Abstract
Environmental DNA (eDNA) metabarcoding is an evolving tool that can provide broader assessments of marine biodiversity than conventional visual census methods. The outputs of this approach are, therefore, crucial to provide data for conservation priorities and to support fisheries management. We conducted a study using eDNA metabarcoding to understand the distribution of marine biodiversity across Indonesia and to investigate the abundance of three major phyla that comprise a majority of marine biodiversity. In this study, a total of 13,819,634 reads corresponding to 23,252 unique sequences belonging to the phyla Chordata, Mollusca, and Echinodermata were generated from COI amplicons obtained from 92 seawater eDNA samples collected from nine locations and 17 sites. Beta diversity differed significantly across locations (PERMANOVA: p < 0.05) based on Bray–Curtis and Jaccard indices. Taxa of interest were not distributed equally and there were no discernible patterns detected across the sampling area. This might be due to the highly variable percentage of sequenced species between families, preventing robust estimation of species richness. Overall, 45% of reads were identified to species level while 55% were classified as unidentified taxa. Interestingly, the percentage of unidentified taxa was similar between two locations with distinct characteristics representing eastern and western extremities of the sampling region. Despite a relatively poor rate of assignment to species level, our results highlight unprecedented levels of marine biodiversity and strong differences in species composition. This further supports the contention that the eDNA approach is a sensitive method that can provide useful data, in particular to detect changes in species composition. Importantly, this method is clearly advantageous to evaluate marine biodiversity on a large scale and can provide data to support region-wide coral reef management strategies. Knowing species diversity and the degree to which various taxa are distributed is a fundamental for advancing our knowledge of marine ecology and can play an important role in forecasting population dynamics and evolution as well as in refining conservation practices.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Environmental DNA (eDNA) metabarcoding is a non-invasive technique focused on the retrieval of DNA materials produced by organisms and released into their environment (Fukumoto et al. 2015; Boussarie et al. 2018; Garlapati et al. 2019). The method can provide broader biodiversity assessments compared to more classic methods (e.g. Smart et al. 2015; Shaw et al. 2016; Evans et al. 2017; Ruppert et al. 2019) due to the minimal reliance on observer expertise and the ability to detect hidden species. The use of eDNA metabarcoding is increasing globally and has been applied to a wide range of fields in a variety of terrestrial and marine ecosystems. For example, eDNA has been used to understand how biodiversity might vary under different degrees of anthropogenic stress (DiBattista et al. 2019, 2020; Alexander et al. 2020), and to identify and genotype molluscs within laboratory settings (e.g. Ardura et al. 2015; Clusa et al. 2017; Holman et al. 2019). This method has also been a crucial to developing biomonitoring tools to identify seasonal patterns of zooplankton in Western Australia (Berry et al. 2019; West et al. 2020). Despite its perceived usefulness and wide applications, in marine environments eDNA applications have tended to be overly wide in focus, with few studies focused on large taxonomic groups responsible for a major percentage of marine biodiversity (e.g. Closek et al. 2019). Although informative, such research can only provide broad estimates of biodiversity without providing an understanding of how each group responds to change, especially anthropogenic stresses. Therefore, research focused on identifying perturbation-sensitive taxa needs to be emphasized as such data could help to provide reliable and comprehensive biodiversity assessments that are useful for refining conservation practices. The Chordata (fish, mammals, birds, reptiles, and amphibians), Mollusca (characterised by a soft body and generally distinguishable head and foot regions including sea snails, clams, octopus and squid) and Echinodermata (characterised by radial symmetry and the body is covered by calcerous plates that can be moveable or fixed including sea stars, sea cucumbers and sea urchins) are three phyla that contribute large percentages to marine biodiversity (Gray 1997; Sala and Knoltown 2006; Tittensor et al. 2010) They play important economic and ecological roles and include many of the species which act as keystone taxa pivotal for the integrity of marine ecosystems (Grassle et al. 1991; Raghukumar and Anil 2003; Strong et al. 2015). The Indonesian seas are reported to contain 10% of the global marine species diversity covering 574 coral species (Veron et al. 2009), 13 seagrass species (Burke and Selig 2002), 45 species of mangrove (Spalding 2010), and 6 species of sea turtle (Ario et al. 2016). Despite this expected high biodiversity, there is still a paucity of genetic records of marine taxa from Indonesia in the public databases and a considerable proportion of existing records are incomplete. For example, a number of locally published studies have recorded immensely rich echinoderm diversity in several sites across Indonesia, including 44 species in the Banda Sea (Setyastuti and Wirawati 2008); 76 species in Lembeh strait (Supono et al. 2014); 692 species in North Natuna (Lane et al. 2000). High molluscan and reef fish diversity has also been reported from eastern Indonesia; for example, the Raja Ampat area is home to at least 699 Molluscs species, 972 reef fishes in 45 observation sites (Allen 2003), and it play important role in the lives of local coastal communities (Nijman 2019). Nonetheless, data from Indonesia are poorly represented in the in BOLD system database (www.v3.boldsystems.org), with only 201 molluscan and 35 echinoderm records, and 1509 fish species belonging to the Actinopterygii (Chordata). Although there may be several other reasons, the limited number and taxonomic scope of Indonesian marine biodiversity records in public databases is most likely related to the relatively low level of genetic research and observations made in this country. This indicates a need to encourage research efforts, particularly using molecular tools (primarily DNA barcoding) combined with morphological identification in order to reliably detect and record new species and to expand the range of taxa with reference barcode database accessions, as well as the use of eDNA methods to detect the magnitude of marine biodiversity.
In some areas, coral reef organisms at their planktonic larva dispersal phase comprise a large proportion of eDNA materials in the ocean. For example, although overall eDNA release is higher in larger Japanese eels (Anguilla japonica), the rate of release (relative to body weight) is higher in early life-stages and there is a significant discharge of eDNA associated with spawning (Takeuchi et al. 2019), while juvenile bluegill sunfish (Lepomis macrochirus) also release eDNA at a higher rate than adult fishes (Maruyama et al. 2014). Many larvae float in the water column or remain close to the air-sea interface; some settle close to their natal habitat, while others drift away and can become widely dispersed. Larval dispersal could theoretically be measured directly, but exact movements are unknown without direct tracking (Saenz-Agudelo et al. 2011; Madduppa et al. 2014; Williamson et al. 2016; Timm et al. 2017). One challenge in direct larval trajectory tracking research is that species level identification is often challenging, preventing a precise estimate of larval movement for specific taxa. To solve this issue eDNA methods can provide an alternative method for identifying larval-stage taxa (Baltazar-Soares et al. 2020; Ratcliffe et al. 2020). eDNA has been used to detect evidence of larval distribution following spawning events in sand eel and clupeid species (Ratcliffe et al. 2020). Albeit arguably still in its infancy, the use of eDNA is a promising tool that can provide species-specific data on larva dispersal and thereby aid in the detection of larval distribution patterns including settlement habitat location and quality as well as life history characteristics of the taxa involved (Cowen et al. 2006; Kool et al. 2011; Treml et al. 2012, 2015).
The high species diversity in the Coral Triangle is widely recognized (Hughes et al., 2002; Tittensor et al. 2010); however, threats to this region include over-exploitation and alarming rates of anthropogenic activities (Burke et al. 2012; Halpern et al. 2015). One response to this situation has been the growing extent of Indonesian coral reefs designated as Marine Protected Areas (MPAs) with the twin goals of protecting biodiversity and ensuring long term sustainability of their use (Mustika and Ratha 2013; Hamdani 2018; Krueck et al. 2017). There are also protection frameworks specifically targeting important taxa such as top predators (e.g. sharks and groupers), migratory species (e.g. marine mammals), and other ecologically important groups (e.g. some molluscs and echinoderms), and economically important species under two Indonesian ministerial decrees (KEPMEN-KP by Ministry of Marine Affairs and Fisheries; PERMEN KLHK by Ministry of Environment and Forestry of Republic of Indonesia). By the first quarter of 2020, around 23 million hectares had been protected by the Indonesian government under a range of MPA designations (0.07% of Indonesian territorial) with targets to protect another nine million hectares by 2030. In addition to MPAs, the Indonesian government is also managing fisheries within eleven Fisheries Management Areas (FMAs). These FMAs, which were designated based on ecological characteristics and the abundance of fish stocks, will serve as units for integrated fisheries management with core functions including conservation and as well as the control and surveillance of fishing and related activities. However, despite ongoing improvements in fisheries management across Indonesia, there is a severe lack of research that can compare species composition between management units. To support future management activities and ensure that the FMAs can achieve their management targets (e.g. sustaining fish stocks and marine biodiversity), reliable estimates of taxonomic composition based on reliable data are needed. Such data could be used to build effective fisheries management plans including conservation areas that are resilient to present and future threats.
Detailed information on the distribution and abundance of species are critical for the design of conservation areas, in particular MPAs, in order to ensure long term benefits from the marine biodiversity conserved (Beger et al. 2015) and maximise the potential to enhance both fisheries management and conservation (Krueck et al. 2017). Therefore, this study was conducted to assess marine biodiversity across the Indonesian coral reef systems and to investigate the relative abundance and distribution of ecologically and economically important coral reef-associated organisms across fisheries management areas in Indonesia using eDNA methods. We focused on the between-site differences in three phyla (Chordata, Mollusca, and Echinodermata) that comprise the majority of taxa in coral reef communities. The data obtained will help to elucidate patterns of biodiversity while providing crucial information for species conservation and fisheries management.
Materials and methods
eDNA seawater sample collection
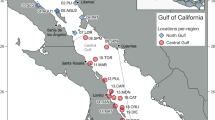
A total of 92 eDNA seawater samples were collected from 17 sites across nine locations in Indonesia (Fig. 1). The sampling locations in this study fall into four Fisheries Management Areas (FMA). FMA 712 was represented by the Seribu Islands in Jakarta and Probolinggo in East Java; FMA 713 by Lombok in West Nusa Tenggara and Selayar in South Sulawesi; FMA 714 by the Wakatobi Archipelago in Southeast Sulawesi; and FMA 715 by sites in Gorontalo, North Maluku, Raja Ampat, and Misool in West Papua. At each site two to four replicate eDNA seawater samples were collected. The eDNA seawater samples were collected directly from the surface or underwater by diving using Self-Contained Underwater Breathing Apparatus (SCUBA) at depths between one and nine meters. This depth was determined by the depth at which coral reefs were mostly observed at the study sites. The seawater samples were collected in 4 L water bottles. Each seawater sample was filtered through a 0.45 µm Pall Corporation sterilized filter paper (47 mm diameter) using a peristaltic pump. The filtration process was interrupted if the flow stopped due to clogging of the filter. A protocol developed by the University of Rhode Island was strictly followed to avoid contamination, using 10% bleach and distilled water to sterilise all equipment used between samples and sampling sites. Filtration was completed at the end of each sampling day. After filtering was completed, each filter paper was placed in a 2 mL cryotube filled with 1.5 mL ZymoBIOMICS DNA/RNA shield.
Nine eDNA seawater sample collection locations across Indonesia: (1) Seribu Islands, Jakarta; (2) Probolinggo, East Java; (3) Lombok, West Nusa Tenggara; (4) Selayar, South Sulawesi; (5) Wakatobi, Southeast Sulawesi; (6) Gorontalo; (7) North Moluccas; (8) Raja Ampat, West Papua; (9) Misool, West Papua. The Indonesian Fisheries Management Area (FMA) boundaries are also shown. The locations represent FMA 712 (locations 1 and 2), 713 (locations 3 and 4), 714 (location 5), and 715 (locations 6, 7, 8 and 9)
eDNA laboratory analysis, library preparation, and next-generation sequencing
DNA extraction was carried out after the field sampling trip (not more than one month after collection) using ZymoBIOMICS DNA extraction kits produced by the Zymo Research Corporation following the manufacturer's guidelines. An approximately 313 bp segment of the mtDNA COI region was amplified using a universal COI primer pair: mICOlintF-adpt as forward (5′-TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG GGW ACW GGW TGA ACW GTW TAY C) and dgHCO2198-adpt as reverse (5′-GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GTA AAC TTC AGG GTG ACC AAA RAA) (Leray et al. 2013). Polymerase Chain Reaction (PCR) reactions contained 2.5 μL 10× buffer, 2.5 μL MgCl2, 0.5 μL mICOlintF-adpt, 0.5 μL dgHCO2198-adpt, 0.5 μL dNTPs, 0.15 μL Taq polymerase, 17.35 μL ddH2O and 1 μL sample template. The PCR profile comprised an initial pre-denaturation stage at 94 °C for 1 min, 30 cycles of denaturation at 94 °C for 30 s, 42 °C for 30 s and extension at 72 °C for 1 min, followed by a final extension at 72 °C for 8 min.
Libraries were purified using AMPure XP beads (Beckman Coulter) according to the Illumina MiSeq 16S metagenomic sequencing library protocol (Illumina 2020). Dual indices and Illumina sequencing adapters from the IDT for Illumina-Nextera DNA Unique Dual Indexes, Set A (catalogue number 20027213) (Illumina, San Diego, USA) were added to the target amplicons in a second PCR step using Kapa HotStart HiFi 2× ReadyMix DNA polymerase (Kapa Biosystems Ltd., London, UK). The PCR profile was 95 °C (3 min), then 9 cycles of 95 °C (30 s), 55 °C (30 s), 72 °C (30 s), followed by a final extension at 72 °C (5 min). Libraries were quantified using the Qubit fluorometric method. The barcode amplicon libraries were combined in equal concentrations into a single pool according to their quantification measurement. The library pool was diluted and denatured according to the Illumina MiSeq library preparation guide. The amplicon library (10 pM) was spiked with 20% denatured and diluted PhiX Illumina control library version 3. The sequencing was conducted on an Illumina MiSeq using the MiSeq reagent kit V3 600 cycle. All library preparation and sequencing was conducted at the Oceanogen Environmental Biotechnology Laboklinikum (Oceanogen) in Bogor. The Oceanogen is a recently established laboratory facility aiming to multiply environmental DNA research effort across Indonesia. All relevant protocols and ongoing projects information are available at www.oceanogen.co.id.
Bioinformatics and data analysis
All eDNA sequences were analysed using the Anacapa pipeline (Curd et al. 2019). Instead of using a publicly available database (e.g., MIDORI, QIIME2 database), a COI database was built using the CRUX package (Creating Reference libraries Using eXisting tools; https://github.com/limeybean/Anacapa/tree/NewMaster/Anacapa_db) to create a de novo sequence reference library combining records from publicly accessible databases, such as European Molecular Biology Laboratory (EMBL) and National Center for Biotechnology Information (NCBI). Upon the completion of database development, Anacapa then de-multiplexed the amplicon read based on the primer sequences mentioned in the laboratory analysis section and trimmed the primers from the reads. Anacapa employed the DADA2 algorithm (Callahan et al. 2016) to denoise and perform error correction on the raw sequence data, merge paired-end reads, and assign high quality reads to Amplicon Sequence Variants (ASVs) through ASV Parsing. In the last step of the pipeline, Anacapa used Bowtie 2 and the Bayesian Least Common Ancestor algorithm to assign ASVs to taxa by using a default likelihood threshold value as suggested by Gao et al. (2017). For most analyses, we excluded singletons to provide the most conservative measure of diversity. To examine patterns of taxonomic diversity, ASV tables output from DADA2 were converted to the BIOM format and imported to R for diversity analyses, statistical testing, and data visualization.
The core-metrics pipeline from the Phyloseq package (McMurdie and Holmes 2013) plugin in R (R development core team) was used to analyse and visualize DNA amplicon data using an even sampling (rarefaction) depth for each sampling locality. The number of reads ranged from 2,361 to 1,054,491 reads per sample (Suppl. 1). For downstream analysis, we excluded taxa within the Orders Primata, Rodentia, and family Bovidae from the Class Mammalia, as well as the Orders Passeriformes and Galliformes from the Class Aves. Furthermore, we also omitted bacterial taxa and rarefied the data to an even depth of 5,219 reads per sample. Using this filtering approach, we discarded two samples with less than 5,000 reads per sample (LBK_175 and RA_107); hence we only used 90 samples from nine locations and 17 sites for further data analysis. This post filtering step provided a total of 469,710 reads with 15,739 ASVs consisting of the Eukaryote Domain and unidentified taxa. The taxonomic composition of each sample was summarized with taxa merged at seven different levels (Kingdom, Phylum, Class, Order, Family, Genus, and Species), and filtered, so that groups representing on average less than 2% (at phylum and order level) and 10% (at species level) of the community were removed. Venn diagrams were constructed through http://bioinformatics.psb.ugent.be/webtools/. We examined patterns of alpha diversity by calculating taxonomic richness for all ASVs and three major phyla (Chordata, Mollusca, and Echinodermata) for each location and Fisheries Management Area (FMA). Multivariate analysis (PERMANOVA) based on the presence/absence of ASVs (Jaccard similarity) and the abundance of reads (Bray–Curtis) was performed in the Vegan package (Oksanen et al. 2018) in R version 3.6.3 (R Development Core Team 2013). Statistical significance was tested using 9999 permutations with a confidence level of α = 0.05. We tested the alpha and beta diversity between locations and FMAs with equal numbers of samples. We analysed seven sampling areas (locations) with four samples each (two locations, East Java and Selayar, had less than four samples). The analysis with an equal number of samples per FMA only included one location per FMA (Seribu Islands, Lombok, Wakatobi, Raja Ampat) with four samples from each location. Principal Coordinate Analyses (PCoA) was implemented using the Ampvis2 package (Andersen et al. 2018) with the phyloseq ordination function for both Jaccard and Bray Curtis dissimilarity matrices using ggplot2 (Wickham, 2009).
Results
Biodiversity assessment using COI marker with eDNA approach
A total of 13,819,634 paired-end reads corresponding to 23,252 unique sequences were generated from the COI amplicons obtained from the 92 samples collected from 17 sites across nine sampling locations. Of the reads used for further analysis, the Eukaryote taxa identified comprised 52% (8,228) of all ASVs and 68% (319,845) of all reads while unidentified taxa composed of 48% (7,511) ASVs and 32% (149,865) reads.
At the kingdom level, taxonomic composition across the sampling locations was dominated by unidentified taxa (Fig. 2). Based on taxon presence-absence, Kingdom Animalia contributed a higher proportion (13%) than Protista (11%), Plantae (5%), and Fungi (3%). Based on read abundance, the Kingdom Fungi was dominant in East Java and prominent in Raja Ampat.
At the phylum level (Fig. 3), based on read abundance, Lombok and Wakatobi had a higher proportion of Chlorophyta than unidentified taxa. East Java had high read abundance of Ascomycota and Bacillariophyta, with the most Chordata detected in Raja Ampat. Based on taxon presence-absence, the phyla Arthropoda and Bacillariophyta, and unidentified taxa were the most common taxonomic groups across all locations.
This study also grouped the locations based on their position in the Fisheries Management Areas (FMAs) (Fig. 4). Comparing the taxonomic composition between the FMAs, FMA 715 had the highest number of unique taxa in this study and FMA 714 the lowest. In particular, FMA 715 sites had more unique species from the Chordata, Mollusca, and Echinodermata. All FMAs shared 45 taxa. The highest species overlap was between FMA 712 FMA 715, even though FMA 715 had the most unique taxa. Interestingly, FMA 714 and FMA 712 had no shared chordate/echinoderm/molluscan species. This result could be due to the ASV approach which considers every unique sequence as a single taxonomic unit.
At the location level, when including all ASVs, Lombok (East Nusa Tenggara) had the highest number of ASVs (3764), followed by Misool (2518 ASVs), Seribu Island (2473 ASVs), and North Mollucas (2204 ASVs) and lowest number was recorded in Selayar (528 ASVs) (Table 1). However, when considering only ASVs from the phyla Chordata, Mollusca, and Echinodermata, Raja Ampat had the highest number of ASVs (140), followed by Gorontalo (138 ASVs) and Seribu Island (101 ASVs) and Wakatobi had the lowest number (14 ASVs). At the FMA level, FMA 715 had the highest number of total ASVs (7722) as well as for ASVs in the three major phyla (350). The lowest number of ASVs was recorded from FMA 714 for both total ASVs (1839) and the three major phyla (14) (Table 2).
Beta diversity analysis showed significant differences in taxonomic composition between locations and between FMAs (PERMANOVA: p < 0.05) based on Bray–Curtis and Jaccard indices. This is consistent with two other tests (equal numbers of samples per location and FMA). PCoA plots (Figs. 5, 6) show a clustering pattern, where Wakatobi sites closely clustered with Lombok sites. Some Raja Ampat sites clustered with Lombok, but Misool in Raja Ampat formed an isolated cluster, while sites in other locations clustered together.
Beta diversity ordination plots by Fisheries Management Area (FMA) based on Bray–Curtis and Jaccard indices: A and B include all samples (9 locations, 90 samples); (C and D) include one location in each FMA (Seribu Islands in FMA 712, Lombok in FMA 713, Wakatobi in FMA 714, and Raja Ampat in FMA 715) with an equal number of samples (four) per location
Assessment of eukaryotic taxa interest using eDNA and a COI barcode
This section describes a snapshot of the distribution of three phyla (Chordata, Mollusca and Echinodermata) based on read abundance across the sampling locations (Fig. 7). Taxa of special interest were included in the dataset for these phyla (Fig. 8), including economically valuable and/or protected bony fish, sharks, and marine mammals (phylum Chordata); giant clams (phylum Mollusca); and echinoderms.
Phylum chordata
The database resulting from this study included 329 ASVs and 22,923 sequence reads, covering five classes, 21 orders, 33 families, 45 genera, and 59 species. The distribution of phylum Chordata at the Order level is shown in Fig. 7A. Within the Phylum Chordata, this study recovered 73 species from 25 orders. There were high read abundances of Pristiformes (sawfishes) at several sites, including Gorontalo and Selayar. Perciformes (ray-finned fish) were observed at all sites with the highest number of reads from Lombok and Wakatobi. Some economically valuable fish genera were detected, including groupers (Plectropomus and Epinephelus), snappers (Lutjanus), and fusiliers (Caesio) (Fig. 7A). For the Class Actinopterygii, relatively common species of interest included Plectropomus areolatus (Seribu Islands, East Java, North Moluccas and Misool); Epinephelus coeruleopunctatus (Seribu Islands, Gorontalo and Misool); Caesio cuning (Lombok and West Papua) and E. aerolatus (Raja Ampat). Another order with high read numbers, the Enterogona (tunicates) was found at all sites but most abundant at Misool. The dataset also captured a single ASV of a rare species, Orcaella brevirostris (Irrawaddy Dolphin) from class Mammalia, with a total of seven reads from a single sample, LBK_190 (Lombok, West Nusa Tenggara).
Phylum mollusca
With 124 ASVs and 17,658 reads, taxa identified as belonging to the phylum Mollusca belonged to 17 orders and one unidentified taxon, and represented four classes (Bivalvia, Gastropoda, Cephalopoda, Polyplacophora) (Fig. 7B). A high percentage of molluscan taxa were unidentified at four sites (North Moluccas, Seribu Islands, Gorontalo and Misool). The location with highest number of reads for the Order Verenoida (bivalves) was Lombok, followed by Raja Ampat and East Java. Read numbers of Nudibranchia (sea slugs) were highest in Raja Ampat and Seribu Islands and Teuthida (squid) reads were highest in Wakatobi and Seribu Islands. Littorinimorpha (gastropods) were also common at most sampling sites.
The tridacnid calms of the subfamily Tridacninae (Class: Bivalvia, Order: Veneroida, Family Cardiidae), previously known as the Family Tridacnidae, are a molluscan taxon of special interest. They are highly sought-after due to their high economic value in and increasing demand from the global aquarium trade as well as for human consumption, curios and other uses (Reksodihardjo-Lilley and Lilley 2007; Yusuf and Moore 2020). Due to the widespread threats and predicated future dwindling population size, all members of this group have been listed in CITES Appendix II to provide protection from international trade (Wells 1997), although CITES listing does not protect these clams from in-country exploitation. With 26 ASVs and 17,002 reads, only two species in the genus Tridacna were recorded: Tridacna maxima (25 unique ASVs) and T. crocea (1 unique ASV) (Fig. 8). Tridacnid clams were identified from three locations with three reads from East Java (T. maxima); 5,373 reads from Lombok (14 T. crocea and 5,359 T. maxima); and 11,083 reads from Raja Ampat (T. maxima) (Fig. 8).
Phylum echinodermata
There were 2,760 reads assigned to the Phylum Echinodermata. These reads comprised 130 ASVs belonging to six classes, ten orders, 20 families, 22 genera, and 21 species, and a group of unidentified taxa (Figs. 7C, 8C). The abundance and taxonomic composition of echinoderm reads obtained varied considerably between teach nine locations. Five out of nine sites had a high proportion of unidentified taxa. The most commonly identified order was Spatangoida (heart urchins) with the highest number of reads from the Seribu Islands followed by Gorontalo, East Java, and Selayar. Among the 10 Orders, only one (Euryalida) was detected in Raja Ampat, represented by a single species (Asrochele pacifica). The most dominant echinoderm group in the Wakatobi was the order Comatulida (crinoids), also identified from five other locations (Seribu Islands, Lombok, Misool, North Moluccas, and East Java). The order Valvatida (starfish) was recorded in Misool and Gorontalo. An order of special interest is the Asphidochirotida (sea cucumbers), detected mostly in Misool, with lesser read numbers from four other sites (Gorontalo, North Moluccas, Seribu Islands, and Selayar).
Discussion
Illuminating marine biodiversity across Indonesian coral reefs and fisheries regions
This study demonstrates the utility of eDNA metabarcoding to illuminate marine biodiversity across coral reef ecosystem in Indonesia. We further highlighted its potential for improving surveillance and conservation of ecologically and economically important taxa, including several taxa supporting coral reef fisheries in Indonesia. eDNA metabarcoding can provide an important complement to traditional approaches (i.e., visual census and morphology-based identification) and generate data on marine biodiversity that previously could not be detected using traditional methods (Taberlet et al. 2012; Deiner et al. 2015; Thomsen and Willerslev 2015; Shaw et al. 2016; Evans and Lamberti 2018; Evans et al. 2017; Porter and Hajibabaei 2018; Ruppert et al. 2019). Nevertheless, the application of this method has been relatively limited in the Coral Triangle (a marine biodiversity hotspot comprising much of Indonesia, Philippines and Papua New Guinea), hindering evaluation on the efficacy of the method in areas with a high concentration of marine biodiversity. As one of the few studies done in the Coral triangle using an eDNA metabarcoding method, this study shows promising results.
The high abundance of unidentified taxa suggests that many of the taxa discovered in this research are not yet recorded in global databases. However, given the high rate of new taxa being described from the area, high marine diversity per square meter and the extent of relatively unexplored regions (Cahyani D., unpublished data) we also argue that some of the unidentified ASVs maybe undescribed marine species as yet unknown to science, potentially leading to an enhanced evaluation of marine biodiversity based on these data after future species identifications have been made. As an example, only 28% of known molluscan species from Raja Ampat (McKenna et al. 2002) could be assigned based on the BOLD database, while much of our eDNA data cannot be assigned to taxa in any of the existing public databases. This suggests that much of the rich biodiversity in Indonesia remains to be explored and highlights a poor representation of taxa from the coral triangle and neighbouring areas in global COI databases. Further, the high abundance of unidentified taxa in this research highlights the gap that needs to be filled in global metazoan databases to support robust taxonomic assignment. Nevertheless, this research has provided important baseline sequence data useful for comparative study with species currently present in these databases and could also be important for future biodiversity and conservation research.
Coral reefs are estimated to harbour approximately 950,000 multicellular species (±40% of estimated global biodiversity), of which only around 10% have been identified (Fisher et al. 2015). These figures reflect the vast gap in our existing knowledge on the biodiversity of coral reefs and highlight how much research remains to be done (Hoeksema 2017). Therefore, research focused on filling the information gaps should be done in areas with enormous biodiversity, but as yet severely underexplored, like the Coral Triangle region, as well as in habitats under high anthropogenic pressure such as many areas in western Indonesia. Although efforts are underway, biodiversity records in the region suffer from, inter alia, methods which are ineffective when used to build comprehensive inventory catalogues in rapid and consistent ways (e.g., biodiversity monitoring mainly uses visual surveys by observers with varying levels of expertise) (Hoeksema 2017). In this context, our study has taken important steps in introducing a new method of biodiversity inventory and has highlighted the magnitude of the gap between what we know and don’t know regarding marine biodiversity in the Coral Triangle. Although still relatively new and arguably not yet developed to its full potential, the application of eDNA metabarcoding can speed up auditing process for species identification by providing a general picture regarding the magnitude of biodiversity. Furthermore, our research has successfully identified several taxa living in interstitial spaces in the coral reef environment (e.g. amphipods and isopods) and taxa that are frequently observed in coral rubble (e.g. bryozoans, sponges, polychaetes and tunicates) (Meesters et al. 1991). These findings show that large components of marine biodiversity dwelling in areas that are hidden and, in many cases, inaccessible using traditional monitoring tools, can potentially be identified using eDNA metabarcoding. These hidden species, and our limitations in observing the life within them, have resulted in poor investigation of the species associated with such habitats; these include species inhabiting reef caves and crevices such as sponges (Reitner et al. 1996; Vicente et al. 2016), some scleractinian corals (Hoeksema 2012) and zoantharians (Irei et al. 2015; Kise and Reimer 2016). Our findings have further highlighted the magnitude of marine biodiversity across regions in Indonesia and the need to undertake further taxonomic studies that aim to provide reference taxonomic information complete with DNA data. The results also further emphasize the importance of eDNA in revealing hidden marine biodiversity, including finding new organisms (Maggioni et al. 2017; Montano et al. 2015; Tornabene et al. 2016), and adding new records to public databases (Valdez-Moreno et al. 2019).
Monitoring of taxonomic composition across space and time in marine biodiversity hotspots like Indonesia remains a significant challenge, especially for less studied, small, rare, and uncommon species (e.g., Madduppa et al. 2020). The Alpha and Beta diversity analyses in our study highlight the significant differences in species richness and biotic community composition between locations. We also show that the percentage of sequenced species among families is highly variable, which is a barrier to robust estimation of species richness. Interestingly, two locations with distinct environmental conditions had a similarly high percentage of unidentified taxa. Raja Ampat is a relatively remote area with pristine reefs in western Papua while the Seribu Islands north of Jakarta is exposed to high anthropogenic pressures. Surprisingly, even though Raja Ampat is well known as an area with particularly high marine biodiversity (Allen 2003; Hukom et al. 2018) our results show that overall the sites in the area had relatively low alpha diversity. This could be due to various ecological reasons (Gray 2001; Stanley 2007), but in the context of our study this could also be due to technical challenges during sampling and laboratory work. Nonetheless, Misool, one of the locations within Raja Ampat, had the highest alpha diversity. The sites in the Seribu Islands, on the other hand, had relatively high alpha diversity compared to the other sites in this study. Our findings contrast with the general idea that areas with historically polluted environmental conditions, like Seribu Islands, necessarily have relatively low biodiversity. Given that this is the first study using an eDNA approach done in these areas, the results suggest that eDNA could potentially reveal hidden and cryptic diversity unobserved by traditional monitoring methods. Therefore, the assessment of biodiversity should not be totally reliant on the environmental state of a given site, as even apparently degraded sites could house unique patterns of biodiversity. The combination of voucher-based DNA monitoring, eDNA, and conventional visual survey methods is therefore highly advisable to aid in understanding the magnitude and patterns of biodiversity.
The relatively comprehensive data from ecological surveillance and visual census of the coral reefs of the Seribu Islands (e.g., Rachello-Dolmen and Cleary 2007; Madduppa et al. 2013; Cleary et al. 2016; Rachman 2020) show that this archipelago suffers from a variety of anthropogenic stresses, including large numbers of introduced and invasive species induced by shipping activities (Huhn et al. 2020; Subhan et al. 2020; Anzani et al. 2019; Utami et al. 2018; Dias et al. 2018). In addition, many of the previous studies in the area are poorly documented in terms of accessions deposited in public DNA sequence databases, (e.g., Polónia et al. 2014; Patantis et al. 2013; Madduppa et al. 2017, 2019). In contrast, habitats in Raja Ampat comprise a high diversity of reef formations, both typical and atypical, with varying coastal and bathymetric profiles, and include unusual types of coral habitat and coral communities (McKenna et al. 2002; Erdmann and Pet 2002). Such features are particularly noticeable in the area around Misool Island, where many of the reefs display none of the classic or predictable coral community zoning patterns. In particular, southeastern Misool has vast variety of unexplored habitat types (Turak and Souhoka 2003) that could be refuges for unexplored and unique biodiversity, which might explain the high percentage of unidentified taxa in this region.
The use of eDNA metabarcoding using standard and taxon-specific COI mini barcodes (up to ~ 400 bp) has been developed to examine degraded DNA materials present in aquatic environments. In this analysis, we successfully identified a variety of taxa from three major phyla (Chordata, Mollusca, and Echinodermata) using short mitochondrial DNA COI sequences. We found that these phyla were not equally distributed over the areas sampled. With respect to sampling methodology, the relatively low number of taxa detected from the water samples might be due to the small amount of DNA material released into the environment by the organisms present, rapid degradation processes following DNA shedding, or local current movements that consistently transport the DNA in the water column.
Unsurprisingly, Raja Ampat, a globally renowned marine biodiversity hotspot (Cox and Bright 2017), had a higher percentage of reads from Chordata than the other locations. The reputation of this area is largely based on a number of studies that have discussed the exceptional richness of chordate taxa, in particular fish (Allen 2003), sharks (Beer 2015, Jaiteh et al. 2017, Sembiring et al. 2015) and marine mammals (Agostini et al. 2012; Ender et al. 2014). However, even though Raja Ampat had the highest chordate species richness recorded in this study, we only detected ~ 20 chordate species, two orders of magnitude lower than the ~ 1,000 species of fish reported from or thought to live in this area (Fenner 2002; McKenna et al. 2002). The dominant fish families of Raja Ampat are usually well represented on coral reefs across the Indo-Western and Central Pacific (Allen 2003). For example, Allen (2003) reported that the five most speciose families in the region are the gobies (Gobiidae; 137 species), damselfishes (Pomacentridae; 114 species), wrasses (Labridae; 109 species), cardinalfishes (Apogonidae; 73 species), and groupers (Serranidae; 58 species). In Raja Ampat, and in the Indonesian region in general, the diversity of sharks and rays (Class Chondrichthyes) can be considered very high, with a total of 207 Chondrichthyes of which 109 are classified into 34 shark families (Fahmi 2010; White et al. 2006). In our study we found 139 shark and ray ASVs with 23 identified species, 60% of which were detected from Raja Ampat. Shark and rays have been exploited for many years and due to their global population status, many have been placed on one of the at risk categories in the IUCN Red List (Suppl. 3). Another interesting finding was a high abundance of Pristiformes reads of at several sites including Gorontalo, Selayar, Lombok and Wakatobi. These reads comprise the first record of Pristiformes in the Lombok and Wakatobi areas.
With respect to the Mollusca, we found a high percentage of unidentified taxa at four sites: North Moluccas, Seribu Islands Gorontalo and Misool. The highest read numbers for the Order Veneroida (bivalve molluscs) were from Lombok, followed by Raja Ampat and East Java, for Nudibranchia (sea slugs) from Raja Ampat and the Seribu Islands and for Teuthida (squids) from Wakatobi and the Seribu Islands. The giant or tridacnid clams are another important taxonomic group identified in this study. Eight species of the former family Tridacnidae (now family Cardiidae, sub-family Tridacninae) are known to live in Indonesian waters (Neo and Low 2018; Borsa et al. 2015). Two tridacnid clam species were recorded at one or more locations in this study: the small giant clam Tridacna maxima (27 unique ASVs) and the boring clam T. crocea (1 unique ASV). The small giant clam T. maxima has the widest distribution among the Tridacninae, and is found throughout the Indo-Pacific region from Africa to island in the eastern Pacific, while T. crocea, the boring clam (also known as the crocus clam, crocea clam or saffron-coloured clam), has a more restricted range in the central Indo-Pacific region (Bin Othman et al. 2010). Our results are in line with other studies reporting T. maxima (the small giant clam) and T. crocea (the boring clam) as the most commonly observed tridacnid clam species in Indonesia (DeBoer et al. 2008; Kochzius and Nuryanto 2008; Nuryanto and Kochzius 2009; Hernawan 2010; Arbi 2017; Yusuf and Moore 2020). The presence of T. crocea may be attributed to the behaviour that earned this species the name of boring clam, as its shell is typically embedded into hard substrate, especially massive coral lifeforms. This boring habit makes it difficult for fishermen to harvest the species, while its relatively small size also makes it a less attractive target compared to other tridacnid clam species. Overexploitation of tridacnid species is reported to occur due to the shell and curio industry (Pasaribu 1988; Wells 1997; Yusuf and Moore 2020), artisanal and subsistence fisheries, cultural practices and (including T. crocea) as feed for high value commodities such as lobsters (Yusuf and Moore 2020) as well as the marine ornamental trade (Reksodihardjo-Lilley and Lilley 2007). Thus far, however, there do not seem to be any records of industrial-scale exploitation of tridacnid clams in Indonesia.
eDNA metabarcoding has proven useful as a tool for species identification in a variety of marine invertebrate taxa, including echinoderms (e.g., Günther et al. 2018). This study detected eleven orders, 21 families, 24 genera, and 23 species belonging to the Phylum Echinodermata. At seven out of nine sites there were unidentified echinoderm taxa. Interestingly, we only identified Euryalida (basket stars) from Raja Ampat. The most frequently identified orders were the Spatangoida (heart urchins) and Ophiurida (brittle stars), while the order Valvatida (starfish) was only identified from Misool and Gorontalo. One order of particular interest was the Asphidochirotida (sea cucumbers); the most abundant reads were from Misool, followed by four other sites (Gorontalo, North Moluccas, Seribu Islands, and Selayar). The Spatangoida (heart urchins) were mostly recorded from the Seribu Islands, Gorontalo, East Java, and Selayar. However, the Comatulida (crinoids) reads were mostly from the Wakatobi with low read numbers from five other locations (Seribu Islands, Lombok, Misool, North Moluccas and East Java). The family-level distribution of echinoderms indicates that echinoderm community composition varied considerably between the nine locations. However, many reads were unidentified, highlighting the need for research to improve reference databases for the phylum Echinodermata.
Implications for conservation and fisheries management
Biodiversity conservation and fisheries management both depend to a varying but considerable extent on effective monitoring of the species present, in particular economically important and endangered taxa, across patchy habitats in large areas (Noss 1990; Regan et al. 2008). Many of the species detected by eDNA metabarcoding in this study are economically important and exploited by small scale fishermen. Across the Indonesian Archipelago, the live reef food-fish trade (LRFFT) tends to concentrate on a narrow range of high value reef fish species (Khasanah et al. 2019a). For example, fishes commonly targeted for sale in the LRFFT include coral trout (Plectropomus leopardus), predominantly in the juvenile or female phase size range (Khasanah et al. 2019a, b), and (mostly) juvenile Napoleon wrasse (Cheilinus undulatus) (Yusuf and Moore 2020). Whenever LRFFT holding facilities in Raja Ampat and Misool were encountered and inspected, they invariably contained a moderate number of these species.
With respect to the Mammalia, a single ASV of the marine mammal Orcaella brevirostris (Irrawaddy dolphin) was detected by eDNA metabarcoding in Lombok, a new species record for Lombok and indeed for West Nusa Tenggara. While often called the Irrawaddy river dolphin, O. brevirostris is not a true river dolphin but rather a euryhaline oceanic dolphin that is often found in brackish water, occurring in discontinuous subpopulations along sea coasts, including estuarine environments and rivers (Minton et al. 2017). The global conservation status of this species based on the latest IUCN Red List assessment is Endangered (EN) (Minton et al. 2017), although five subpopulations are classified as Critically Endangered (CR), including the Mahakam River population in East Kalimantan, Indonesia (Jefferson et al. 2008). Entanglement in fishnets and habitat degradation are given as the main threats to Irrawaddy dolphin populations. No range-wide survey has been conducted for this endangered species, and direct observation and identification of these organisms is always difficult and costly (Bogich et al. 2008). However, in Indonesia Irrawaddy dolphins are frequently observed in the freshwater reaches of the Mahakam River (Kreb 1999; Oktaviani et al. 2007) and Banten Bay (Khalifa et al. 2014). Conservation efforts are being made at international and national levels to alleviate threats to this species; however, the Irrawaddy dolphin's proximity to developing communities makes conservation difficult in practice (Beasley et al. 2013). Nonetheless, a conservation program, entitled Conservation Foundation for the Protection of Rare Aquatic Species of Indonesia, is focused on protecting the Irrawaddy dolphin population and their habitat in the Mahakam River (http://www.ykrasi.org/). The program not only educates and surveys the public, but also monitors the dolphins and their habitat. A prime example of this is the establishment of patrols in several villages. The detection of Irrawaddy dolphin sequences from Lombok calls for verification and eventually targeted conservation measures if the record is confirmed.
Shark and rays identified in this study can help managers in prioritizing areas and species within regions and in particular within fisheries management areas (FMAs). Among the taxa detected, two species are categorized as critically endangered (CE) in the IUCN Red List (IUCN 2021), three as endangered (EN), four as vulnerable (VU), nine as near threatened (NT), two as least concern (LC), one as data deficient (DD) and one species has not yet been evaluated (NE) (Suppl. 3).Sharks are cartilaginous fish of the Class Chondrichthyes and are mostly top predators in the ocean food chain (Fahmi and Dharmadi 2013). Sharks play an important role in maintaining the stability of the food chain through their predation on other organisms (Roff et al. 2016; Heupel et al. 2014). Therefore, it is important to manage shark populations sustainably, especially in areas such as Lombok which are known to be hotspots for shark fisheries (Sembiring et al. 2015).
The results of this study indicate that the eDNA approach used is not (at least as yet) capable of reliably identifying all the species actually present in a given area. However, this method can provide insights into which taxonomic groups occur or predominate in specific locations. This study reinforces the contention that this approach can be very useful in evaluating marine biodiversity, despite the many caveats for implementing metabarcoding analysis for eDNA samples using COI markers (Stat et al 2017; Thomsen and Willerslev 2015; Cristescu and Hebert 2018). However, these caveats or restrictions limit or even prohibit the use of metabarcoding approaches for characterizing whole fish assemblages by direct species classification. This study also highlighted the scarcity of genetic reference sequences for Indonesia in extant databases, as evidenced by the high percentage of unidentified taxa at all locations. Largely due to a lack of genetic sampling and data submission, this lack is especially critical for species-rich ecosystems such as the marine biodiversity hotspots in Indonesia (Juhel et al. 2020). One reason for the limited genetic sampling could be the difficulties involved in biodiversity sampling at many Indonesian sites by classical methods and even with the advent of organism-based DNA barcoding. A second challenge is that even when sequences are deposited in the public reference repositories they may have been collected by people who are not experts in the taxonomy of the species in question and therefore identification of the specimens from which the sequences were extracted may not always be reliable. The lack of sequence coverage highlights the immense gap that needs to be filled in the building of robust online repositories, and the large number of species which still need to be identified and barcoded. Therefore, all unidentified sequences from this study remain potentially useful for further research and are visualized to demonstrate their similarity with other taxa. In addition to their value as useful genetic data in the future, they can serve to raise awareness, especially in the Indonesian scientific community and government.
Contributions of eDNA metabarcoding to support the management effectiveness of Marine Protected Areas (MPAs) could include monitoring the presence of targeted and important species within MPAs and the surrounding region. MPAs are seen as an important tool for achieving sustainable use and avoiding over-exploitation of living marine resources (Palumbi 2001). Protected areas have been widely promoted as an important tool for biodiversity conservation and management (Brooks 2010; Venter et al. 2014). Species and habitat protection support the achievement of MPA goals (Beger et al. 2003, Williams et al. 2009, Hart 2003, Péron et al. 2013), including fisheries management (e.g., enhanced productivity, species diversity, and spillover effect) (Russ and Alcala 2010; Hamilton et al. 2011; Edgar et al. 2014; Smith and Garcia 2014), and the provision of recreational and educational opportunities (Ballantine 2014). Consequently, MPAs are highly recommended as a management tool for marine fisheries in Indonesia (Mous et al. 2005). It is suggested that the spatial distribution should suit the dispersal capabilities of the species to be protected to ensure connectivity between MPAs (Palumbi 2003). Two decades ago, Indonesia had established 51 MPAs with coral reefs covering an area of about 58,000 km2 (Spalding et al. 2001), representing around 1% of the Indonesian Archipelago’s total marine area. Since then, many more MPAs have been established under four MPA categories that are “Suaka Alam Perairan” (Marine Wildlife Reserve), “Taman Nasional Perairan” (Marine National Park), “Taman Wisata Perairan” (Marine Recreational Park) and “Suaka Perikanan” (Fisheries Refugee) (Wiadnya et al. 2011). Monitoring these vast areas is clearly a major challenge. Using eDNA protocols to effectively inform management regarding the presence of certain important species could be an efficient way to support the achievement of MPA objectives. While financial support for genetic research in Indonesia is an important issue, it is not the only aspect where resources need to be increased, human resources are also crucial. Environmental DNA (eDNA) metabarcoding surveys have proven useful for many ecosystems and can potentially provide an efficient and cost-effective complement to traditional ecosystem surveys, such as visual underwater censuses. This could be beneficial for marine conservation managers and fisheries officers to monitor the areas for which they are responsible. However, the managers need to understand the principles; and relevant people within (or collaborating with) these agencies need to be trained in eDNA sampling protocols, so that they can collect seawater sample from their area. These could then be used for metabarcoding a wide range of species, depending on the taxa of interest within their locations, using general and/or specific primers with high-throughput sequencing.
Conclusion
eDNA metabarcoding is a sensitive, efficient technique for studying marine organisms with large scale spatial distribution patterns. The use of eDNA methods can complement traditional methods. Increased availability of reference sequences, especially from similar geographic areas, will increase eDNA identification scope and confidence. eDNA metabarcoding could potentially be an excellent tool for species identification, tracking habitats, biodiversity surveys and to support environmental conservation at large spatial extents and in remote areas.
Data availability
Raw data are available in the electronic supplementary material.
References
Agostini VN, Grantham HS, Wilson J, Mangubhai S, Rotinsulu C, Hidayat N, Muljadi A, Muhajir MM, Darmawan A, Rumetna L, Erdmann MV (2012) Achieving fisheries and conservation objectives within marine protected areas: zoning the Raja Ampat network. The Nature Conservancy Indonesia Marine Program, Denpasar, Indonesia
Alexander JB, Bunce M, White N, Wilkinson SP, Adam AA, Berry T, Stat M, Thomas L, Newman SJ, Dugal L, Richards ZT (2020) Development of a multi-assay approach for monitoring coral diversity using eDNA metabarcoding. Coral Reefs 39(1):159–171
Allen GR (2003) Coral reef fishes of the Raja Ampat Islands. Report on a rapid ecological assessment of the Raja Ampat Islands, Papua, Eastern Indonesia held October 30–November 22, 2002, p 42
Andersen KS, Kirkegaard RH, Karst, SM, Albertsen M (2018) Ampvis2: an R package to analyse and visualize 16S rRNA amplicon data, bioRxiv: p. 299537
Anzani L, Madduppa HH, Nurjaya IW, Dias PJ (2019) Molecular identification of white sea squirt didemnum sp. (Tunicata, Ascidiacea) colonies growing over corals in Raja Ampat Islands Indonesia. Biodivers J Biol Divers 20(3):636–642
Arbi UY (2017) Kepadatan dan kondisi habitat kerang kima (Cardiidae: Tridacninae) di beberapa lokasi di Perairan Sulawesi Utara. BAWAL Widya Riset Perikanan Tangkap 3(2):139–148
Ardura A, Zaiko A, Martinez JL, Samuiloviene A, Borrell Y, Garcia-Vazquez E (2015) Environmental DNA evidence of transfer of North Sea molluscs across tropical waters through ballast water. J Molluscan Stud 81(4):495–501
Ario R, Wibowo E, Pratikto I, Fajar S (2016) Turtle habitat conservation from threaten at turtle conservation and educatiuon center (TCEC). Bali Jurnal Kelautan Tropis 19(1):60–68
Ballantine B (2014) Fifty years on: lessons from marine reserves in New Zealand and principles for a worldwide network. Biol Cons 176:297–307
Baltazar-Soares M, Pinder AC, Harrison AJ, Oliver W, Picken J, Britton JR, Andreou D (2020) A non-invasive eDNA tool for detecting sea lamprey larvae in river sediments: analytical validation and field testing in a low abundance ecosystem, bioRxiv.
Beasley I, Pollock K, Jefferson TA, Arnold P, Morse L, Yim S, Lor Kim S, Marsh H (2013) Likely future extirpation of another Asian river dolphin: the critically endangered population of the Irrawaddy dolphin in the Mekong River is small and declining. Mar Mamm Sci 29(3):E226–E252
Beer AJE (2015) Diversity and abundance of sharks in no-take and fished sites in the marine protected area network of Raja Ampat, West Papua, Indonesia, using baited remote underwater video (BRUVs). Royal Roads University, Canada
Beger M, Jones GP, Munday PL (2003) Conservation of coral reef biodiversity: a comparison of reserve selection procedures for corals and fishes. Biol Cons 111(1):53–62
Beger M, McGowan J, Treml EA, Green AL, White AT, Wolff NH, Klein CJ, Mumby PJ, Possingham HP (2015) Integrating regional conservation priorities for multiple objectives into national policy. Nat Commun 6:8208
Berry TE, Saunders BJ, Coghlan ML, Stat M, Jarman S, Richardson AJ, Davies CH, Berry O, Harvey ES, Bunce M (2019) Marine environmental DNA biomonitoring reveals seasonal patterns in biodiversity and identifies ecosystem responses to anomalous climatic events. PLoS Genet 15(2):e1007943
Bin Othman AS, Goh GHS, Todd PA (2010) The distribution and status of giant clams (Family Tridacnidae)—a short review. Raffles Bull Zool 58(1):103–111
Bogich TL, Liebhold AM, Shea K (2008) To sample or eradicate? A cost minimization model for monitoring and managing an invasive species. J Appl Ecol 45(4):1134–1142
Borsa P, Fauvelot C, Andréfouët S, Chai TT, Kubo H, Liu LL (2015) On the validity of Noah’s giant clam Tridacna noae (Röding, 1798) and its synonymy with Ningaloo giant clam Tridacna ningaloo Penny and Willan, 2014. Raffles Bull Zool 63:484–489
Boussarie G, Bakker J, Wangensteen OS, Mariani S, Bonnin L, Juhel JB, Kiszka JJ, Kulbicki M, Manel S, Robbins WD, Vigliola L (2018) Environmental DNA illuminates the dark diversity of sharks. Sci Adv 4(5):eaap9661
Brooks T (2010) Conservation planning and priorities. Conservation biology for all, pp 199–219
Burke L, Reytar K, Spalding K, Perry A (2012) Reefs at risk revisited in the coral triangle: world resources institute. In The nature conservancy. World-Fish Center, International Coral Reef Action Network. UNEP world conservation monitoring centre and global coral reef monitoring network Washington, DC
Burke L, Selig L (2002) Reefs at risk in Southeast Asia. World resource institute, Washington
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13(7):581–583
Closek CJ, Santora JA, Starks HA, Schroeder ID, Andruszkiewicz EA, Sakuma KM, Bograd SJ, Hazen EL, Field JC, Boehm AB (2019) Marine vertebrate biodiversity and distribution within the central California current using environmental DNA (eDNA) metabarcoding and ecosystem surveys. Front Mar Sci 6:732
Cleary DFR, Polónia ARM, Renema W, Hoeksema BW, Rachello-Dolmen PG, Moolenbeek RG, Budiyanto A, Tuti Y, Draisma SGA, Prud’homme van Reine WF, Hariyanto R (2016) Variation in the composition of corals, fishes, sponges, echinoderms, ascidians, molluscs, foraminifera and macroalgae across a pronounced in-to-offshore environmental gradient in the Jakarta Bay-Thousand Islands coral reef complex. Mar Pollut Bull 110(2):701–717
Clusa L, Miralles L, Basanta A, Escot C, Garcia-Vazquez E (2017) eDNA for detection of five highly invasive molluscs. A case study in urban rivers from the Iberian Peninsula. PLoS ONE 12(11):e0188126
Cowen RK, Paris CB, Srinivasan A (2006) Scaling of connectivity in marine populations. Science 311(5760):522–527
Cox K, Bright J (2017) Raja ampat: a biodiversity hot spot and the future of marine conservation. Fisheries 42(9):462–467
Cristescu ME, Hebert PD (2018) Uses and misuses of environmental DNA in biodiversity science and conservation. Annu Rev Ecol Evol Syst 49:209–230
Curd EE, Gold Z, Kandlikar GS, Gomer J, Ogden M, O’Connell T, Pipes L, Schweizer TM, Rabichow L, Lin M, Shi B (2019) Anacapa Toolkit: an environmental DNA toolkit for processing multilocus metabarcode datasets. Methods Ecol Evol 10(9):1469–1475
DeBoer TS, Subia MD, Ambariyanto EMV, Kovitvongsa K, Barber PH (2008) Phylogeography and limited genetic connectivity in the endangered boring giant clam across the coral triangle. Conserv Biol 22(5):1255–1266
Deiner K, Walser JC, Mächler E, Altermatt F (2015) Choice of capture and extraction methods affect detection of freshwater biodiversity from environmental DNA. Biol Cons 183:53–63
Dias PJ, Gilg MR, Lukehurst SS, Kennington WJ, Huhn M, Madduppa HH, McKirdy SJ, de Lestang P, Teo SL, Lee SSC, McDonald JI (2018) Genetic diversity of a hitchhiker and prized food source in the Anthropocene: the Asian green mussel Perna viridis (Mollusca, Mytilidae). Biol Invasions 20(7):1749–1770
DiBattista JD, Reimer JD, Stat M, Masucci GD, Biondi P, De Brauwer M, Bunce M (2019) Digging for DNA at depth: rapid universal metabarcoding surveys (RUMS) as a tool to detect coral reef biodiversity across a depth gradient. PeerJ 7:e6379
DiBattista JD, Reimer JD, Stat M, Masucci GD, Biondi P, De Brauwer M, Wilkinson SP, Chariton AA, Bunce M (2020) Environmental DNA can act as a biodiversity barometer of anthropogenic pressures in coastal ecosystems. Sci Rep 10(1):1–15
Edgar GJ, Stuart-Smith RD, Willis TJ, Kininmonth S, Baker SC, Banks S, Barrett NS, Becerro MA, Bernard AT, Berkhout J, Buxton CD (2014) Global conservation outcomes depend on marine protected areas with five key features. Nature 506(7487):216–220
Ender AI, Mangubhai S, Wilson JR, Muljadi A (2014) Cetaceans in the global centre of marine biodiversity. Marine Biodivers Records 7:1–9
Erdmann MV, Pet JS (2002) A rapid marine survey of the northern Raja Ampat Islands. Report from Henry Foundation/The Nature Conservancy/NRM/EPIQ
Evans NT, Lamberti GA (2018) Freshwater fisheries assessment using environmental DNA: a primer on the method, its potential, and shortcomings as a conservation tool. Fish Res 197:60–66
Evans NT, Shirey PD, Wieringa JG, Mahon AR, Lamberti GA (2017) Comparative cost and effort of fish distribution detection via environmental DNA analysis and electrofishing. Fisheries 42(2):90–99
Fahmi (2010) Shark and rays in Indonesia. Mar Res Indones 22(4):225–242
Fenner D (2002) Reef corals of the Raja Ampat Islands, Papua Province, Indonesia. Part II. comparison of individual survey sites. Appendix 2. coral species recorded at individual sites in the Raja Ampat Islands. A marine rapid assessment of the Raja Ampat Islands, Papua Province, Indonesia. RAP Bull Biol Assessment 22:29–36
Fisher R, O’Leary RA, Low-Choy S, Mengersen K, Knowlton N, Brainard RE, Caley MJ (2015) Species richness on coral reefs and the pursuit of convergent global estimates. Curr Biol 25(4):500–505
Fukumoto S, Ushimaru A, Minamoto T (2015) A basin-scale application of environmental DNA assessment for rare endemic species and closely related exotic species in rivers: a case study of giant salamanders in Japan. J Appl Ecol 52(2):358–365
Gao X, Lin H, Revanna K, Dong Q (2017) A Bayesian taxonomic classification method for 16S rRNA gene sequences with improved species-level accuracy. BMC Bioinform 18(1):247
Garlapati D, Charankumar B, Ramu K, Madeswaran P, Murthy MR (2019) A review on the applications and recent advances in environmental DNA (eDNA) metagenomics. Rev Environ Sci BioTechnol 18(3):389–411
Grassle JF, Lassere P, McIntyre AD, Ray GC (1991) Marine biodiversity and ecosystem function. Biol Int Special 23:1–9
Gray JS (1997) Marine biodiversity: patterns, threats and conservation needs. Biodivers Conserv 6(1):153–175
Gray JS (2001) Marine diversity: the paradigms in patterns of species richness examined. Sci Mar 65(S2):41–56
Günther B, Knebelsberger T, Neumann H, Laakmann S, Arbizu PM (2018) Metabarcoding of marine environmental DNA based on mitochondrial and nuclear genes. Sci Rep 8(1):1–13
Halpern BS, Frazier M, Potapenko J, Casey KS, Koenig K, Longo C, Lowndes JS, Rockwood RC, Selig ER, Selkoe KA, Walbridge S (2015) Spatial and temporal changes in cumulative human impacts on the world’s ocean. Nat Commun 6(1):1–7
Hamdani B (2018) Threats, challenges and opportunities to marine protected areas in the coral triangle area: a case study of Indonesia sea
Hamilton RJ, Potuku T, Montambault JR (2011) Community-based conservation results in the recovery of reef fish spawning aggregations in the coral triangle. Biol Cons 144(6):1850–1858
Hart T (2003) Rules of engagement for conservation lessons from the democratic republic of congo. Conserv Pract 4(1):14–22
Hernawan UE (2010) Study on giant clams (Cardiidae) population in Kei Kecil waters Southeast-Maluku. Widyariset 13(3):101–108
Heupel MR, Knip DM, Simpfendorfer CA, Dulvy NK (2014) Sizing up the ecological role of sharks as predators. MEPS 495:291–298. https://doi.org/10.3354/meps10597
Hoeksema BW (2012) Forever in the dark: the cave-dwelling azooxanthellate reef coral Leptoseris troglodyta sp. n. (Scleractinia, Agariciidae). ZooKeys 228:21
Hoeksema BW (2017) The hidden biodiversity of tropical coral reefs. Biodiversity 18(1):8–12
Holman LE, Hollenbeck CM, Ashton TJ, Johnston IA (2019) Demonstration of the use of environmental DNA (eDNA) for the non-invasive genotyping of a bivalve mollusc, the European Flat Oyster (Ostrea edulis). Front Genet 10:1159
Hughes TP, Bellwood DR, Connolly SR (2002) Biodiversity hotspots, centres of endemicity, and the conservation of coral reefs. Ecol Lett 5(6):775–784
Huhn M, Madduppa HH, Khair M, Sabrian A, Irawati Y, Anggraini NP, Wilkinson SP, Simpson T, Iwasaki K, Setiamarga DH, Dias PJ (2020) Keeping up with introduced marine species at a remote biodiversity hotspot: awareness, training and collaboration across different sectors is key. Biol Invasions 22(2):749–771
Hukom FD, Yulianda F, Bengen DG, Kamal MM (2018) Reef fishes in the marine protected area of Dampier strait, Raja Ampat Islands, West Papua Province, Indonesia. Int J Fisheries Aqua Stud 6(6):131–135
Irei Y, Sinniger F, Reimer JD (2015) Descriptions of two azooxanthellate Palythoa species (subclass Hexacorallia, order Zoantharia) from the Ryukyu Archipelago, southern Japan. ZooKeys 478:1–26
IUCN (2021) The IUCN red list of threatened species. Version 2021-1. Available at: www.iucnredlist.org. Accessed 1 June 2021
Jaiteh VF, Loneragan NR, Warren C (2017) The end of shark finning? Impacts of declining catches and fin demand on coastal community livelihoods. Mar Policy 82:224–233
Jefferson TA, Karczmarski L, Kreb D, Laidre K, O’Corry-Crowe G, Reeves R, Rojas-Bracho L, Secchi E, Slooten E, Smith BD, Wang JY, Zhou K (2008) Orcaella brevirostris (Mahakam River subpopulation) (errata version published in 2016). The IUCN Red List of Threatened Species 2008: e.T39428A98842174
Juhel JB, Utama RS, Marques V, Vimono IB, Sugeha HY (1930) Accumulation curves of environmental DNA sequences predict coastal fish diversity in the coral triangle. Proc R Soc B 287:20200248
Khalifa MA, Kamal MM, Adiwilaga EM, Sunuddin A (2014) Preliminary study on the distribution of irrawaddy dolphin, Orcaella brevirostris, in Banten Bay. Open J Marine Sci 4(04):338–343
Khasanah M, Kadir NN, Jompa J (2019) Reproductive biology of three important threatened/near-threatened groupers (Plectropomus leopardus, Epinephelus polyphekadion and Plectropomus areolatus) in Eastern Indonesia and implications for management. Animals 9(643):1–18
Krueck NC, Ahmadia GN, Green A, Jones GP, Possingham HP, Riginos C, Treml EA, Mumby PJ (2017) Incorporating larval dispersal into MPA design for both conservation and fisheries. Ecol Appl 27(3):925–941
Kasanah M, Nurdin N, Sadovy de Mitcheson Y, Jompa J (2019) Management of the grouper export trade in Indonesia. Rev Fish Sci Aquac 28(1):1–15
Kise H, Reimer JD (2016) Unexpected diversity and a new species of Epizoanthus (Anthozoa, Hexacorallia) attached to eunicid worm tubes from the Pacific Ocean. ZooKeys 562:49–71
Kochzius M, Nuryanto A (2008) Strong genetic population structure in the boring giant clam, Tridacna crocea, across the Indo-Malay Archipelago: implications related to evolutionary processes and connectivity. Mol Ecol 17(17):3775–3787
Kool JT, Paris CB, Barber PH, Cowen RK (2011) Connectivity and the development of population genetic structure in Indo-West Pacific coral reef communities. Glob Ecol Biogeogr 20(5):695–706
Kreb D (1999) Facultative river dolphins: conservation and social ecology of freshwater and coastal Irrawaddy. University of Amsterdam, Thesis
Lane DJ, Marsh LM, VandenSpiegel D, Rowe FW (2000) Echinoderm fauna of the South China Sea: an inventory and analysis of distribution patterns. Raffles Bull Zool 48:459–494
Leray M, Yang JY, Meyer CP, Mills SC, Agudelo N, Ranwez V, Boehm JT, Machida RJ (2013) A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents. Front Zool 10(1):34
Madduppa HH, Bahri S, Subhan B, Anggraini NP, Ohoiulun H, Abdillah T, Arafat D, Santoso P, Sangadji IM (2019) DNA barcoding of sea turtles (Dermochelyidae and Cheloniidae) and its protocol using different tissues quality: implication to conservation managers. IOP Conf Series Earth Environ Sci 278(1):012041
Madduppa H, Putri AS, Wicaksono RZ, Subhan B, Akbar N, Ismail F, Arafat D, Prabuning D, Sani LMI, Srimariana ES, Baksir A (2020) Morphometric and DNA Barcoding of endemic Halmaheran walking shark (Hemiscyllium halmahera, Allen, 2013) in North Maluku Indonesia. Biodiv J Biol Div 21(7):3331–3342
Madduppa H, Subhan B, Anggraini NP, Fadillah R, Tarman K (2017) DNA barcoding reveals vulnerable and not evaluated species of sea cucumbers (Holothuroidea and Stichopodidae) from Kepulauan Seribu reefs Indonesia. Biodiv J Biol Div 18(3):893–898
Madduppa HH, Subhan B, Suparyani E, Siregar AM, Arafat D, Tarigan SA, Alimuddin A, Khairudi D, Rahmawati F, Brahmandito A (2013) Dynamics of fish diversity across an environmental gradient in the Seribu Islands reefs off Jakarta. Biodivers J Biol Div 14(1):17–24
Madduppa HH, Timm J, Kochzius M (2014) Interspecific, spatial and temporal variability of self-recruitment in anemonefishes. PLoS ONE 9(2):e90648
Maggioni D, Montano S, Arrigoni R, Galli P, Puce S, Pica D, Berumen ML (2017) Genetic diversity of the Acropora-associated hydrozoans: new insight from the Red Sea. Mar Biodivers 47(4):1045–1055
Maruyama A, Nakamura K, Yamanaka H, Kondoh M, Minamoto T (2014) The release rate of environmental DNA from juvenile and adult fish. PLoS ONE 9(12):e114639
McKenna SA, Allen GR, Suryadi S (2002) A marine rapid assessment of the Raja Ampat islands, Papua Province, Indonesia. Conservation International, Center for Applied Biodiversity Science, Department of Conservation Biology
McMurdie PJ, Holmes S (2013) Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217
Meesters E, Knijn R, Willemsen P, Pennartz R, Roebers G, van Soest RV (1991) Sub-rubble communities of Curaçao and Bonaire coral reefs. Coral Reefs 10(4):189–197
Minton G, Smith BD, Braulik GT, Kreb D, Sutaria D, Reeves R (2017). Orcaella brevirostris (errata version published in 2018). The IUCN Red List of Threatened Species 2017: e.T15419A123790805. https://doi.org/10.2305/IUCN.UK.2017-3.RLTS.T15419A50367860.en.
Montano S, Seveso D, Galli P, Puce S, Hoeksema BW (2015) Mushroom corals as newly recorded hosts of the hydrozoan symbiont Zanclea sp. Mar Biol Res 11(7):773–779
Mous PJ, Pet JS, Arifin Z, Djohani R, Erdmann MV, Halim A, Knight M, Pet-Soede L, Wiadnya G (2005) Policy needs to improve marine capture fisheries management and to define a role for marine protected areas in Indonesia. Fish Manage Ecol 12(4):259–268
Mustika PL, Ratha I (2013) Towards the bali MPA network. In Bali Marine Rapid Assessment Program 2011. Conservation International
Neo ML, Low JK (2018) First observations of Tridacna noae (Röding, 1798) (Bivalvia: Heterodonta: Cardiidae) in christmas island (Indian Ocean). Mar Biodivers 48(4):2183–2185
Noss RF (1990) Indicators for monitoring biodiversity: a hierarchical approach. Conserv Biol 4(4):355–364
Nijman V (2019) Wildlife trade, CITES and the protection of marine molluscs in Indonesia. Molluscan Res 39(3):195–204
Nuryanto A, Kochzius M (2009) Highly restricted gene flow and deep evolutionary lineages in the giant clam Tridacna maxima. Coral Reefs 28(3):607–619
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin .R, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H (2018) Vegan: ecological diversity R package, Version 2.5-4
Oktaviani D, Aisyah A, Dharmadi D (2007) Pemantauan status populasi pesut (Orcaella brevirostris) di sungai pella (Daerah Aliran Sungai Mahakam), kalimantan timur. BAWAL Widya Riset Perikanan Tangkap 1(6):209–214
Palumbi SR (2001) The ecology of marine protected areas. Marine ecology, the new synthesis 509–530
Palumbi SR (2003) Population genetics, demographic connectivity, and the design of marine reserves. Ecol Appl 13(sp1):146–158
Pasaribu BP (1988) Status of giant clams in Indonesia. Giant clams in Asia and the Pacific. Australian Centre for International Agricultural Research, Canberra
Patantis G, Rahmadara G, Elfidasari D, Chasanah E (2013) Molecular identification of sponges obtained from Seribu Islands National Park and their associated bacteria. Squalen Bull Marine and Fish Postharvest Biotechnol 8(1):29–36
Péron C, Grémillet D, Prudor A, Pettex E, Saraux C, Soriano-Redondo A, Authier M, Fort J (2013) Importance of coastal marine protected areas for the conservation of pelagic seabirds: the case of Vulnerable yelkouan shearwaters in the Mediterranean Sea. Biol Cons 168:210–221
Polónia AR, Cleary DF, Duarte LN, de Voogd NJ, Gomes NC (2014) Composition of Archaea in seawater, sediment, and sponges in the Kepulauan Seribu reef system Indonesia. Microbial Ecol 67(3):553–567
Porter TM, Hajibabaei M (2018) Automated high throughput animal CO1 metabarcode classification. Sci Rep 8(1):1–10
Rachello-Dolmen PG, Cleary DFR (2007) Relating coral species traits to environmental conditions in the Jakarta Bay/Pulau Seribu reef system, Indonesia. Estuar Coast Shelf Sci 73(3–4):816–826
Rachman A (2020) Checklist and estimation of total number of phytoplankton species in Pari, Tidung, and Payung Islands Indonesia. Biodivers J Biol Div 21(6):2446–2458
Raghukumar S, Anil AC (2003) Marine biodiversity and ecosystem functioning: a perspective. Curr Sci 84(7):884–892
Ratcliffe FC, Uren Webster TM, Garcia de Leaniz C, Consuegra S (2020) A drop in the ocean: monitoring fish communities in spawning areas using environmental DNA. Environ DNA 3(1):43–54
Regan HM, Hierl LA, Franklin J, Deutschman DH, Schmalbach HL, Winchell CS, Johnson BS (2008) Species prioritization for monitoring and management in regional multiple species conservation plans. Divers Distrib 14(3):462–471
Reitner J, Wörheide G, Thiel V, Gautret P (1996) Reef caves and cryptic habitats of Indo-Pacific reefs—distribution patterns of coralline sponges and microbialites. Gott Arb Geol Palaont 2:91–100
Reksodihardjo-Lilley G, Lilley R (2007) Towards a sustainable marine aquarium trade: an Indonesian perspective. SPC Live Reef Fish Inform Bull 17:11–19
Roff G, Doropoulos C, Rogers A, Bozec YM, Krueck NC, Aurellado E, Priest M, Birrell C, Mumby PJ (2016) The ecological role of sharks on coral reefs. Trends Ecol Evol 31(5):395–407
Ruppert KM, Kline RJ, Rahman MS (2019) Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: A systematic review in methods, monitoring, and applications of global eDNA. Global Ecol Conserv 17:e00547
Russ GR, Alcala AC (2010) Decadal-scale rebuilding of predator biomass in Philippine marine reserves. Oecologia 163(4):1103–1106
Saenz-Agudelo P, Jones GP, Thorrold SR, Planes S (2011) Connectivity Dominates Larval Replenishment in a Coastal Reef Fish Metapopulation. Proc Royal Soc B Biol Sci 278(1720):2954–2961
Sembiring A, Pertiwi NPD, Mahardini A, Wulandari R, Kurniasih EM, Kuncoro AW, Cahyani ND, Anggoro AW, Ulfa M, Madduppa H, Carpenter KE (2015) DNA barcoding reveals targeted fisheries for endangered sharks in Indonesia. Fish Res 164:130–134
Setyastuti A, Wirawati I (2008) The banda sea: a hotspot of deep-sea echinoderms diversity in Indonesia. IOP Conf Ser Earth Environ 184:012003
Shaw JL, Clarke LJ, Wedderburn SD, Barnes TC, Weyrich LS, Cooper A (2016) Comparison of environmental DNA metabarcoding and conventional fish survey methods in a river system. Biol Cons 197:131–138
Smart AS, Tingley R, Weeks AR, Van Rooyen AR, McCarthy MA (2015) Environmental DNA sampling is more sensitive than a traditional survey technique for detecting an aquatic invader. Ecol Appl 25(7):1944–1952
Smith AD, Garcia SM (2014) Fishery management: contrasts in the Mediterranean and the Atlantic. Curr Biol 24(17):R810–R812
Spalding M (2010) World atlas of mangroves. Routledge
Spalding M, Spalding MD, Ravilious C, Green EP (2001) World atlas of coral reefs. University of California Press
Stanley SM (2007) An analysis of the history of marine animal diversity. Paleobiology 33:1–55
Stat M, Huggett MJ, Bernasconi R, DiBattista JD, Berry TE, Newman SJ, Harvey ES, Bunce M (2017) Ecosystem biomonitoring with eDNA: metabarcoding across the tree of life in a tropical marine environment. Sci Rep 7(1):1–11
Strong JA, Andonegi E, Bizsel KC, Danovaro R, Elliott M, Franco A, Garces E, Little S, Mazik K, Moncheva S, Papadopoulou N (2015) Marine biodiversity and ecosystem function relationships: the potential for practical monitoring applications. Estuar Coast Shelf Sci 161:46–64
Subhan B, Arafat D, Rahmawati F, Dasmasela YH, Royhan QM, Madduppa H, Santoso P, Prabowo B (2020) Coral disease at Mansuar Island, Raja Ampat, Indonesia. IOP Conf Series Earth Environ Sci 429(1):012027
Supono, Lane DJ, Susetiono (2014) Echinoderm fauna of the Lembeh Strait, North Sulawesi: inventory and distribution review. Marine Res Indones 39(2):51–611
Taberlet P, Coissac E, Pompanon F, Brochmann C, Willerslev E (2012) Towards next-generation biodiversity assessment using DNA metabarcoding. Mol Ecol 21(8):2045–2050
Takeuchi A, Iijima T, Kakuzen W, Watanabe S, Yamada Y, Okamura A, Horie N, Mikawa N, Miller MJ, Kojima T, Tsukamoto K (2019) Release of eDNA by different life history stages and during spawning activities of laboratory-reared Japanese eels for interpretation of oceanic survey data. Sci Rep 9(1):1–9
Thomsen PF, Willerslev E (2015) Environmental DNA–An emerging tool in conservation for monitoring past and present biodiversity. Biol Cons 183:4–18
Timm J, Kochzius M, Madduppa HH, Neuhaus AI, Dohna T (2017) Small scale genetic population structure of coral reef organisms in Spermonde Archipelago Indonesia. Front Marine Sci 4:294
Tittensor DP, Mora C, Jetz W, Lotze HK, Ricard D, Berghe EV, Worm B (2010) Global patterns and predictors of marine biodiversity across taxa. Nature 466(7310):1098–1101
Tornabene L, Valdez S, Erdmann MV, Pezold FL (2016) Multi-locus sequence data reveal a new species of coral reef goby (Teleostei: Gobiidae: Eviota), and evidence of Pliocene vicariance across the Coral Triangle. J Fish Biol 88(5):1811–1834
Treml EA, Roberts J, Halpin PN, Possingham HP, Riginos C (2015) The emergent geography of biophysical dispersal barriers across the Indo-West Pacific. Divers Distrib 21(4):465–476
Treml EA, Roberts JJ, Chao Y, Halpin PN, Possingham HP, Riginos C (2012) Reproductive output and duration of the pelagic larval stage determine seascape-wide connectivity of marine populations. Integr Comp Biol 52(4):525–537
Turak E, Souhoka J (2003) Coral diversity and the status of coral reefs in the Raja Ampat Islands. In: Report on a rapid ecological assessment of the Raja Ampat Islands, Papua, Pastern Indonesia held, vol 30. pp 59–83
Utami RT, Zamani NP, Madduppa HH (2018) Molecular identification, abundance and distribution of the coral-killing sponge Terpios hoshinota in Bengkulu and Seribu Islands Indonesia. Biodiver J Biol Divers 19(6):2238–2246
Valdez-Moreno M, Ivanova NV, Elías-Gutiérrez M, Pedersen SL, Bessonov K, Hebert PDN (2019) Using eDNA to biomonitor the fish community in a tropical oligotrophic lake. PlosOne. https://doi.org/10.1371/journal.pone.0215505
Venter O, Fuller RA, Segan DB, Carwardine J, Brooks T, Butchart SH, Di Marco M, Iwamura T, Joseph L, O’Grady D, Possingham HP (2014) Targeting global protected area expansion for imperilled biodiversity. PLoS Biol 12(6):e1001891
Veron JE, Devantier LM, Turak E, Green AL, Kininmonth S, Stafford-Smith M, Peterson N (2009) Delineating the coral triangle. Galaxea J Coral Reef Stud 11(2):91–100
Vicente J, Zea S, Hill RT (2016) Sponge epizoism in the Caribbean and the discovery of new Plakortis and Haliclona species, and polymorphism of Xestospongia deweerdtae (Porifera). Zootaxa 4178(2):209–233
Wells S (1997) Giant clams: status, trade and mariculture, and the role of CITES in management. IUCN/SSC Wildlife Trade Programme, Gland, Switzerland and Cambridge, UK
West KM, Stat M, Harvey ES, Skepper CL, DiBattista JD, Richards ZT, Travers MJ, Newman SJ, Bunce M (2020) eDNA metabarcoding survey reveals fine-scale coral reef community variation across a remote, tropical island ecosystem. Mol Ecol 29(6):1069–1086
White WT, Last PR, Stevens JD, Yearsley GK, Fahmi D (2006) Economically important sharks and rays of Indonesia. Lamb Print, Perth
Wiadnya DG, Syafaat R, Susilo E, Setyohadi D, Arifin Z, Wiryawan B (2011) Recent development of marine protected areas (MPAs) in Indonesia: policies and governance. J Appl Environ Biol Sci 1(12):608–613
Wickham H (2009) Elegant graphics for data analysis (ggplot2). Springer
Williams SE, Williams YM, VanDerWal J, Isaac JL, Shoo LP, Johnson CN (2009) Ecological specialization and population size in a biodiversity hotspot: how rare species avoid extinction. Proc Natl Acad Sci 106(Supplement 2):19737–19741
Williamson DH, Harrison HB, Almany GR, Berumen ML, Bode M, Bonin MC, Choukroun S, Doherty PJ, Frisch AJ, Saenz-Agudelo P, Jones GP (2016) Large-scale, multidirectional larval connectivity among coral reef fish populations in the Great Barrier Reef Marine Park. Mol Ecol 25(24):6039–6054
Yusuf S, Moore AM (2020) Hunting in the seas: population status and community perspectives on giant clams (Tridacnidae) and Napoleon wrasse (Cheilinus undulatus), endangered marine taxa of the Wallacea Region, Indonesia. IOP Conf Series Earth Environ Sci 473:012061. https://doi.org/10.1088/1755-1315/473/1/012061
Acknowledgements
The authors thank the Ministry of Research and Technology (Ristek/BRIN) for supporting the research entitled “Population genomics to estimate distribution pattern and larval distances of economically fish using environmental DNA to support food security and fisheries conservation”; the Institute of Research and Community Service (LPPM) IPB University for undertaking the research administration; the Laboratory of Marine Biodiversity and Biosystematics IPB University, Scientific Diving Laboratory IPB University, and Oceanogen Laboklinikum for the use of their research logistics and facilities.
Funding
This study was funded by the Ministry of Research and Technology (Ristek/BRIN) through a grant to Hawis Madduppa under Agreement No. 3988/IT3.L1/PN/2020.
Author information
Authors and Affiliations
Contributions
HM conceived the research idea; HM, BS and DGB designed the methodology; HM, DA, EJ, LMIS, NA and BS collected field data; HM and LMIS performed laboratorium analysis; HM, NKDC, AWA performed all statistical analyses; HM led the writing of the manuscript. DGB helped with statistical advice, edited and improved the manuscript together with NKDC, AWA, and LMIS. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures followed the guidelines of the Ministry of Research and Technology (Ristek/BRIN) under Agreement No. 3988/IT3.L1/PN/2020. The research was approved by the ethical committee at the Institute of Research and Community Service (LPPM) IPB University prior to the research agreement.
Additional information
Communicated by Angus Jackson.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article belongs to the Topical Collection: Coastal and marine biodiversity
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Madduppa, H., Cahyani, N.K.D., Anggoro, A.W. et al. eDNA metabarcoding illuminates species diversity and composition of three phyla (chordata, mollusca and echinodermata) across Indonesian coral reefs. Biodivers Conserv 30, 3087–3114 (2021). https://doi.org/10.1007/s10531-021-02237-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10531-021-02237-0