–26. https://doi.org/10.1016/j.pocean.2007.01.013
Macher TH, Beermann AJ, Leese F (2021) TaxonTableTools: A comprehensive, platform-independent graphical user interface software to explore and visualise DNA metabarcoding data. Mol Ecol Resour 21:1705–1714. https://doi.org/10.1111/1755-0998.13358
Manu S, Umapathy G (2021) A novel metagenomic workflow for biomonitoring across the tree of life using PCR-free ultra-deep sequencing of extracellular eDNA. Authorea Preprints. https://doi.org/10.22541/au.161401815.51766652/v1
Marques V, Guérin PÉ, Rocle M et al (2020) Blind assessment of vertebrate taxonomic diversity across spatial scales by clustering environmental DNA metabarcoding sequences. Ecography 43:1779–1790. https://doi.org/10.1111/ecog.05049
Miya M, Nishida M (2014) The mitogenomic contributions to molecular phylogenetics and evolution of fishes: a 15-year retrospect. Ichthyol Res 62:29–71. https://doi.org/10.1007/s10228-014-0440-9
Mora C, Tittensor DP, Adl S et al (2011) How many species are there on Earth and in the ocean? PLoS Biol 9:e1001127. https://doi.org/10.1371/journal.pbio.1001127
Morzaria-Luna HN, Cruz-Piñón G, Brusca RC et al (2018) Biodiversity hotspots are not congruent with conservation areas in the Gulf of California. Biodivers Conserv 27:3819–3842. https://doi.org/10.1007/s10531-018-1631-x
Mugnai F, Meglécz E, Abbiati M et al (2021) Are well-studied marine biodiversity hotspots still blackspots for animal barcoding? Global Ecol Conserv 32:e01909. https://doi.org/10.1016/j.gecco.2021.e01909
Munguia-Vega A, Green AL, Suarez-Castillo AN et al (2018) Ecological guidelines for designing networks of marine reserves in the unique biophysical environment of the Gulf of California. Rev Fish Biol Fish 28:749–776. https://doi.org/10.1007/s11160-018-9529-y
Munguía-Vega A, Torre J, Turk-Boyer P et al (2015) PANGAS: an interdisciplinary ecosystem-based research framework for small-scale fisheries in the Northern Gulf of California. J Southwest 57:337–390. https://doi.org/10.1353/jsw.2015.0003
Nichols PK, Marko PB (2019) Rapid assessment of coral cover from environmental DNA in Hawai’i. Environ DNA 1:40–53. https://doi.org/10.1002/edn3.8
Nishitsuji K, Nagata T, Narisoko H et al (2023) An environmental DNA metabarcoding survey reveals generic-level occurrence of scleractinian corals at reef slopes of Okinawa Island. Proc R Soc B: Biol Sci 290:20230026. https://doi.org/10.1098/rspb.2023.0026
Ogle DH, Doll JC, Wheeler AP, Dinno A (2023) FSA: Simple fisheries stock assessment methods. R package version 0.9.5. https://CRAN.R-project.org/package=FSA
Oksanen J, Blanchet FG, Friendly M et al (2018) Vegan: Community ecology package. R package version 2.5-2. https://CRAN.R-project.org/package=vegan
Olivier D, Loiseau N, Petatán-Ramírez D et al (2018) Functional-biogeography of the reef fishes of the islands of the Gulf of California : integrating functional divergence into marine conservation. Global Ecol Conserv 16:e00506. https://doi.org/10.1016/j.gecco.2018.e00506
Patin NV, Goodwin KD (2022) Long-read sequencing improves recovery of picoeukaryotic genomes and zooplankton marker genes from marine metagenomes. mSystems 7: e00595-22. https://doi.org/10.1128/msystems.00595-22
Pawlowski J, Kelly-Quinn M, Altermatt F et al (2018) The future of biotic indices in the ecogenomic era: integrating (e)DNA metabarcoding in biological assessment of aquatic ecosystems. Sci Total Environ 637–638:1295–1310. https://doi.org/10.1016/j.scitotenv.2018.05.002
Peguero-Icaza M, Sánchez-Velasco L, Lavín MF, Marinone SG (2008) Larval fish assemblages, environment and circulation in a semienclosed sea (Gulf of California, Mexico). Estuar Coast Shelf Sci 79:277–288. https://doi.org/10.1016/j.ecss.2008.04.008
Pinsky ML, Selden RL, Kitchel ZJ (2020) Climate-driven shifts in marine species ranges: scaling from organisms to communities. Ann Rev Mar Sci 12:153–179. https://doi.org/10.1146/annurev-marine-010419-010916
Pollock LJ, Connor LMJO, Mokany K et al (2020) Protecting biodiversity ( in all its complexity ): new models and methods. Trends Ecol Evol 35:1119–1128. https://doi.org/10.1016/j.tree.2020.08.015
Pörtner H-O, Roberts DC, Masson-Delmotte V et al (2019) Summary for Policymakers. In: IPCC Special Report on the Ocean and Cryosphere in a changing Climate. IPCC Report 2019 35. http://www.ipcc.ch/publications_and_data/ar4/wg2/en/spm.html
Pruitt KD, Tatusova T, Maglott DR (2005) NCBI Reference Sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 33:501–504. https://doi.org/10.1093/nar/gki025
QGIS Development Team (2009) QGIS Geographic Information System. http://qgis.osgeo.org
Questel JM, Hopcroft RR, DeHart HM et al (2021) Metabarcoding of zooplankton diversity within the Chukchi Borderland, Arctic Ocean: improved resolution from multi-gene markers and region-specific DNA databases. Mar Biodivers 51:4. https://doi.org/10.1007/s12526-020-01136-x
Ratnasingham S, Hebert PDN (2007) BOLD: The barcode of life data system. Mol Ecol Notes 7:355–364. https://doi.org/10.1111/j.1471-8286.2007.01678.x
R Core team (2018) R: A language and environment for statistical computing . R Foundation for Statistical Computing , Vienna, Austria. ISBN 3-900051-07-0. http://www.r-project.org/
Ribeiro B, Velazco S, Guidoni-Martins K et al (2022) BDC: biodiversity data cleaning. Version 1.0.0. https://github.com/brunobrr/bdc
Roberts CM, McClean CJ, Veron JEN et al (2002) Marine biodiversity hotspots and conservation priorities for tropical reefs. Science 295:1280–1285. https://doi.org/10.1126/science.1067728
Robinson KM, Prentice C, Clemente-Carvalho R et al (2023) Paired environmental DNA and dive surveys provide distinct but complementary snapshots of marine biodiversity in a temperate fjord. Environ DNA 5:597–612. https://doi.org/10.1002/edn3.423
Rodríguez-Ezpeleta N, Zinger L, Kinziger A et al (2021) Biodiversity monitoring using environmental DNA. Mol Ecol Resour 21:1405–1409. https://doi.org/10.1111/1755-0998.13399
Rogers AD, Appeltans W, Assis J et al (2022) Discovering marine biodiversity in the 21st century. Adv Mar Biol 93:23–115. https://doi.org/10.1016/bs.amb.2022.09.002
Ruggiero MA, Gordon DP, Orrell TM et al (2015) A higher level classification of all living organisms. PLoS One 10:e0119248. https://doi.org/10.1371/journal.pone.0119248
Ruppert KM, Kline RJ, Rahman MS (2019) Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: a systematic review in methods, monitoring, and applications of global eDNA. Global Ecol Conserv 17:e00547. https://doi.org/10.1016/j.gecco.2019.e00547
Sánchez-Velasco L, Beier E, Godínez VM et al (2017) Hydrographic and fish larvae distribution during the “Godzilla El Niño 2015–2016” in the northern end of the shallow oxygen minimum zone of the Eastern Tropical Pacific Ocean. J Geophys Res C: Oceans 122:2156–2170. https://doi.org/10.1002/2016jc012622
Santiago-García MW, Marinone SG, Velasco-Fuentes OU (2014) Three-dimensional connectivity in the Gulf of California based on a numerical model. Prog Oceanogr 123:64–73. https://doi.org/10.1016/j.pocean.2014.02.002
Seymour M, Edwards FK, Cosby BJ et al (2020) Executing multi-taxa eDNA ecological assessment via traditional metrics and interactive networks. Sci Total Environ 729:138801. https://doi.org/10.1016/j.scitotenv.2020.138801
Stat M, Huggett MJ, Bernasconi R et al (2017) Ecosystem biomonitoring with eDNA: metabarcoding across the tree of life in a tropical marine environment. Sci Rep 7:12240. https://doi.org/10.1038/s41598-017-12501-5
Taberlet P, Bonin A, Zinger L, Coissac E (2018) Environmental DNA: for biodiversity research and monitoring. Oxford University Press
Taberlet P, Coissac E, Pompanom F et al (2012) Towards next-generation biodiversity assessment using DNA metabarcoding. Mol Ecol 21:2045–2050. https://doi.org/10.1111/j.1365-294X.2012.05470.x
Takahashi M, Saccò M, Kestel JH et al (2023) Aquatic environmental DNA: a review of the macro-organismal biomonitoring revolution. Sci Total Environ 873:162322. https://doi.org/10.1016/j.scitotenv.2023.162322
Thomson DA, Findley LT, Kerstitch AN (2000) Reef fishes of the sea of cortez: the rocky-shore fishes of the Gulf of California. University of Texas Press, Austin
Tsuji S, Inui R, Nakao R et al (2022) Quantitative environmental DNA metabarcoding shows high potential as a novel approach to quantitatively assess fish community. Sci Rep 12:21594. https://doi.org/10.1038/s41598-022-25274-3
Ulate K, Sánchez C, Sánchez-Rodríguez A et al (2016) Latitudinal regionalization of epibenthic macroinvertebrate communities on rocky reefs in the Gulf of California. Mar Biol Res 12:389–401. https://doi.org/10.1080/17451000.2016.1143105
Valdivia-Carrillo T, Rocha-Olivares A, Reyes-Bonilla H et al (2021) Integrating eDNA metabarcoding and simultaneous underwater visual surveys to describe complex fish communities in a marine biodiversity hotspot. Mol Ecol Resour 21:1558–1574. https://doi.org/10.1111/1755-0998.13375
Wangensteen OS, Palacín C, Guardiola M, Turon X (2018) DNA metabarcoding of littoral hardbottom communities: high diversity and database gaps revealed by two molecular markers. PeerJ 6:e4705. https://doi.org/10.7717/peerj.4705
West KM, Adam AAS, White N et al (2022) The applicability of eDNA metabarcoding approaches for sessile benthic surveying in the Kimberley region, north-western Australia. Environ DNA 4:34–49. https://doi.org/10.1002/edn3.184
Wood SA, Biessy L, Latchford JL et al (2020) Release and degradation of environmental DNA and RNA in a marine system. Sci Total Environ 704:135314. https://doi.org/10.1016/j.scitotenv.2019.135314
Yamamoto S, Masuda R, Sato Y et al (2017) Environmental DNA metabarcoding reveals local fish communities in a species-rich coastal sea. Sci Rep 7:40368. https://doi.org/10.1038/srep40368
Yates MC, Glaser D, Post J et al (2020) Allometric scaling strengthens the relationship between eDNA particle concentration and organism abundance in nature. Mol Ecol 30:3068–3082. https://doi.org/10.1111/mec.15543
Zhang Y, Pavlovska M, Stoica E et al (2020) Holistic pelagic biodiversity monitoring of the Black Sea via eDNA metabarcoding approach: from bacteria to marine mammals. Environ Int 135:105307. https://doi.org/10.1016/j.envint.2019.105307
Acknowledgements
We thank the support from Consejo Nacional de Ciencia y Tecnología, México (institutional fund CONACyT Fronteras de la Ciencia 292/2016), The University of Arizona—CONACYT Binational Consortium for the Regional Scientific Development and Innovation (CAZMEX), Comunidad y Biodiversidad A.C., The Nature Conservancy Mexico. AMV received support from National Geographic Society NGS-62186R-19. We received a grant from Langebio (CINVESTAV) and Biotech del Norte for Illumina sequencing. We specially thank Eldridge Wisely for helping filtering seawater and two anonymous reviewers for comments that improved the manuscript.
Funding
Partial financial support was received from Consejo Nacional de Ciencia y Tecnología, México (institutional fund CONACyT Fronteras de la Ciencia 292/2016), The University of Arizona—CONACYT Binational Consortium for the Regional Scientific Development and Innovation (CAZMEX 2019 & 2020) and National Geographic Society NGS-62186R-19 to AMV.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval
No animal testing, capture, or handling was performed during this study.
Sampling and field studies
All necessary permits for sampling and observational field studies have been obtained by the authors.
Data availability
Raw sequences were deposited in NCBI’s Short Read Archive under Bioproject No.PRJNA919092 (https://www.ncbi.nlm.nih.gov/sra/PRJNA919092).
Author contribution
AMV and HRB conceived the project. AMV, HRB, and TVC conducted sampling. AMV and TVC performed laboratory procedures. CML performed bioinformatic and ecological analyses, and wrote the first draft. AMV, FVQ, and RCB supervised the project. All authors critically reviewed the analyses and contributed to the final manuscript.
Additional information
Communicated by H. Stuckas
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mac Loughlin, C., Valdivia-Carrillo, T., Valenzuela-Quiñonez, F. et al. eDNA metabarcoding warms up a hotspot of marine biodiversity: revealing underrepresented taxa in visual surveys and historical records from the Gulf of California. Mar. Biodivers. 54, 22 (2024). https://doi.org/10.1007/s12526-024-01415-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12526-024-01415-x