Abstract
Forest canopy density (FCD) is one of the important parameters for forest mapping and monitoring. Terrestrial Laser Scanning (TLS) is one of the most accurate tools used in field data collection. Hence, the present study aimed to predict forest aboveground biomass (AGB) by integrating TLS data, satellite data-derived FCD and spectral indices. FCD Mapper was used to classify Landsat-8 OLI data into FCD classes, which were validated using field-measured FCD. In the field, point cloud data were collected from each FCD class using TLS. The diameter at breast height (dbh) and height of individual trees were retrieved from the point cloud data and validated with field-measured dbh and height. AGB was estimated from the TLS-derived dbh and height and modelled as a function of Landsat-8 OLI-derived FCD classes and spectral indices. A total of 11 FCD classes were generated, which showed a strong positive correlation (r = 0.96) with the field-measured FCD. From the TLS point cloud data, 96% of individual trees were extracted. Positive correlations were found between TLS-measured dbh and field-measured dbh (r = 0.99), and TLS-measured height and field-measured height (r = 0.96). A linear function best fitted between TLS-estimated AGB and FCD classes was established. Because of the low variability in AGB due to absolute FCD classes, the model was further extended using a few spectral indices. Using a multiple linear model, the average AGB (374 Mg ha−1) and total AGB (3,024,550 Mg) of the study area were predicted. The study highlighted that the combined application of TLS and satellite data-derived FCD and spectral indices can be one of the fastest and accurate methods in forest AGB prediction.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Forest cover has been changing rapidly around the globe, increasing through plantation and conservation, and decreasing through deforestation and forest degradation. Different methods have been devised to measure and monitor these changes and quantify the contribution of forests to climate change mitigation through biomass production and carbon sequestration. Forest biomass has been estimated and predicted using various models and equations at the individual tree-to-stand levels (Bhandari and Chherti, 2020; Bhandari & Neupane, 2014; Chave et al., 2014; Sharma et al., 2017; Shrestha et al., 2018; Xing et al., 2019; Virgulino-Júnior et al., 2020). Satellite remote sensing (RS) data have also been used to assess forest biomass at various spatial scales (Dang et al., 2019; Kalita et al., 2022; Kushwaha et al., 2014; López-Serrano et al., 2019; Nandy et al., 2019; 2021; Nandy & Kushwaha, 2021; Nuthammachot et al., 2020; Sousa et al., 2015; Yadav & Nandy, 2015).
Forest canopy density (FCD) is an important parameter for forest mapping (FSI, 2021) as well as forest degradation status assessment (Mon et al., 2012; Nandy et al., 2011). It can also be reliably used for the prediction of forest biomass at local, regional, and global levels. FCD can be estimated using instruments like spherical densiometer, angle count method, qualitative ocular method, photographic recording of the canopy density, aerial photograph interpretation, and biophysical spectral response modelling using FCD Mapper (Abdollahnejad et al., 2017; Chandrashekhar et al., 2005; Goodenough & Goodenough, 2012; Keane et al., 2005; Korhonen et al., 2006; Nandy et al., 2003). Biophysical spectral response modelling is one of the most appropriate and rapid methods for canopy density mapping using satellite data (Chandrashekhar et al., 2005; Nandy et al., 2003). Unlike conventional methods, biophysical spectral response modelling indicates growth and changes in forest conditions over time including degradation (Rikimaru et al., 2002). To correlate the FCD with biomass and carbon, a precise forest inventory is required at the ground level. The diameter at breast height (dbh) and the height of individual trees are the most common variables measured in forest inventories. These variables also serve as input in growth and yield models (Burkhart et al., 1972; Curtis et al., 1981; Wykoff et al., 1982). The measurements made manually in the field are subjective and can be prone to errors.
Terrestrial Laser Scanner (TLS), also known as ground-based LiDAR, is an active remote sensing technique based on a ranging sensor that is mounted on a ground-based platform (Kelbe, 2015). It has gained high popularity across numerous application domains (Tansey et al., 2009). It can provide forest inventory parameters like dbh and height with high accuracy which is generally used for tree volume, biomass, and carbon estimation (Disney et al., 2019; Liang et al., 2012; Wassihun et al., 2019). The dbh can be estimated by fitting a circle, cylinder, or free-form curve to a scattering point cloud at breast height (Kalwar, 2015; Tesfai, 2015). The positional accuracy of TLS is within 0.5–10 cm which is more than the accuracy of airborne LiDAR, 0.1–1 m (Yang et al., 2013). The TLS has the potential to improve forest inventories by providing faster and more detailed information on the forest structure than the time-consuming manual techniques (Dassot et al., 2011). TLS can be a better option for collecting ground inventory data with considerable accuracy (Wang et al., 2014). Hence, the present study aims to develop a model that can predict aboveground biomass (AGB) using TLS-derived inventory data, and Landsat 8 OLI-derived FCD and spectral indices.
Materials and Methods
Study Area
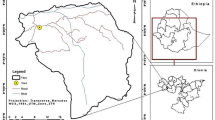
The study was conducted in Barkot Reserve Forest (30° 03′ 52″–30° 10′ 43″ N and 78° 09′ 49″–78° 17′ 09″ E) (Fig. 1) of Uttarakhand, India. The study area has a monsoon-influenced subtropical humid climate. The temperature ranges from 1.5 to 40.5 °C and the average annual rainfall is 1250 mm, the area lies in the moist deciduous plant functional type in the northwest Himalayan foothills (Srinet et al., 2020). The forest type present in the study area is Northern Tropical Moist Deciduous Forest (Champion & Seth, 1968). Shorea robusta is the dominant tree species in the overwood, forming nearly pure stands (Nandy et al., 2017). Its common associates are Lagerstroemia parviflora, Terminalia belerica, Syzygium cumini, Adina cordifolia, and Tectona grandis (plantation). The common underwood species are Mallotus philippensis, Ougeinia oojeinensis, Ehretia laevis, and Cassia fistula.
Methodology
The methodology used in this study involves image analysis, ground truth data collection and the collection of TLS point cloud data. The detailed methodology of the study is given in Fig. 2.
Forest Canopy Density Classification
Landsat-8 OLI satellite imagery of 12 February 2015 was used to generate FCD classes using FCD Mapper software (Rikimaru et al., 2002). FCD Mapper is a semi-expert system that combines four indices, viz., Advanced Vegetation Index, Bare Soil Index, Shadow Index, and Thermal Index, to classify forest canopy density into 10 classes with 10% intervals (Nandy et al., 2003). From each FCD class, a minimum of two sample points were selected to collect the ground truth data. To ensure the collection of ground truth data from the same FCD classes, 3 × 3 cells were selected wherever possible. At each sample location, FCD was measured at five points (four corners and one at the centre) in each 30 m × 30 m plot using a spherical densiometer. The correlation coefficient was used to validate the FCD classes generated by FCD Mapper with FCD classes measured in the field.
Terrestrial Laser Scanning
From each FCD class (generated by FCD Mapper), sample points were selected purposively to collect the TLS data. At each point, a square plot of size 10 m × 10 m was established. Undergrowth was cleared wherever it obstructed the line of sight of TLS. Each tree of the sample plot was tagged by using white numbered paper (Fig. 3a). The point cloud LiDAR data acquisition was carried out from four positions in each plot i.e. one from the centre of the plot and three from outside the boundary of the plot using a Riegl VZ-400 TLS (RIEGL Laser Measurement Systems GmbH). For central scanning, 0.03 mrad angular resolution at 30–130° vertical angle and 0–360° horizontal angle was used. For outer scanning, the angular resolution and vertical angle were similar to central scanning but the horizontal angle was reduced to 0–120°. Three circular reflectors were fixed inside the plot ensuring that these reflectors are visible from three outer scan positions and one central scan position of TLS. The three outer scan positions were fixed at 10–15 m away from the boundary of the plot to capture the point cloud of the whole tree. A Nikon camera was mounted at the top of the Riegl VZ-400 TLS. This camera was set in such a way that it automatically took images after the completion of each scan (13 images in the central scan and 4 images in the outer scan). These images were used to colourize the point cloud data for better visualization during processing. Circumference at breast height (CBH) of each tree within each plot was measured at 1.37 m above the ground level using linear tape and the height of each tree was measured using a laser range finder.
TLS Data Processing
The point cloud data collected from 3 outer scanning positions were registered with the point cloud data of the central scanning position using RiSCAN PRO V1.8.1 software. The error during the registration of point cloud data of 3 outer scanning with central scanning is given as standard deviations in Table 1. An error of up to 2 m in the form of a standard deviation is acceptable (RiSCAN PRO, 2013). The minimum and maximum average standard deviation in multiple scan registration were 0.004 m and 0.054 m, respectively (Table 1). The overall average error of multiple registrations was 0.018 m which is slightly higher than the average error (0.016) reported by Kalwar (2015). The registered point cloud data of each scan position was exported to ASCII format. All exported ASCII format point cloud data were merged to form a single data in CloudCompare V 2.6.3 software.
Plot and Tree Extraction and Measurement
The plot of 10 m × 10 m was extracted from the merged point cloud data using the cross-section and segmentation tool in CloudCompare V 2.6.3. The length and breadth of the plot were measured by the point-picking tool. Plot extraction was facilitated by tagging trees using a white numbered plate. However, the white numbered plate tags were visible only in the colourized (RGB) mode of point cloud data. The extracted plot in the scalar field and RGB is shown in Fig. 3b, c. Similarly, each tree was extracted from each plot by using the cross-section and segmentation tool in CloudCompare V 2.6.3. After the extraction of a tree from the plot, a connected component was used to separate the component of other trees and branches which were intermixed with the tree of interest. Then the tree was exported to Point Cloud Library format.
The dbh and total height of each tree were measured using the 3D Forest software. The different parameters (tree position, tree dbh, tree height/tree length, stem curve, and tree planner projection) can be measured from the extracted trees (Trochta et al., 2017). In 3D Forest, two approaches are possible for the measurement of the dbh of the extracted trees. The first is randomized Hough transformation (RHT) and the second one is the least square regression (LSR). The LSR method may include the outlying points and lead to an overestimation of the dbh in comparison to RHT. RHT is considered a more accurate method than LSR (Krůček et al., 2015), and therefore, we used this method (RHT) in this study. The reason behind the difference in the measurement between LSR and RHT methods is that the subset of points from which dbh is calculated includes overhanging branches or points which do not belong to the point cloud of a particular tree. This error can be minimized by using the tree cloud edit function or the dbh cloud edit function. The reliability of dbh and height measured from point cloud data was validated with the dbh measured by tape and height measured by laser range finder during field inventory.
Aboveground Biomass Estimation
The validated dbh was used to calculate the volume of each tree by using the site and species-specific volumetric equations (FSI, 1996). Dbh was used as a predictor variable, and volume was used as the dependent variable to calculate the volume of individual trees. By multiplying the volume of the individual trees with the species-specific wood density (FRI, 2002) and biomass expansion factor (Haripriya, 2000), the AGB of the individual trees was calculated. The AGB of all trees inside the plot was summed to get the plot-level AGB. The plot-level biomass was further converted to per ha AGB.
Aboveground Biomass Prediction Model
Different AGB prediction models (linear, logarithmic, quadratic, power, sigmoid, and exponential) were fitted to the data (Table 2). These models were evaluated based on the values of coefficient of determination (R2), root mean squared error (RMSE) (Montgomery et al., 2021), and Akaike Information Criterion (AIC) (Akaike, 1972), and the best model was selected. The parameters and fit statistics for each model were estimated in R (R Core Team, 2017). The variation in AGB as a function of FCD classes was very low because of the absolute FCD classes. Therefore, different spectral indices such as Enhanced Vegetation Index (EVI) (Huete et al., 2002), Moisture Stress Index (MSI) (Hunt & Rock, 1989), and Normalized Difference Moisture Index (NDMI) (Gao, 1996) were also used to predict AGB. A model representing AGB as a function of the EVI, MSI, FCD, and NDMI was developed for predicting the spatial distribution of AGB in the study area.
Results
FCD Classes
A total of 11 FCD classes were generated (Fig. 4). The total area classified by FCD Mapper was 8429.3 ha. Of this, 22.22% (21% was covered by non-canopy class and 1.22% area had less than 10% canopy cover) was non-forest area and the remaining 77.78% was forest area. The distribution of the different canopy density classes is presented in Fig. 5a. Among the FCD classes, the maximum area was under class 8 (70–80%) and the minimum area was under class 1 (1–10%). Density classes generated from FCD Mapper and field measurement showed a strong positive correlation (r = 0.96) (Fig. 5b).
Tree Extraction and Measurement
We extracted 96% of individual trees from the LiDAR point cloud. The numbering of each tree was found useful in identification and extraction. The top view was found useful in plotting and individual tree extraction, whereas the front view, back view, and side views in CloudCompare V 2.6.3 were found useful in tree identification. The extracted tree is shown in Figs. 3d, e, and f. The correlation between TLS-measured dbh and field-measured dbh was 0.99, whereas the correlation between TLS-measured height and field-measured height was 0.96. The TLS-measured dbh and height were strongly and positively correlated with field-measured dbh and height (Fig. 5c, d).
Model Development
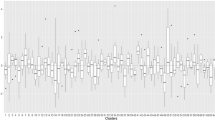
All the parameters of all the models were significant at a 95% confidence interval except the quadratic form of the model. Hence, the quadratic form of the model was excluded from further analysis because of insignificant parameters (p > 0.05). The logarithmic form of the model described 81% of the variation in total AGB with the highest RMSE and AIC than other models (Table 2). The power and exponential form of the model overestimated the AGB for lower FCD classes (Fig. 6). Therefore, the logarithmic, power, and exponential form of models were also excluded from further analysis. Out of the remaining two models, the linear model had the highest value of R2 (0.88) and the lowest value of RMSE (90.60 Mg ha−1) and AIC (145.28) (Table 2). Considering fit statistics, we selected the linear form of the model as the best AGB prediction model although the difference between the linear and sigmoidal forms of the model was very small.
Because of low variability in AGB due to the absolute FCD classes, the model was further extended using a few spectral indices (Enhanced Vegetation Index (EVI), Moisture Stress Index (MSI), and Normalized Difference Moisture Index (NDMI)) generated from Landsat-8 OLI satellite data. The multiple linear regression model developed by using additional indices described more than 90% (R2 = 0.91) variation in AGB (Eq. 1).
The AGB map (Fig. 7) of the study area was generated from the developed multiple linear regression model. The average AGB of the study area was found to be 374 Mg ha−1 and the total AGB was 30,24,550 Mg.
Discussion
FCD Classes
FCD classification using satellite images is one of the fastest and most cost-effective methods of forest canopy density classification. FCD classification generated 11 canopy density classes, where 10 classes were the forest canopy classes and one class was a non-canopy class. The 10 forest canopy classes ranged from 1 to 100 at an interval of 10 per cent (Fig. 4). However, the area with less than 10 per cent canopy cover was not considered as forest (FSI, 2021). Hence, the two classes were combined (FCD class 0 and 1) as non-forest classes. The number of forest canopy density classes generated by this method is more than those generated by other methods (Nandy et al., 2003). This study showed a correlation coefficient (measured between canopy density classes of FCD mapper and field inventory) of 0.96 (Fig. 5b) which is higher than other similar studies (Chandrashekhar et al., 2005; Nandy et al., 2003). The correlation coefficients between canopy density classes of FCD mapper and field inventory were 0.95 (Chandrashekhar et al., 2005) and 0.92 (Nandy et al., 2003). FCD Mapper can stratify forest canopy density into 10 classes and can detect small changes in the forest canopy and hence, can be used as a superior method for forest canopy density classification (Nandy et al., 2003).
Tree Extraction and Measurement
In this study, 96% of individual trees were extracted from the TLS data which is higher than in several other studies. Thies et al. (2004), Tesfai (2015), Kalwar (2015), and Othmani et al. (2011) extracted 52%, 87%, 89%, and 90.6% of the trees, respectively. Four reasons may explain a higher percentage of individual tree extraction in the present study. The first reason: in this study, only trees with dbh > 5 cm were considered. Larger trees are easier to detect and extract from the TLS point cloud data. The second reason: each tree of the plot was numbered and tagged. The numbering and tagging of the individual trees significantly improved the identification after extraction. The third reason: four scans were carried out for each plot. This increased the probability of coverage of the individual trees in scanning. The fourth reason: the forest had relatively larger-sized trees and the stand density was low. The low stand density provided higher visibility to the laser scanner. In a plantation with a stand density of 16,552 stems ha−1, tree detection was only 61% (Seidel et al., 2013). Another reason could be the smaller size of the plot (10 m × 10 m). However, Seidel et al. (2013) detected only 61% of individual trees from 2 m × 2 m plots. In contrast, Bienert et al. (2007) detected 97.4% of trees. Hence, the higher tree detection rate, in this case, was due to the tree tagging, large size trees, a large number of scans (4 scans), and low density per unit area. The tree detection rate can be increased by increasing the number of scanning but it also depends on the degree of density of stand growth, slope, and extent of undergrowth (Tesfai, 2015).
The accuracy of tree detection, extraction, and measurement are also displayed by the higher degree of correlation between TLS-measured dbh and field-measured dbh (r = 0.99) (Fig. 5c). In a study using TLS and field inventory in Dehradun, India, Haldar (2016) found the same degree of correlation (r = 0.99) between TLS-measured dbh and field-measured dbh. In a similar study, Sium (2015) found a correlation of r = 0.98 between TLS-measured dbh and field-measured dbh. Similarly, Kalwar (2015) found a correlation of r = 0.97 between TLS-measured dbh and field-measured dbh while Kelbe (2015) found a correlation of r = 0.89. This study found a high degree of correlation between TLS-measured height and field-measured height (r = 0.96) (Fig. 5d) which is higher than the correlation found by Sium (2015) (r = 0.86) and Kalwar (2015) (r = 0.87), but, lower than the correlation (r = 0.99) found by Haldar (2016).
Model Development and Selection of the Best Model
The biomass of individual trees generally follows the power function with individual tree variables (Bhandari & Chhetri, 2020; Kebede & Soromessa, 2018; Pastor et al., 1984; Sharma et al., 2017; Shrestha et al., 2018). However, stand-level AGB followed a linear function with FCD classes in this study. Higher FCD classes either have a higher stand density or bigger-sized trees than the lower FCD classes. High stand densities and big-sized trees always contribute to forming a linear relationship between stand-level AGB and FCD classes.
Multiple Linear Regression Model and Biomass Map
FCD classes ranged from 1 to 10 in absolute values. This raised an issue of low variability in AGB if only FCD classes were used as predictor variables. This limitation of the model was addressed by including spectral indices in the model which resulted in a multiple linear AGB prediction model (R2 = 0.88, RMSE = 72 Mg ha−1). The developed model predicted an average AGB of 374 Mg ha−1 and a total of 30,24,550 Mg AGB of the study area (Fig. 7) which is slightly lower than the observed AGB in the field (458 Mg ha−1).
TLS has been used for biomass prediction in a variety of forest conditions and species. In a study, Takoudjou et al. (2017) used TLS data to predict large tropical tree biomass and calibrate allometric models. In a similar study, Calders et al. (2015) compared TLS-derived biomass with destructively estimated biomass and found a high degree of correlation. Estimation of individual tree biomass is possible with high automation and accuracy by reconstructing the stem from TLS point cloud data (Yu et al., 2013). Data retrieved from TLS improved the predictive capacity of the biomass model, especially for branch biomass (Kankare et al., 2013). They also suggested the use of stem curves and crown size geometry from TLS for allometric biomass models rather than statistical 3D point metrics. Branch biomass was predicted using the TLS-measured data with higher accuracy than conventional allometric models (Hauglin et al., 2013). All these studies developed a model to predict the biomass of individual trees using TLS. These models still require the measurement of individual tree parameters (dbh, height, or crown) to predict the individual tree biomass. These models have limited scope to be applied in a larger area as this will increase the cost and time. The AGB prediction model developed in this study used FCD classes and spectral indices which can easily be derived from satellite imagery. The advantage of using such models is that they provide a cost-effective and less time-consuming method of biomass prediction at local, regional, and global levels.
Conclusion
TLS is one of the most accurate ground-based RS tools for forest inventory data collection. On the other hand, FCD Mapper is one of the fastest tools for the classification of forest canopy density. It can classify the FCD into a larger number of classes with a higher level of accuracy than other RS-based methods. The combined application of TLS and FCD Mapper along with satellite data-derived spectral indices may be one of the effective methods for forest AGB estimation. The results of this study were based on a case study from a particular forest, and therefore the application of the model developed in this study should be restricted to similar forest areas. Further studies in the estimation of biomass using FCD classes and TLS data from a larger number of TLS plots and a larger geographical region are recommended to make the model applicable to a larger geographical area.
References
Abdollahnejad, A., Panagiotidis, D., & Surový, P. (2017). Forest canopy density assessment using different approaches—Review. Journal of Forest Science, 63(3), 107–116. https://doi.org/10.17221/110/2016-JFS
Akaike, H. (1974). A new look at the statistical model identification. IEEE Transactions on Automatic Control, 19(6), 716–723. https://doi.org/10.1109/TAC.1974.1100705
Bhandari, S. K., & Chhetri, B. B. K. (2020). Individual-based modelling for predicting height and biomass of juveniles of Shorea robusta. Austrian Journal of Forest Science, 137(2), 133–160.
Bhandari, S. K., & Neupane, H. (2014). Allometric equations for estimating the above-ground biomass of Castanopsis indica at juvenile stage. Banko Janakari, 24(1), 14–22.
Bienert, A., Scheller, S., Keane, E., Mohan, F., & Nugent, C. (2007). Tree detection and diameter estimations by analysis of forest terrestrial laserscanner point clouds. In ISPRS workshop on laser scanning (Vol. 36, pp. 50–55). IAPRS.
Burkhart, H. E., Parker, R. C., Strub, M. R., & Oderwald, R. G. (1992). Yield of oldfield loblolly pine plantations. School of Forestry and Wildlife Resources, Virginia Polytechnic Institute and State University, Blacksburg. Publ. FWS-3-72.
Calders, K., Newnham, G., Burt, A., Murphy, S., Raumonen, P., Herold, M., Culvenor, D., Avitabile, V., Disney, M., Armston, J., & Kaasalainen, M. (2015). Nondestructive estimates of above-ground biomass using terrestrial laser scanning. Methods in Ecology and Evolution, 6(2), 198–208. https://doi.org/10.1111/2041-210X.12301
Champion, H. G., & Seth, S. K. (1968). A revised survey of the forest types of India. The Manager of Publications, Delhi.
Chandrashekhar, M. B., Saran, S., Raju, P. L. N., & Roy, P. S. (2005). Forest canopy density stratification: How relevant is biophysical spectral response modelling approach? Geocarto International, 20(1), 15–21. https://doi.org/10.1080/10106040508542332
Chave, J., Réjou-Méchain, M., Búrquez, A., Chidumayo, E., Colgan, M. S., Delitti, W. B. C., Duque, A., Eid, T., Fearnside, P. M., Goodman, R. C., Henry, M., Martínez-Yrízar, A., Mugasha, W. A., Muller-Landau, H. C., Mencuccini, M., Nelson, B. W., Ngomanda, A., Nogueira, E. M., Ortiz-Malavassi, E., … Vieilledent, G. (2014). Improved allometric models to estimate the aboveground biomass of tropical trees. Global Change Biology, 20(10), 3177–3190. https://doi.org/10.1111/gcb.12629
Curtis, R. O., Clendenen, G. W., & De Mars, D. J. (1981). A new stand simulator for coast Douglas-fir: DFSIM user’s guide. USDA forest service. Pacific Northwest Forest Experiment Station, General Technic Report. PNW-135.
Dang, A. T. N., Nandy, S., Srinet, R., Luong, N. V., Ghosh, S., & Kumar, A. S. (2019). Forest aboveground biomass estimation using machine learning regression algorithm in Yok Don National Park, Vietnam. Ecological Informatics, 50, 24–32. https://doi.org/10.1016/j.ecoinf.2018.12.010
Dassot, M., Constant, T., & Fournier, M. (2011). The use of terrestrial LiDAR technology in forest science: Application fields, benefits and challenges. Annals of Forest Science, 68(5), 959–974. https://doi.org/10.1007/s13595-011-0102-2
Disney, M., Burt, A., Calders, K., Schaaf, C., & Stovall, A. (2019). Innovations in ground and airborne technologies as reference and for training and validation: Terrestrial laser scanning (TLS). Surveys in Geophysics, 40(4), 937–958. https://doi.org/10.1007/s10712-019-09527-x
FRI. (2002). Indian woods: Their identification, properties and uses (Vol. I–VI, Revised Ed.). Forest Research Institute, Indian Council of Forestry Research and Education, Ministry of Environment and Forests, Government of India.
FSI. (1996). Volume equations for forests of India, Nepal and Bhutan. Forest Survey of India, Ministry of Environment and Forests, Government of India, Dehradun.
FSI. (2021). India State of Forest report 2021. Forest Survey of India, Ministry of Environment, Forest and Climate Change, Government of India, Dehradun.
Gao, B. C. (1996). NDWI—A normalized difference water index for remote sensing of vegetation liquid water from space. Remote Sensing of Environment, 58(3), 257–266. https://doi.org/10.1016/S0034-4257(96)00067-3
Goodenough, A. E., & Goodenough, A. S. (2012). Development of a rapid and precise method of digital image analysis to quantify canopy density and structural complexity. International Scholarly Research Notices, 2012, 1–11. https://doi.org/10.5402/2012/619842
Haldar, A.K. (2016). Terrestrial laser scanning based modelling for forest structural parameter retrieval. Master’s thesis, University of Twente.
Haripriya, G. S. (2000). Estimates of biomass in Indian forests. Biomass and Bioenergy, 19(4), 245–258. https://doi.org/10.1016/S0961-9534(00)00040-4
Hauglin, M., Astrup, R., Gobakken, T., & Næsset, E. (2013). Estimating single-tree branch biomass of Norway spruce with terrestrial laser scanning using voxel-based and crown dimension features. Scandinavian Journal of Forest Research, 28(5), 456–469. https://doi.org/10.1080/02827581.2013.777772
Huete, A., Didan, K., Miura, T., Rodriguez, E. P., Gao, X., & Ferreira, L. G. (2002). Overview of the radiometric and biophysical performance of the MODIS vegetation indices. Remote Sensing of Environment, 83(1–2), 195–213. https://doi.org/10.1016/S0034-4257(02)00096-2
Hunt, E. R., Jr., & Rock, B. N. (1989). Detection of changes in leaf water content using near-and middle-infrared reflectances. Remote Sensing of Environment, 30(1), 43–54. https://doi.org/10.1016/0034-4257(89)90046-1
Kalita, R. M., Nandy, S., Srinet, R., Nath, A. J., & Das, A. K. (2022). Mapping the spatial distribution of aboveground biomass of tea agroforestry systems using random forest algorithm in Barak valley, Northeast India. Agroforestry Systems, 96, 1175–1188. https://doi.org/10.1007/s10457-022-00776-1
Kalwar, O. P. P. (2015). Derivation of forest plot inventory parameters from terrestrial LiDAR data for carbon estimation. Master’s thesis, University of Twente.
Kankare, V., Holopainen, M., Vastaranta, M., Puttonen, E., Yu, X., Hyyppä, J., Vaaja, M., Hyyppä, H., & Alho, P. (2013). Individual tree biomass estimation using terrestrial laser scanning. ISPRS Journal of Photogrammetry and Remote Sensing, 75, 64–75. https://doi.org/10.1016/j.isprsjprs.2012.10.003
Keane, R. E., Reinhardt, E. D., Scott, J., Gray, K., & Reardon, J. (2005). Estimating forest canopy bulk density using six indirect methods. Canadian Journal of Forest Research, 35(3), 724–739. https://doi.org/10.1139/x04-213
Kebede, B., & Soromessa, T. (2018). Allometric equations for aboveground biomass estimation of Olea europaea L. subsp. cuspidata in Mana Angetu forest. Ecosystem Health and Sustainability, 4(1), 1–12. https://doi.org/10.1080/20964129.2018.1433951
Kelbe, D. (2015). Forest structure from terrestrial laser scanning–in support of remote sensing calibration/ validation and operational inventory. Ph.D. Thesis, Rochester Institute of Technology.
Korhonen, L., Korhonen, K. T., Rautiainen, M., & Stenberg, P. (2006). Estimation of forest canopy cover: a comparison of field measurement techniques. Silva Fennica, 40(4), 577–588. https://doi.org/10.14214/sf.315
Krůček, M., Trochta, J., & Kral, K. (2015). 3D forest user guide: Tool for processing of point clouds acquired by terrestrial laser scanning in forest. Silva Tarouca Research Institute.
Kushwaha, S. P. S., Nandy, S., & Gupta, M. (2014). Growing stock and woody biomass assessment in Asola-Bhatti Wildlife Sanctuary, Delhi, India. Environmental Monitoring and Assessment, 186(9), 5911–5920. https://doi.org/10.1007/s10661-014-3828-0
Liang, X., Litkey, P., Hyyppa, J., Kaartinen, H., Vastaranta, M., & Holopainen, M. (2012). Automatic stem mapping using single-scan terrestrial laser scanning. IEEE Transactions on Geoscience and Remote Sensing, 50(2), 661–670. https://doi.org/10.1109/TGRS.2011.2161613
López-Serrano, P. M., Cárdenas Domínguez, J. L., Corral-Rivas, J. J., Jiménez, E., López-Sánchez, C. A., & Vega-Nieva, D. J. (2019). Modeling of aboveground biomass with Landsat 8 OLI and machine learning in temperate forests. Forests, 11(1), 11. https://doi.org/10.3390/f11010011
Mon, M. S., Mizoue, N., Htun, N. Z., Kajisa, T., & Yoshida, S. (2012). Estimating forest canopy density of tropical mixed deciduous vegetation using Landsat data: A comparison of three classification approaches. International Journal of Remote Sensing, 33(4), 1042–1057. https://doi.org/10.1080/01431161.2010.549851
Montgomery, D. C., Peck, E. A., & Vining, G. G. (2021). Introduction to linear regression analysis. Wiley.
Nandy, S., Ghosh, S., Kushwaha, S. P. S., & Kumar, A. S. (2019). Remote sensing-based forest biomass assessment in northwest Himalayan landscape. In R. R. Navalgund, A. Senthil Kumar, & S. Nandy (Eds.), Remote sensing of Northwest Himalayan ecosystems (pp. 285–311). Springer. https://doi.org/10.1007/978-981-13-2128-3_13
Nandy, S., Joshi, P. K., & Das, K. K. (2003). Forest canopy density stratification using biophysical modeling. Journal of the Indian Society of Remote Sensing, 31(4), 291–297. https://doi.org/10.1007/BF03007349
Nandy, S., & Kushwaha, S. P. (2021). Forest biomass assessment integrating field inventory and optical remote sensing data: A systematic review. International Journal of Plant and Environment, 7(3), 181–186. https://doi.org/10.18811/ijpen.v7i03.1
Nandy, S., Kushwaha, S. P. S., & Dadhwal, V. K. (2011). Forest degradation assessment in the upper catchment of the river Tons using remote sensing and GIS. Ecological Indicators, 11(2), 509–513. https://doi.org/10.1016/j.ecolind.2010.07.006
Nandy, S., Singh, R., Ghosh, S., Watham, T., Kushwaha, S. P. S., Kumar, A. S., & Dadhwal, V. K. (2017). Neural network-based modelling for forest biomass assessment. Carbon Management, 8(4), 305–317. https://doi.org/10.1080/17583004.2017.1357402
Nandy, S., Srinet, R., & Padalia, H. (2021). Mapping forest height and aboveground biomass by integrating ICESat-2, Sentinel-1 and Sentinel-2 data using Random Forest algorithm in northwest Himalayan foothills of India. Geophysical Research Letters, 48(14), e2021GL093799. https://doi.org/10.1029/2021GL093799
Nuthammachot, N., Askar, A., Stratoulias, D., & Wicaksono, P. (2020). Combined use of sentinel-1 and sentinel-2 data for improving above-ground biomass estimation. Geocarto International, 37(2), 366–376. https://doi.org/10.1080/10106049.2020.1726507
Othmani, A., Piboule, A., Krebs, M., Stolz, C., & Voon, L. L. Y. (2011). Towards automated and operational forest inventories with T-Lidar. In 11th international conference on LiDAR applications for assessing forest ecosystems (SilviLaser 2011), Oct 2011, Hobart, Australia.
Pastor, J., Aber, J. D., & Melillo, J. M. (1984). Biomass prediction using generalized allometric regressions for some northeast tree species. Forest Ecology and Management, 7(4), 265–274. https://doi.org/10.1016/0378-1127(84)90003-3
R Core Team. 2017. R: A language and environment for statistical computing. https://www.r-project.org/
Rikimaru, A., Roy, P. S., & Miyatake, S. (2002). Tropical forest cover density mapping. Tropical Ecology, 43(1), 39–47.
RiSCAN PRO. (2013). Manual of RiSCAN PRO operating and processing software for REIGL 3D laser scanners. Version 1.8.1.
Seidel, D., Albert, K., Ammer, C., Fehrmann, L., & Kleinn, C. (2013). Using terrestrial laser scanning to support biomass estimation in densely stocked young tree plantations. International Journal of Remote Sensing, 34(24), 8699–8709. https://doi.org/10.1080/01431161.2013.848308
Sharma, R. P., Bhandari, S. K., & Ram Bahadur, B. K. (2017). Allometric bark biomass model for Daphne bholua in the Mid-hills of Nepal. Mountain Research and Development, 37(2), 206–215. https://doi.org/10.1659/MRD-JOURNAL-D-16-00052.1
Shrestha, D. B., Sharma, R. P., & Bhandari, S. K. (2018). Individual tree aboveground biomass for Castanopsis indica in the mid-hills of Nepal. Agroforestry Systems, 92(6), 1611–1623. https://doi.org/10.1007/s10457-017-0109-2
Sium, M.T. (2015). Application of very high resolution imagery and terrestrial laser scanning for estimating carbon stock in tropical rain forest of Royal Belum, Malaysia. Master’s thesis, University of Twente, The Netherlands.
Sousa, A. M., Gonçalves, A. C., Mesquita, P., & da Silva, J. R. M. (2015). Biomass estimation with high resolution satellite images: A case study of Quercus rotundifolia. ISPRS Journal of Photogrammetry and Remote Sensing, 101, 69–79. https://doi.org/10.1016/j.isprsjprs.2014.12.004
Srinet, R., Nandy, S., Padalia, H., Ghosh, S., Watham, T., Patel, N. R., & Chauhan, P. (2020). Mapping plant functional types in Northwest Himalayan foothills of India using random forest algorithm in Google Earth Engine. International Journal of Remote Sensing, 41(18), 7296–7309. https://doi.org/10.1080/01431161.2020.1766147
Takoudjou, S. M., Ploton, P., Sonké, B., Hackenberg, J., Griffon, S., de Coligny, F., Kamdem, N. G., Libalah, M., Mofack, G., Moguédec, G. L., Pélissier, R., & Barbier, N. (2017). Using terrestrial laser scanning data to estimate large tropical trees biomass and calibrate allometric models: A comparison with traditional destructive approach. Methods in Ecology and Evolution, 9(4), 905–916. https://doi.org/10.1111/2041-210X.12933
Tansey, K., Selmes, N., Anstee, A., Tate, N. J., & Denniss, A. (2009). Estimating tree and stand variables in a Corsican Pine woodland from terrestrial laser scanner data. International Journal of Remote Sensing, 30(19), 5195–5209. https://doi.org/10.1080/01431160902882587
Tesfai, A. S. (2015). Upscaling estimation of tropical rain forest biomass/carbon stock using TLS and Landsat-8 ETM+ data: A case study from Royal Belum, Malaysia. Master’s thesis, University of Twente.
Thies, M., & Spiecker, H. (2004). Evaluation and future prospects of terrestrial laser scanning for standardized laser scanning for standardized forest inventories. International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences, 36(8), 192–197.
Trochta, J., Krůček, M., Vrška, T., & Král, K. (2017). 3D Forest: An application for descriptions of three-dimensional forest structures using terrestrial LiDAR. PLoS ONE, 12(5), e0176871. https://doi.org/10.1371/journal.pone.0176871
Virgulino-Júnior, P. C. C., Carneiro, D. N., Nascimento, W. R., Jr., Cougo, M. F., & Fernandes, M. E. B. (2020). Biomass and carbon estimation for scrub mangrove forests and examination of their allometric associated uncertainties. PLoS ONE, 15(3), e0230008. https://doi.org/10.1371/journal.pone.0230008
Wang, W., Zhao, W., Huang, L., Vimarlund, V., & Wang, Z. (2014). Applications of terrestrial laser scanning for tunnels: A review. Journal of Traffic and Transportation Engineering, 1(5), 325–337. https://doi.org/10.1016/S2095-7564(15)30279-8
Wassihun, A. N., Hussin, Y. A., Van Leeuwen, L. M., & Latif, Z. A. (2019). Effect of forest stand density on the estimation of above ground biomass/carbon stock using airborne and terrestrial LIDAR derived tree parameters in tropical rain forest, Malaysia. Environmental Systems Research, 8, 27. https://doi.org/10.1186/s40068-019-0155-z
Wykoff, W. R., Crookston, N. L., & Stage, A. R. (1982). User’s guide to the stand prognosis model, USDA forest service - general technical report INT-133.
Xing, D., Bergeron, J. C., Solarik, K. A., Tomm, B., Macdonald, S. E., Spence, J. R., & He, F. (2019). Challenges in estimating forest biomass: Use of allometric equations for three boreal tree species. Canadian Journal of Forest Research, 49(12), 1613–1622. https://doi.org/10.1139/cjfr-2019-0258
Yadav, B. K. V., & Nandy, S. (2015). Mapping aboveground woody biomass using forest inventory, remote sensing and geostatistical techniques. Environmental Monitoring and Assessment, 187(5), 308. https://doi.org/10.1007/s10661-015-4551-1
Yang, X., Strahler, A. H., Schaaf, C. B., Jupp, D. L., Yao, T., Zhao, F., Wang, Z., Culvenor, D. S., Newnham, G. J., Lovell, J. L., Dubayah, R. O., Woodcock, C. E., & Ni-Meister, W. (2013). Three-dimensional forest reconstruction and structural parameter retrievals using a terrestrial full-waveform lidar instrument (Echidna®). Remote Sensing of Environment, 135, 36–51. https://doi.org/10.1016/j.rse.2013.03.020
Yu, X., Liang, X., Hyyppä, J., Kankare, V., Vastaranta, M., & Holopainen, M. (2013). Stem biomass estimation based on stem reconstruction from terrestrial laser scanning point clouds. Remote Sensing Letters, 4(4), 344–353. https://doi.org/10.1080/2150704X.2012.734931
Acknowledgements
The first author gratefully acknowledges the Centre for Space Science and Technology Education in Asia and the Pacific (CSSTEAP) for financial support during the study. The authors are thankful to the Head, Forestry and Ecology Department of the Indian Institute of Remote Sensing (IIRS), Director, IIRS and CSSTEAP for their encouragement and support in this study. The authors wish to acknowledge the Divisional Forest Officer, Dehradun Forest Division and staff of Barkot Forest Range, Dehradun Forest Division, Government of Uttarakhand, India and field staff of Barkot Flux Research Site for field support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
About this article
Cite this article
Bhandari, S.K., Nandy, S. Forest Aboveground Biomass Prediction by Integrating Terrestrial Laser Scanning Data, Landsat 8 OLI-Derived Forest Canopy Density and Spectral Indices. J Indian Soc Remote Sens 52, 813–824 (2024). https://doi.org/10.1007/s12524-023-01687-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12524-023-01687-z