Abstract
In this paper, we propose a new sparsity-based approach for the spectral–spatial classification of hyperspectral imagery. The proposed approach exploits the sparse representations of the spectral and spatial information contained in the data to generate an accurate classification map; specifically, we use all the spectral information (reflectance registered in the bands) and extended multiattribute profiles to extract spatial features. Hyperspectral image classification with sparse representations is based on the study that a pixel can be sparsely represented by a linear combination of a few learning examples from a structured dictionary. Then, by giving the set of training samples, any given sample may be sparsely represented by solving a sparsity-constrained optimization problem and thus classified in the class that minimizes a residual function. In this paper, we propose a new residual function which combines the sparse representations of the spectral features and the sparse representations of the spatial features to determine the class label of the test sample. Experiments are conducted on the familiar AVIRIS “Indian Pines” data set. It was found that the proposed method provided more accurate classification results than SVM with composite kernel.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Advances on remote sensors allow having images with high spectral resolution: the hyperspectral images. Each pixel in a hyperspectral image is represented by hundreds of different narrow wavelengths resulting, that, a high-dimensional vector with more dedicated spectral information. This property makes hyperspectral images usually used in various applications, such as military (Manolakis and Shaw 2002; Stein et al. 2002), agriculture (Patel et al. 2001; Datt et al. 2003) and environmental protection (Benediktsson et al. 2005). An important application of hyperspectral images is the supervised classification. The main goal of this application is to categorize the pixels in the image into one of several classes with representative training samples. In the literature, different hyperspectral image classification techniques have been proposed, including independent component analysis (ICA) (Palmason et al. 2005), artificial neural networks (ANN) (Goel et al. 2003) and multinomial logistic regression (MLR) (Krishnapuram et al. 2005). Among these techniques, support vector machine (SVM) has shown a good performance refer to their powerful to solve classification problems for high-dimensional data (Ben Salem et al. 2016). However, SVM needs an important number of training samples to ensure an accurate classification which is not available in most hyperspectral image classification problems due to the complexity of the collection of labeled samples.
Recently, sparse representation has been widely used in various fields, such as face recognition (Wright et al. 2009), image denoising (Elad and Aharon 2006) and hyperspectral image classification (Chen et al. 2011a). Sparse representation classification is a nonparametric learning method which does not need a training process but does need a few set of training data, and can directly assign a class label to a test sample. This property boosts researchers to adopt the sparse representations for hyperspectral image classification. Accordingly, various sparsity-based classification approaches have been proposed such as the nonlocal weighted joint sparse representation classification method (Zhang et al. 2014) based on the use of the simultaneous orthogonal matching pursuit technique, the classification approach proposed in Song et al. (2014) that exploits sparse representations of morphological attribute profiles, the two manifold-based sparse representation algorithms proposed for hyperspectral image classification (Tang et al. 2014), the class-dependent sparse representation classifier presented in Cui et al. (2015) that exploits the residual and Euclidean distance information to determinate the class of a test pixel, and the classification approach proposed in Pan et al. (2017) that uses low-rank, sparse representation and spectral consistency constraint.
Recently, the high spatial resolution of hyperspectral images encourages researchers to not only focus on analyzing the spectral features to generate the classification map with sparse representation, but also to take into account information in the spatially domain. Previous methods have registered a good performance by incorporating the contextual information. In Chen et al. (2011b), the local smoothing constraint and the joint sparsity model are used to integrate the spatial information in the classification with sparse representation, in Chen et al. (2011a), the joint sparsity model is introduced to the kernel sparse representation classifier, and in Liu et al. (2013), a spectral–spatial kernel sparse representation has been proposed to incorporate the spatial features.
In this paper, we propose a sparsity-based approach for the spectral–spatial classification of hyperspectral imagery based on the uses of spectral and spatial features to exploit the high resolution of these images. Toward this goal, a unified class membership function is developed, which utilizes, simultaneously, the spectral and the spatial residual. In doing so, each pixel in the image must be defined by two vectors: a spectral vector including the reflectance registered in bands and a spatial vector composed by features extracted by EMAPs. Two sparse representations must be used to define the unified class membership function: a spectral sparse representation training the spectral features aims at finding the spectral residual and a spatial sparse representation processes the spatial features to determine the spatial residual. Experimental results based on several real-world hyperspectral data sets demonstrate that the proposed approach increases the classification performance of traditional sparse representation classifier and outperforms kernel-based SVM.
The remainder of this paper is organized as follows. In “Proposed Approach” section, we present the different steps of the proposed approach. In “Experimental Results” section, we experimentally demonstrate the efficacy of the proposed methods and compare them with several state-of-the-art algorithms by using two hyperspectral data sets. In “Conclusion” section, we summarize the results and provide concluding remarks.
Proposed Approach
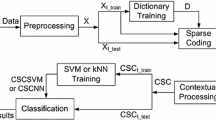
In this paper, we propose new sparsity-based approach for the spectral–spatial classification of hyperspectral imagery based on combining a spectral sparse representation and a spatial sparse representation to find the label of a test sample (Fig. 1).
The proposed approach implements the following three main steps: (1) spectral and spatial characterization which introduces all the spectral information to present each pixel in the spectral domain and EMAPs to extract spatial features, (2) spectral and spatial sparse representations that aim at finding the spectral and the spatial residuals and (3) classification by using the proposed unified class membership function which combines the two residuals.
Spectral and Spatial Characterization
The high resolution of hyperspectral image in the spectral and spatial domains increases the possibility to distinguish between spectrally similar materials. Different techniques of spectral and spatial characterizing hyperspectral pixels have been widely applied in the literature. Among these, for the spectral features extraction, authors usually used all the spectral information or techniques of dimensionality reduction to extract the most informative data such as independent component analysis (ICA) and principal component analysis (PCA). For the spatial characterization, various means have been adopted like morphological filters, features provided from the neighborhood of the pixel and attribute filters.
In this paper, we used all the spectral information for the spectral characterization and we implemented EMAP using attribute filters for the spatial features extraction.
EMAP (Mura et al. 2010): Extended multiattribute profiles aim at modeling the structural information contained in the considered image. They provide a multilevel characterization of the image by using a sequence of morphological attribute filters. EMAP is a vector which stacked the different extended attribute profiles (EAPs) of the image resulted from the using of many types of attribute. The EAP is resulted by generating an attribute profile (AP) on each of the first p principal components resulted by PCA (AP is obtained by applying a sequence of attribute filters using various thresholds).
Spectral and Spatial Sparse Representation
Sparse representation classification is a nonparametric learning method that presents an unknown test pixel as a linear combination of training pixels from all classes.
Let \( t \in {\mathbb{R}}^{B \times 1} \) the feature vector of a test pixel and \( D = [D_{1} \ldots D_{i} \ldots D_{C} ] \in {\mathbb{R}}^{M \times N} \) a structural dictionary composed by the feature vectors of the N training pixels (atoms of D) where \( D_{i} \in {\mathbb{R}}^{{M \times N_{i} }} \) the ith class sub-dictionary presenting the training samples in the ith class, C is the number of classes, Ni is the number of atoms in sub-dictionary Di (number of the training samples in class (i)) and \( N = \sum\nolimits_{i = 1}^{C} {N_{i} } \) is the total number of atoms in D. The test pixel t can be sparsely presented as:
where \( \alpha \in {\mathbb{R}}^{N \times 1} \) is a sparse coefficient vector.Given the dictionary of training samples D, α can be recovered by solving:
where K is a given upper bound on the sparsity level that represents the maximum number of selected atoms in the dictionary. K corresponds to the nonzero coefficients in \( \hat{\alpha } \). The problem (2) is a nondeterministic polynomial-time hard (NP-hard) (Davis et al. 1997), but it can be approximately solved by greedy pursuit algorithms such as orthogonal matching pursuit (OMP) (Tan et al. 2012). Accordingly, for a test pixel t, the main goal of the OMP algorithm is to find a representative atom at each iteration based on the correlation between the dictionary D and the residual vector R, where R = t − Dα. In fact, at each iteration, the OMP algorithm consists to (Fang et al. 2014):
-
1.
Calculate the residue correlation vector \( E \in {\mathbb{R}}^{N \times 1} \)
$$ E = D^{T} R $$(3) -
2.
Select a new representative atom (index j) based on the current residual correlation vector:
$$ \hat{j} = \hbox{max} \left\| {E_{j} } \right\|,\quad \, j = 1, \ldots ,N $$(4) -
3.
Add the newly selected atom’s index \( \hat{j} \) with the previously selected atom’s index set I.
-
4.
Estimate the sparse coefficient α by projecting the test samples t on DI :
$$ \hat{\alpha } = \left( {D_{I}^{T} D_{I} } \right)^{ - 1} D_{I}^{T} t $$(5)where DI is found by using the selected atoms.
The class of t can be determined by the characteristics of the sparse coefficient vector \( \hat{\alpha } \). In fact, it can be found by the minimal representation error between t and its approximation from the sub-dictionary of each class:
where \( \hat{\alpha }_{i} \) denotes the portion of the recovered sparse coefficients corresponding to the training samples in the ith class.
In this paper, we present a new sparse-based classification approach based on the incorporating of the spatial features to ameliorate the accuracy of the classification. Specifically, the proposed approach consists to recover the spectral sparse coefficient vector \( \hat{\alpha }_{\text{spect}} \) by using the spectral features vectors of pixels and to compute the spatial sparse coefficient vector \( \hat{\alpha }_{\text{spat}} \) by using the spatial features vectors. Accordingly, we used the spectral signatures of pixels to determine \( \hat{\alpha }_{\text{spect}} \) and features extracted by EMAPs to calculate \( \hat{\alpha }_{\text{spat}} \).
Classification
To determine the class of a test pixel, we define, in this paper, a new unified class function that combines the spectral and the spatial sparse representation (7).
where tspect is the spectral features vector of the test sample, \( D_{{{\text{spect}}_{i} }} \) is the ith class sub-dictionary presenting the spectral features vectors of training samples in the ith class, \( \hat{\alpha }_{{{\text{spect}}_{i} }} \) presents the portion of the spectral recovered sparse coefficients corresponding to the training samples in the ith class, tspect is the spatial features vector of the test sample, \( D_{{{\text{spect}}_{i} }} \) is the ith class sub-dictionary presenting the spatial features vectors of training samples in the ith class, \( \hat{\alpha }_{{{\text{spect}}_{i} }} \) presents the portion of the spatial recovered sparse coefficients corresponding to the training samples in the ith class.
Experimental Results
In this section, we evaluate the proposed approach according to the classification of a real hyperspectral data set: AVIRIS Indian pines. For that, we use OMP algorithm to approximately solve the sparse recovery problems for each test sample and then find the class by adopting the proposed unified class function. The classification results are then compared to those obtained by the spectral–spatial classifier SVMs with composite kernels (SVM-CK) that combine the spectral and spatial information via a weighted kernel summation, which have shown high performances in hyperspectral classification (Li et al. 2013).
To evaluate the effectiveness of the uses of the spectral and spatial information, the spectral and the spatial classification performance using, respectively, the spectral and the spatial features with the sparse representation classifier is included (OMPspect, OMPspat).
To build EMAP, it should be noted that we use the principal components which contain more than 98% of the total variance of the hyperspectral image and two attributes: the area with a threshold values ranging from 50 to 500 with a stepwise of 50 and the standard deviation with a threshold values in the range {2.5%, 20%} with a stepwise of 2.5%.
In all conducted experiments, the training dictionary is found by randomly selected samples from the available reference data, and the remaining samples are used for test. To evaluate the performance of the classification, we used the overall accuracy (OA), average accuracy (AA) and the kappa statistic. Different numbers of training samples have been used in our experiments to evaluate their impact on the classification accuracy.
AVIRIS “Indian pines”Footnote 1 is a 145×145 image that illustrates the Indian Pines region in Northwestern Indiana. It is collected by AVIRIS sensor in June 1992. The scene has 220 spectral bands range from 0.4 to 2.5 μm with a nominal spectral resolution of 10 nm. We used in the experiments 200 radiance channels (20 noisy bands covering the region of water absorption have been removed). The reference map for the scene is presented in Fig. 2, and it contains 16 classes characterized by their spectral similarity. The number of pixels in each class is reported in Table 1. For each class, we randomly select 10% of the labeled samples for training and use the rest for testing.
In our first experiment, we illustrate the advantage of using sparse representation in spectral and spatial domains for classification purposes by comparing the classification accuracies obtained by the proposed approach with that obtained by other classification approaches. Table 2 illustrates the individual class accuracies, the overall accuracy (OA), average accuracy (AA) and the kappa statistic coefficient (k) using different classifiers. As observed in Table 2, the use of the spectral and spatial sparse representations to determine the labels of test samples (OMPspect–spat) leads to have an accurate classification comparing with the spectral sparse representation classifier (OMPspect) and the spatial sparse representation classifier (OMPspat), and it allowed having an OA equal to 95.39%, 10.7% larger than OMPspat and 20.6% larger than OMPspect, which reflect the importance of incorporating spectral and contextual information for hyperspectral image classification purposes. Comparing with SVM-CK, the proposed approach has showed high performance (OA 1.5% larger than SVM-CK), which illustrates the great potential of the proposed sparsity-based classification approach to discriminate similar spectral classes. Focusing on the individual class accuracies, we note that, in all cases, OMPspect–spat provides the best results when compared with other methods. This is because the exploitation of the spectral and spatial features in the classification based on sparse representation greatly improves the class discriminability.
For illustrative purposes, Fig. 3 shows the classification maps obtained for the experiments reported in Table 2.
In the second experiment, we evaluate the impact of the size of the training dictionary. Figure 4 illustrates the obtained accuracies for the different implemented classification methods (OMPspect, OMPspat, SVM-CK and OMPspect–spat) as a function of the number of training samples. Several observations are shown in Fig. 4. First, the best classification accuracies are obtained by the OMPspect–spat approach even with limited training samples (training samples size < 10%) which shows the performance of the proposed classification method. The spectral–spatial classifications (OMPspect–spat and SVM-CK) outperform the classification when we used the spectral information only (OMPspect) and the spatial classification (OMPspat). Comparing OMPspect–spat and SVM-CK classification accuracies, we note that the advantage of OMPspect–spat is smaller with a high number of training samples. This is because the SVM is a discriminative approach based on the estimation of a separator plan in the transformed kernel space. Therefore, it is reasonable to have competitive classification accuracies when we used an important number of labeled samples. This observation reveals the importance of using sparse representation technique in the case of the availability of limited training samples.
Conclusion
In this paper, we have developed a new spectral–spatial sparsity-based classification approach which combines the spectral recovered sparse coefficients (when we used spectral features) and the spatial recovered sparse coefficients (when we used spatial features) via a proposed unified class function to exploit the wealth of hyperspectral images. The proposed approach is based on the use of sparse representation to overcome the problem of the availability of a limited number of training samples. By using all the spectral information, EMAPs to extract spatial attributes and OMP to solve the sparsity problem, the proposed method provides good accuracies when compared with the spectral and the spatial classification. It also exhibits robustness to the SVM classification with composite kernels (it allowed to have an OA 1.5% larger than SVM with composite kernels in the classification of the Indian Pines data set). Although our experimental results are competitive and encouraging when dealing with a limited training samples, further work should be focused on incorporating textural features to present pixels in the spatial domain.
Notes
Available at: http://engineering.purdue.edu/~biehl/MultiSpec/.
References
Ben Salem, R., Saheb, E. K., & Hamdi, M. A. (2016). Spectral–spatial classification of hyperspectral image based on oversampling and multi-feature kernels. Graphics, Vision and Image Processing Journal, 16(2), 31–40.
Benediktsson, J. A., Palmason, J. A., & Sveinsson, J. R. (2005). Classification of hyperspectral data from urban areas based on extended morphological profiles. IEEE Transactions on Geoscience and Remote Sensing, 43(3), 480–491.
Chen, Y., Nasrabadi, N., & Tran, T. (2011a). Hyperspectral image classification using dictionary-based sparse representation. IEEE Transactions on Geoscience and Remote Sensing, 49(10), 3973–3985.
Chen, Y., Nasrabadi, N. & Tran, T. (2011b). Hyperspectral image classification via kernel sparse representation. In Proceedings of ICIP, pp. 1233–1236.
Cui, M., & Prasad, S. (2015). Class-dependent sparse representation classifier for robust hyperspectral image classification. IEEE Geoscience and Remote Sensing Society, 53(5), 2683–2695.
Datt, B., McVicar, T. R., Van Niel, T. G., Jupp, D., & Pearlman, J. (2003). Preprocessing eo-1 hyperion hyperspectral data to support the application of agricultural indexes. IEEE Transactions on Geoscience and Remote Sensing, 41(6), 1246–1259.
Davis, G., Mallat, S., & Avellaneda, M. (1997). Adaptive greedy approximations. Constructive Approximation, 13(1), 57–98.
Elad, M., & Aharon, M. (2006). Image denoising via sparse and redundant representations over learned dictionaries. IEEE Transaction on Image Processing, 15(12), 3736–3745.
Fang, L., Li, S., Kang, X., & Benediktsson, J. A. (2014). Spectral–spatial hyperspectral image classification via multiscale adaptive sparse representation. IEEE Geoscience and Remote Sensing Society, 52(12), 7738–7749.
Goel, P. K., Prasher, S. O., Patel, R. M., Landry, J. A., Bonnell, R. B., & Viau, A. A. (2003). Classification of hyperspectral data by decision trees and artificial neural networks to identify weed stress and nitrogen status of corn. Computers and Electronics in Agriculture, 39(2), 67–93.
Krishnapuram, B., Carin, L., Figueiredo, M., & Hartemink, A. (2005). Sparse multinomial logistic regression: fast algorithms and generalization bounds. IEEE Transactions on Pattern Analysis and Machine Intelligence, 27(6), 957–968.
Li, J., Marpu, P. R., Plaza, A., Bioucas-Dias, J. M., & Benediktsson, J. A. (2013). Generalized composite kernel framework for hyperspectral image classification. IEEE Geoscience and Remote Sensing Society, 51(9), 4816–4829.
Liu, J., Wu, Z., Wei, Z., Xiao, L., & Sun, L. (2013). Spatial-spectral kernel sparse representation for hyperspectral image classification. IEEE Geoscience & Remote Sensing Society, 6(6), 2462–2471.
Manolakis, D., & Shaw, G. (2002). Detection algorithms for hyperspectral imaging applications. IEEE Signal Processing Magazine, 19(1), 29–43.
Mura, M. D., Benediktsson, J. A., Waske, B., & Bruzzone, L. (2010). Extended profiles with morphological attribute filters for the analyses of hyperspectral data. International Journal of Remote Sensing, 31(22), 5975–5991.
Palmason, J. A., Benediktsson, J. A., Sveinsson, J. R. & Chanussot, J. (2005). Classification of hyperspectral data from urban areas using morphological preprocessing and independent component analysis. In Proceedings of IGARSS, pp. 25–29.
Pan, L., Li, H., Meng, H., Li, W., Du, Q., & Emery, W. J. (2017). Hyperspectral image classification via low-rank and sparse representation with spectral consistency constraint. IEEE Geoscience and Remote Sensing Society, 14(11), 2117–2121.
Patel, N., Patnaik, C., Dutta, S., Shekh, A., & Dave, A. (2001). Study of crop growth parameters using airborne imaging spectrometer data. International Journal of Remote Sensing, 22(12), 2401–2411.
Song, B., Li, J., Mura, M. D., Li, P., Plaza, A., Bioucas-Dias, J. M., et al. (2014). Remotely sensed image classification using sparse representations of morphological attribute profiles. IEEE Transactions on Geoscience and Remote Sensing, 52(8), 5122–5136.
Stein, D., Beaven, S., Hoff, L., Winter, E., Schaum, A., & Stocker, A. (2002). Anomaly detection from hyperspectral imagery. IEEE Signal Processing Magazine, 19(1), 58–69.
Tan, M., Tsang, I. W., Wang, L. & Zhang, X. (2012). Convex matching pursuit for large-scale sparse coding and subset selection. In Proceedings of AAAI conference on artificial intelligence, pp. 1119–1125.
Tang, Y. Y., Yuan, H., & Li, L. (2014). Manifold-based sparse representation for hyperspectral image classification. IEEE Geoscience and Remote Sensing Society, 52(12), 7606–7618.
Wright, J., Yang, A., Ganesh, A., Sastry, S., & Ma, Y. (2009). Robust face recognition via sparse representation. IEEE Transactions on Pattern Analysis and Machine Intelligence, 31(2), 210–227.
Zhang, H., Li, J., Huang, Y., & Zhang, L. (2014). A nonlocal weighted joint sparse representation classification method for hyperspectral imagery. IEEE Journal of Selected Topics in Applied Earth Observations and Remote Sensing, 7(6), 2056–2065.
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Hamdi, M.A., Ben Salem, R. Sparse Representations for the Spectral–Spatial Classification of Hyperspectral Image. J Indian Soc Remote Sens 47, 923–929 (2019). https://doi.org/10.1007/s12524-018-0908-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12524-018-0908-6