Abstract
High soil salinity possesses a major challenge for plant growth and productivity. Plants have evolved various mechanisms to withstand the adverse effects of salt stress, including E3 ubiquitin ligases that label salt-responsive proteins for degradation. Here, we characterized the mechanisms RING E3 ubiquitin ligase OsSIRH2-3 (Oryza sativa Salt Induced RING H2-type-3 E3 ligase) used to facilitate salt tolerance in rice. OsSIRH2-3 expression was upregulated under high NaCl concentrations and upon abscisic acid (ABA) treatment. OsSIRH2-3 was primarily found in the nucleus of rice protoplasts. The OsSIRH2-3 protein contains an H2-type-RING domain that confers E3 ligase activity. OsSIRH2-3 overexpression was also found to be associated with enhanced salt tolerance in transgenic plants, decreased Na+ accumulation in both roots and leaves, decreased Na+ transport activity in the xylem sap, increased levels of proline and soluble sugars, elevated activity of reactive oxygen species scavenging enzymes, and altered expression of Na+/K+ transporters. Furthermore, OsSIRH2-3-overexpressing plants also exhibited high sensitivity to exogenous ABA treatment. Our findings demonstrate that OsSIRH2-3 enhances salt tolerance by regulating Na+/K+ homeostasis and modulating Na+/K+ transporter expression. This study illuminates the molecular mechanisms involved in RING E3 ubiquitin ligase-mediated salt tolerance in rice and provides a potential strategy for enhancing crop productivity in saline environments.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The worldwide food demand is projected to double by 2050 to sustain a global population estimated to reach 9.7 billion (Fukase and Martin 2020). Enhancing crop productivity is thus crucial in this regard. However, abiotic stresses including salinity, drought, cold, heat, and flooding, significantly reduce crop productivity. Among these stresses, salinity represents the primary constraint on plant growth and productivity worldwide (Janz and Polle 2012; Polle and Chen 2015).The effects of salt stress have been compounded by rising sea levels due to global warming, affecting approximately six percent of the Earth’s total land mass, equivalent to 800 million hectares of agricultural land (Munns and Tester 2008). Compared to other major crops such as wheat (Triticum aestivum L.), sorghum (Sorghum bicolor L.), barley (Hordeum vulgare L.), and rice (Oryza sativa L.) has been shown to be highly sensitive to salt (i.e., display “salt hypersensitivity”). For instance, wheat and barley productivity was found to decline by 7% and 5% under 60 and 80 mM NaCl, respectively, while rice showed a productivity decrease of approximately 12%, even at 30 mM NaCl (Chinnusamy et al. 2005). Elucidating the mechanisms of salt tolerance thus holds promise for boosting productivity in rice.
During periods of salt stress, plant growth is restricted due to an imbalance between sodium ions (Na+) and other ions. Plants have thus developed various mechanisms to withstand salt stress (Horie et al. 2007; Su et al. 2015). Excessive amounts of external Na+ interfere with the absorption of potassium ions (K+), leading to detrimental effects such as inhibition of essential enzymes and photosynthesis, and ultimately Na+ toxicity and plant mortality (Horie et al. 2007). Several mechanisms have been previously discovered to reduce Na+ toxicity, including reduced influx and increased efflux of Na+ influx, and sequestering of Na+ into the cell vacuoles (Su et al. 2015). Recently, various Na+/K+ channels and transporters were shown to be essential in maintaining Na+ homeostasis in plant roots during salt stress, including non-selective cation channels (Demidchik and Tester 2002), salt-overly sensitive 1 (SOS1; Ji et al. 2013), Na+/H+ antiporters (NHXs; Bassil et al. 2011), and high-affinity potassium transporters1 (HKT1s; Horie et al. 2007; Waters et al. 2013). In addition, the HKT2 family, including OsHKT2;1 and OsHKT2;2 in rice (Horie et al. 2007; Yao et al. 2010) and TaHKT2;1 in wheat (Ariyarathna et al. 2014) were identified as Na+ transporters as well.
Na+ accumulation in plants under salt stress further leads to ionic stress that disrupts protein synthesis, photosynthesis, and enzymatic activities. Osmotic stress promotes the accumulation of reactive oxygen species (ROS), which leads to the degradation of lipids, proteins, RNA, and DNA, ultimately resulting in plant mortality (Mittler 2002). Plants have evolved antioxidant defense systems that employ ROS-scavenging mechanisms to counteract such unfavorable conditions and maintain redox homeostasis. Antioxidant enzymes such as superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT) are utilized to alleviate ROS-induced damage (Hossain and Dietz 2016).
Furthermore, osmotic stress caused by salt stress triggers the production of the phytohormone abscisic acid (ABA), which is essential for successful regulation of plant physiological processes in response to various abiotic stresses (Finkelstein et al. 2002; Jia et al. 2002; Xiong et al. 2002; Wei et al. 2020). ABA serves not only as a mediator of salinity signals but also as a protective agent against salt stress by regulating the expression of stress-responsive genes (Danquah et al. 2014; Yoshida et al. 2014).
Plants respond to stress conditions via changes in intracellular metabolic processes, regulation of protein synthesis and degradation, and expression of transcription factors. Ubiquitination is a post-translational modification mechanism that involves the attachment of small ubiquitin tags to target proteins. The resulting ubiquitin-substrate complex is recognized and degraded by the 26S proteasome. The ubiquitin/26S proteasome system is crucial for the regulation of various physiological processes in plants, including growth, disease resistance, and stress responses. RING (Really Interesting New Gene) proteins, which are characterized by a zinc-binding motif (Cys-X2-Cys-X9-39-Cys-X1-3-H-X2-3-Cys-X2-Cys-X4-48-Cys-X2-Cys), are involved in multiple steps of ubiquitin activation, conjugation, and ligase activity, and function in conjunction with the ubiquitin activation enzyme (E1), ubiquitin conjugation enzyme (E2), and ubiquitin ligase (E3) (Deshaies and Joazeiro 2009). In addition, RING protein plays significant role in response to abiotic stress conditions in rice species in particular (Chapagain et al. 2017). For example, the rice protein Oryza sativa salt-induced RING H2-14 (OsSIRH2-14) interacts with the Na+ transporter OsHKT2;1. OsSIRH2-14-overexpressing plants exhibit lower levels of Na+ accumulation than wild-type (WT) plants (Park et al. 2019).
A previous study examined the transcriptional patterns of 47 RING finger proteins in rice under various abiotic stress conditions. Among these proteins, OsSIRH2-3 (Oryza sativa salt-induced RING finger protein H2-3), an H2-type RING protein gene (previously called OsRFPH2-3), is upregulated under salt stress conditions (Lim et al. 2013). However, the precise molecular function of OsSIRH2-3 remains elusive. Here, we performed a phenotypic analysis of OsSIRH2-3-overexpressing plants in terms of Na+ accumulation and transcription analysis of Na+ or K+ transporter genes to gain further insights into the function of OsSIRH2-3.
Material and Methods
Generation of Transgenic Plants
The coding sequence of OsSIRH2-3 (LOC_Os01g74040) was amplified from rice cDNA using Q5 polymerase 2X Master Mix (NEB, Massachusetts, USA) and the appropriate primers. PCR products were cloned into a pGA3425 binary plasmid vector tagged with the pUbi promoter. OsSIRH2-3-overexpressing plants were generated at the National Institute of Agriculture Sciences (Suwon, Korea). The cloned pUbi:OsSIRH2-3 recombinant plasmid was transferred using the heat shock method (Froger and Hall 2007), into Agrobacterium tumefaciens strain EHA105. Overexpressing plants were transformed using a previously described method (Toki et al. 2006), and rice seeds were sterilized using 2% sodium hypochlorite with Tween-20 (Sigma-Aldrich, Missouri, USA) and grown on 2N6D medium at 30 °C for 5 d. Precultured calli were infected with EHA105 cells, including the pUbi:OsSIRH2-3 plasmid and cultured at 25 °C for 3 d under dark conditions. The infected calli were washed five times with sterilized water containing 500 mg L−1 carbenicillin, transferred to 2N6D medium containing 50 mg L−1 hygromycin and 400 mg L−1 carbenicillin, and incubated for one month under light conditions. Then, selected calli were transferred to a shoot induction medium containing 0.1 mg L−1 naphthaleneacetic acid (Duchefa Biochemie, Haarlem, Netherlands) and 2 mg L−1 kinetin (Duchefa Biochemie) and incubated at 32 °C for 3 weeks under light conditions. Transformed plants were grown in a well-controlled greenhouse. Seeds of two independent T3 homozygous plants were selected for further experiments.
Plant Growth Conditions and Phenotypic Assay
OsSIRH2-3-overexpressing and WT seeds were germinated at 30 °C in the dark for 2 d. The germinated seeds were transferred to plastic containers containing half-strength Kimura B nutrient solution (pH 5.6) and incubated in a growth chamber under 16/8 h light/dark conditions with 70% relative humidity. Ten-day-old seedlings were then treated with 100 mM NaCl. Phenotype characteristics, including shoot length, root length, shoot fresh weight, and fresh root weight, were measured in triplicate (each sample number = 15). Plant leaf and root tissues were harvested, ground using liquid nitrogen, and stored at – 70 °C for further study.
RNA Extraction and Gene Expression Analysis
Total RNA was extracted using TRIzol® reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s protocol. To avoid genomic DNA contamination, total RNA was treated with DNase I (Sigma-Aldrich) and 1 µg of total RNA was used for cDNA synthesis using a PrimeScript™ 1st-Strand cDNA Synthesis Kit (Takara Bio, Ohtsu, Japan). Quantitative real-time PCR (qPCR) was performed using the CFX ConnectTM Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) with SYBR® Green TOP real qPCR 2X PreMIX (EnzynomicsTM, Daejeon, Korea), 25 ng of cDNA, and appropriate gene-specific primers. Primers were designed using the Beacon Designer™ (PRIMER Biosoft, Palo Alto, CA, USA) and synthesized by a commercial service Macrogen (Macrogen, Seoul, Korea). The amplification conditions were as follows: pre-denaturation at 95 °C for 10 m, 40 cycles of denaturation at 95 °C for 10 s, annealing at the appropriate annealing temperature for each primer pair (Table S1), and extension at 70 °C for 30 s. OseEF-1a (LOC_Os03g08010) was used as an internal control. Relative expression levels were calculated using the 2−ΔΔCT method (Livak and Schmittgen 2001).
E3 Ligase Activity Analysis of OsSIRH2-3
To verify the E3 ubiquitin ligase activity of OsSIRH2-3, the full-length OsSIRH2-3 gene was amplified using Q5 DNA polymerase (New England Biolabs, Ipswich, MA, USA), and the PCR products were cloned into the pMAL-c5X-MBP tagged vector (New England BioLabs). To generate a point mutation, an amino acid substitution (OsSIRH2-3C149A) in the RING finger domain was made using Q5® Site-Directed Mutagenesis Kit (New England BioLabs) and cloned into the pMAL-c5X-MBP tagged vector. Recombinant MBP-OsSIRH2-3, MBP-OsSIRH2-3C149A, and non-recombinant pMAL (negative control) were transformed into Escherichia coli strain BL21 (DE3) and purified by affinity chromatography using an amylose resin column (New England BioLabs). E2 (Arabidopsis UBC10, At5g53300) was cloned into a pET-28a ( +)-6 X-His-tagged vector (Novagen, Gibbstown, NJ, USA) and transformed into E. coli strain BL21 (DE3). Recombinant AtUBC10 was purified using a Ni–NTA Purification System (Invitrogen Life Technologies). For the in vitro ubiquitination assay, purified MBP-OsSIRH2-3, and mutated MBP-OsSIRH2-3C149A (300 ng) each was mixed with 50 ng of human E1 (Sigma-Aldrich), 150 ng purified E2 (AtUBC10), bovine ubiquitin (Sigma-Aldrich), and reaction buffer containing 50 mM Tris–HCl (pH 7.5); 100 mM MgCl2; 40 mM DTT; and 40 mM ATP; The reaction mixture was incubated 4 h at 30 °C, the reaction was stopped by mixing 5X sample buffer and boiling at 95 °C for 5 m. The samples were loaded onto an 8% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred to nitrocellulose membranes. Ubiquitinated OsSIRH2-3 proteins were detected by western blotting using primary anti-ubiquitin in rabbit (U5379; Sigma-Aldrich) and anti-MBP monoclonal antibody (E8032; NEB) and a secondary anti-rabbit lgG (A0545; Sigma-Aldrich) and anti-mouse lgG (A8924; Sigma-Aldrich) and photographed using a ChemiDoc™XRS imaging system (Bio-Rad).
Rice Protoplast Extraction and Subcellular Localization
Rice seeds were germinated and grown in half Kimura B solution in a growth chamber at 30/25 °C (16/8 h light/dark) with 70% of relative humidity. Protoplasts were isolated from 1-week-old WT leaf sheaths. Approximately 30 seedlings were chopped with a razor blade and incubated in 30 mL of filtered enzyme solution containing 0.25% macerozyme R-10 (Yakult Honsa), 1% cellulase R-10 (Yakult Honsa Co. Ltd., Tokyo, Japan), 500 mM mannitol (Sigma-Aldrich), 1 mM CaCl2 (Sigma-Aldrich), 0.1% BSA (Sigma-Aldrich), and 10 mM MES (Duchefa Biochemie), at pH 5.6, the mixture was incubated in the dark condition with gentle rocking at 25 °C for 6 h. The mixture was then filtered using 40 μm nylon mesh filters (SPL, Gyeonggi-do, Korea). The protoplasts were collected by centrifugation at 300 × g for 4 m. The protoplast pellets were mixed in 30 mL of W5 solution containing 154 mM NaCl (Duchefa Biochemie), 125 mM CaCl2 (Sigma-Aldrich), 5 mM KCl (Sigma-Aldrich), 5 mM glucose (Duchefa Biochemie), and 1.5 mM MES (Duchefa Biochemie), and then incubated at 4 °C for 3 h. After incubation, the pelleted protoplasts were resuspended in MMG solution containing 0.4 M mannitol (Duchefa Biochemie), 15 mM MgCl2 (Sigma-Aldrich), and 4.7 mM MES (Duchefa Biochemie), pH 5.6. The recombinant vector 35 s:OsSIRH2-3-YFP (10 μg) was transfected into protoplasts with MMG solution, and mixed. The mixture was added to a prepared PEG solution containing 40% PEG 4000 (Sigma-Aldrich), 100 mM Ca(NO3)2, and 400 mM mannitol and gently mixed. The prepared samples were incubated at 25 °C for 30 m in dark. After incubation, the protoplast pellets were collected by centrifugation at 300 × g for 4 m. The pelleted protoplasts were gently resuspended in 1 mL of W5 solution and incubated overnight for 16 h at room temperature in dark. The protoplasts were photographed using a confocal laser scanning microscope at the Central Laboratory of Kangwon National University (Chuncheon, Korea). Photographs were acquired using ZNE (ZEISS, Oberkochen, Germany).
Visualization of Na+ Accumulation
To visualize Na+ accumulation in the roots, the Na+-ion-specific fluorescent dye CoroNa Green (Invitrogen Life Technologies) was used to visualize Na+ accumulation in the roots. Ten-day-old seedlings of OsSIRH2-3-transgenic and WT plants were grown in 0 or 100 mM NaCl for 24 h. The root tips were cut into approximately 2 cm pieces and washed with distilled water. The root tips were then stained with 25 μM CoroNa Green staining solution dissolved in dimethyl sulfoxide and incubated for 3 h at room temperature. After washing the tissue with distilled water, the prepared samples were placed on a sliding glass and Na+ accumulation was detected using the green fluorescence signal SR-CLSM (LSM880, Carl Zeiss). Photographs were acquired using a ZNE microscope (ZEISS).
Measurement of Chlorophyll Content
Ten-day-old rice leaves were used to measure chlorophyll content. Fresh rice leaf tissue (200 mg) was ground in liquid nitrogen and 80% acetone (1.5 mL) was added. The samples were then incubated in the dark at 4 °C for 30 m. The incubated samples were collected by centrifugation at 13,000 × g at 4 °C for 20 m, and the supernatants were transferred to fresh microtubes. The absorbance of the prepared samples was measured at 663 and 645 nm using a microplate reader (SPECTROstar Nano, BMG Labtech) and the chlorophyll content was calculated as previously described (Ni et al. 2009).
Measurement of H2O2 Contents
H2O2 content was measured as previously described (Gay and Gebicki 2000), each root and leaf sample (100 mg each) was mixed with 0.1% trichloroacetic acid (Sigma-Aldrich), incubated at 4 °C for 30 m, and centrifuged at 12,000 × g at 4 °C for 30 m. The supernatant was transferred to a new microtube and allowed to react with solutions A (2.5 M H2SO4 and 25 mM ammonium ferrous II sulfate) and B (100 mM sorbitol and 125 μM xylenol orange). The absorbance of the samples was measured at 560 nm using a microplate reader (SPECTROstar Nano; BMG LabTech).
Measurement of SOD, POD, CAT, and MDA Contents
The root and leaf blade tissues (100 mg) were harvested using an enzyme extraction buffer composed of 50 mM sodium phosphate buffer (pH 7.0), 5 mM-mercaptoethanol, 2 mM EDTA, and 4% PV-40. The harvested tissues were then homogenized in the extraction buffer and centrifuged at 15,000 rpm for 40 m at 4 °C. The resulting supernatant was collected for further analysis of enzyme activity. The protein concentration in the extracted samples was determined using a QubitTM Protein Assay Kit (Invitrogen), which provides a quantitative measurement of protein content. To measure the activities of superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT), the extracted samples were subjected to specific assays following a previously described method (Kim et al. 2023) with some modifications. Modifications made to the assay protocol may include changes in the reaction conditions or additional steps to meet the experimental requirements. To determine the malondialdehyde (MDA) content in root and leaf blade tissues, the following procedure was conducted based on a previously described method (Sun et al. 2020) with some modifications. First, 100 mg of root or leaf blade tissue was weighed and ground in 5% (w/v) trichloroacetic acid (TCA). The homogenate was centrifuged at 12,000 × g for 15 m and the supernatant was carefully transferred to a new tube. Next, 1 mL of a solution containing 0.5% thiobarbituric acid (TBA) and 20% TCA was added to the supernatant and the mixture was thoroughly mixed. The tube was then incubated in a water bath at 95 °C for 20 m. Finally, after cooling the liquid to 4 °C, the absorbance of the mixture was measured at 450 nm, 532 nm, and 600 nm using a microplate reader (SPECTROstar Nano).
Measurement of Proline and Soluble Sugar Contents
The proline contents of the root and leaf tissues were measured as previously described (Carillo and Gibon 2011). Leaf and root tissues (100 mg) were mixed with 1 mL 70% ethanol and incubated at room temperature for 20 m. Thereafter, 50 μL of the extract was mixed with 100 μl of the reaction solution [20% ethanol and 1% ninhydrin (w/v) in 60% acetic acid (v/v)] and boiled at 95 °C for 20 m in a water bath. The prepared samples were immediately cooled to room temperature and the absorbance was measured at 520 nm using a microplate reader (SPECTROstar Nano). Soluble sugar content was measured as previously described (Song et al. 2011)(Song et al. 2011)(Song et al. 2011)(Song et al. 2011)(Song et al. 2011). Rice leaf and root tissues (100 mg) were mixed with 1 mL of 80% ethanol and incubated at 80 °C for 30 m. Thereafter, the volume was adjusted to 10 mL with 80% ethanol. To determine soluble sugar content, 1 mL of the extract was mixed with 5 mL of the reaction solution (anthrone reagent dissolved in 0.2% H2SO4). The mixed samples were then boiled at 95 °C for 15 m. The sample was then cooled to room temperature, and the absorbance was measured at 620 nm using a microplate reader (SPECTROstar Nano).
Measurement of Ionic Compounds
Ten-day-old seedlings of OsSIRH2-3-overexpressing and WT plants were exposed to stress conditions with either 0 or 100 mM NaCl. After the incubation period, the root and leaf tissues were harvested and dried in an oven at 60 °C for 2 d. The dried tissues were used to measure the levels of Na+ and K+ using inductively coupled plasma-optical emission spectroscopy (ICP-OES) with an Optima 700 DV instrument (Perkin Elmer, located at the Central Laboratory of Kangwon National University). In addition, the leaf sheaths of ten-day-old rice seedlings from OsSIRH2-3-overexpressing and WT plants were treated with either 0 or 100 mM NaCl. The lower part of the leaf sheath was cut, and xylem sap samples that overflowed from the surface of the leaf sheath were collected using a micropipette over a 12-h period. The collected samples were then analyzed using ICP-OES to measure the content of Na+ and K+.
Germination Assay of Exogenous ABA Treatment
To evaluate the growth performance in response to ABA treatment, seeds of OsSIRH2-3-overexpressing and WT plants were subjected to different concentrations of ABA. The seeds were placed on half-strength MS medium supplemented with 0 μM (control), 3 μM, or 5 μM ABA. The growth chamber provided optimal conditions for seed germination, including a 16 h/8 h (light/dark) period and 70% humidity. The number of germinated seeds was recorded daily for 5 d at 24-h intervals.
Statistical Analysis
All statistical data were evaluated using a two-tailed Student’s t-test (Millar 2001)(Millar 2001)(Millar 2001)(Millar 2001)(Millar 2001), and the significance of the data was determined using three different P-values (95%, * P < 0.05; **99%, P < 0.01; and ***99.9%, P < 0.001). The resulting data were analyzed using Microsoft Excel 2016 (Microsoft).
Results
OsSIRH2-3 Expression Under Abiotic Stress
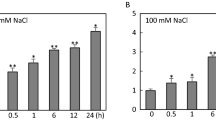
We evaluated the expression of OsSIRH2-3 in rice upon treatment with 100 mM NaCl, 20% of PEG, and 50 µM of ABA at 0, 1, 6, 12, and 24 h (Fig. 1). OsSIRH2-3 expression was found to increase between 6 h (4.7-fold) and 24 h (51-fold) under salt stress (100 mM NaCl) (Fig. 1a). The expression of OsNAC10 (salinity), OsSalT (drought), and OsRAB16A (ABA) gradually increased under each stress treatment from 0 to 24 h (Fig. S1). Following 20% PEG treatment, OsSIRH2-3 expression rapidly decreased by up to 0.01-fold after 24 h (Fig. 1b). Upon 50 µM ABA treatment, OsSIRH2-3 expression increased by up to 2.2-fold after 24 h (Fig. 1c).
Gene expression pattern of OsSIRH2-3 under two abiotic stresses (100 mM NaCl and 20% PEG) and abscisic acid (ABA) treatment in wild-type plants. Ten-day-old seedlings were treated with abiotic stresses and hormone. The qRT-PCR analysis of OsSIRH2-3 was performed under 100 mM NaCl (a), 20% PEG (b), and 50 μM ABA (c) in leaves. Relative expression levels are measured compared to non-treatment controls. The expressional values are calculated with three biological replicates (each sample number = 10). The OseEF-1a (LOC_Os03g08010) gene was used as an internal control. The asterisks indicate a significant difference according to the student’s t-test; NS = non-significant ns: non-significant; *P < 0.05, **P < 0.01 or ***P < 0.001
OsSIRH2-3 Shows E3 Ubiquitin Ligase Activity
Next, we performed an analysis of protein domain sequences and conducted an in vitro ubiquitination assay to characterize the function of OsSIRH2-3. The domains of OsSIRH2-3, which encodes 335 amino acids, were identified and analyzed using the National Center for Biotechnology Information Conserved Domain Database. A comparison of the RING domains of OsSIRH2-3 and its orthologs in Arabidopsis thaliana (AT2G39720), Sorghum bicolor (Sobic.009g010400), and Triticum aestivum (Traes_1DS_06B2330A5) revealed that OsSIRH2-3 harbors a highly conserved C3H2-type RING domain (131–171 residues) (Fig. 2a). OsSIRH2-3 and its mutant OsSIRH2-3C149A in which the third cysteine in RING is replaced with alanine, were subjected to an in vitro ubiquitination assay with an empty vector as a negative control (Fig. 2b). The negative control and mutant OsSIRH2-3C149A was not found to contain polyubiquitinated chains, where OsSIRH2-3 exhibited polyubiquitin chain. These results demonstrate the E3-ubiquitin ligase function of the RING domain of OsSIRH2-3.
Molecular characterization of OsSIRH2-3. a Multiple amino acid sequence alignment of the RING domain between the OsSIRH2-3 and orthologs from Arabidopsis thaliana (AT2G39720) Sorghum bicolor (Sobic. 009G010400) and Triticum aestivum (Traes_1DS_06B2330A5). b In vitro ubiquitin assay of the wild-type OsSIRH2-3, mutated OsSIRH2-3 C149A, and pMAL-EV proteins. The purified MBP- OsSIRH2-3 and MBP- OsSIRH2-3 C149A proteins were incubated with E1 (human), E2 (Arabidopsis, UBC10), ubiquitin, and 20 × Z-buffer for 3 h at 30℃. Poly ub-chains were detected using an anti-MBP or an anti-ubiquitin antibody. c Subcellular localization of the 35S: OsSIRH2-3-YFP in rice protoplasts. 35S:YFP was used as a negative control. In rice protoplasts, the co-transfection of 35S: OsSIRH2-3-YFP and nucleus protein (OsMeCP-RFP) was employed. Scale bar = 5 μm
Subcellular Localization of OsSRIH2-3
To identify the subcellular localization of OsSIRH2-3 in rice, we used 35S: EYFP as a positive control and the 35S: OsSIRH2-3-YFP recombinant vector. 35S: EYFP was localized to the cytosol, whereas 35S: OsSIRH2-3-YFP was localized only to the nucleus. Furthermore, the nuclear marker gene (35S: OsMeCP-DsRed2) was co-transfected into rice protoplasts. These results clearly showed co-localization of 35S: OsSIRH2-3-YFP and 35S: OsMeCP-DsRed2.
Phenotypical Characterization of OsSIRH2-3-Overexpressing Plants to Salt Stress
Arabidopsis OsSIRH2–3-overexpressing plants were previously shown to exhibit significantly increased salt tolerance (Lim et al. 2013). To further determine the effect of OsSIRH2-3 overexpression on phenotype, we developed transgenic rice plants. First, a qPCR analysis of five independent overexpressing transgenic rice lines was performed, and the results were compared with those of WT plants (Fig. S2). The expression levels of OsSIRH2–3 was significantly higher in the four independent transgenic lines than those in the WT line. Thus, we chose the #10–3 (693-fold) and #17–1 (3,371-fold) overexpressing lines for further studies. Phenotypic analyses such as the lengths and fresh weights of shoots and roots, were evaluated after growth in Kimura B solution for 10 d (Fig. 3a, b). No significant difference in shoot and root length, and weight was observed between the transgenic and WT plants under non-treatment (Fig. 3c–f). At 100 mM NaCl, higher shoot length, shoot weight, and root weight were observed in the transgenic plants than in the WT plants (Fig. 3c–f). However, there was no significant difference in root length under 100 mM NaCl (Fig. 3e). These results indicate that OsSIRH2-3 overexpression improved the tolerance to salt stress. Salt stress negatively affects chloroplast membranes and chlorophyll content, and thereby reduces photosynthetic efficiency (Ali et al. 2004). To test whether the same effect is observed on OsSIRH-2–3 overexpressing rice, the chlorophyll content was measured in the leaves of transgenic and WT plants grown with (100 mM) or without NaCl. The total chlorophyll content, including chlorophyll a and b, was significantly higher in the transgenic plants than in the WT plants under both conditions (Fig. 3g–I and Fig. S3).
Phenotypes of OsSIRH2-3-overexpressing and WT plants. a, b Images of rice seedlings plants grown for 10 d. Ten-day-old seedlings were treated with 0 (a) or 100 mM NaCl (b) for 7 d. c Shoot length, d Shoot weight, e Root length, and f Root weight were measured when grown in 0 or 100 mM NaCl. g–i The quantification of chlorophyll (a), chlorophyll (b), and chlorophyll (a + b). The experiments were conducted on three biological replicates (each sample number = 15). The asterisks indicate a significant difference according to the student’s t-test; ns: non-significant**P < 0.01, or ***P < 0.001
Antioxidant Enzyme Activity of OsSIRH2-3-Overexpressing Plants to Salinity Stress
High salinity induces the production of ROS such as H2O2 in plants (Pang and Wang 2008). Therefore, plants have developed various molecular mechanisms to avoid ROS-induced stress. For example, increase in activities of antioxidant enzymes, such as SOD, POD, and CAT, is necessary to improve the tolerance to salt stress in rice (Monsur et al. 2020). We thus measured the SOD, POD, and CAT content in the leaves and roots of transgenic and WT plants grown with (100 mM) and without NaCl treatment. There were no significant differences in SOD, POD, or CAT levels without NaCl treatment. However, transgenic plants exhibited significantly lower SOD, POD, and CAT content than WT plants in both leaves and roots under 100 mM NaCl (Fig. 4a–c and f–h). In contrast, H2O2 levels were significantly higher in the shoots and roots of transgenic plants than in those of WT plants (Fig. 4d, i). Malondialdehyde (MDA) content is a commonly used indicator of lipid peroxidation and oxidative damage in plants (Gerona et al. 2019). We found no significant differences in the MDA content between tissues in the absence of NaCl (Fig. 4e, j). However, when plants were exposed to 100 mM NaCl, the leaf and root tissues of OsSIRH2-3-overexpressing plants showed lower MDA content than those of WT plants (Fig. 4e, j). These results suggest that overexpression of OsSIRH2-3 improves antioxidant capacity and reduces lipid peroxidation under salt stress, and thus alleviates oxidative damage in rice.
Antioxidant activity in the leaves and roots of OsSIRH2-3-overexpressing and WT plants under 0 or 100 mM NaCl. The seedlings were grown for 10 d and treated with 0 or 100 mM NaCl for 7 d. Antioxidant activity was measured in leaves (a–e) and roots (f–j). a, f The content of Superoxide dismutase (SOD), b, g peroxidase (POD), c, h catalase (CAT), d, i H2O2, and e, j Malondialdehyde (MDA). All experiments were conducted in three biological replicates (each sample number = 15). The asterisks indicate a significant difference according to the student’s t-test; ns, non-significant **P < 0.01, or ***P < 0.001
Measurement of Soluble Sugar and Proline Content
Soluble sugar and proline accumulation play important roles in response to various abiotic stressors (Nemati et al. 2011; Hayat et al. 2012). Significantly higher levels of proline and soluble sugars were observed in the leaf and root tissues of OsSIRH2-3-overexpressing plants than in WT plants under 100 mM NaCl, compared to the absence of NaCl (Fig. 5a-d). This suggests that the overexpression of OsSIRH2-3 increases the accumulation of these osmoprotectants.
OsSIRH2-3-Overexpression Increased ABA Sensitivity in Rice
The expression of OsSIRH2-3 was found to increase after exogenous ABA treatment (Fig. 1c). To further investigate the role of OsSIRH2-3 in the ABA response, a germination assay was conducted using OsSIRH2-3-overexpressing and WT seeds at different ABA concentrations. Notably, OsSIRH2-3-overexpressing seeds exhibited severe sensitivity to ABA during germination, as evidenced by significantly lower germination rates compared to WT seeds under 3 μM or 5 μM ABA treatment (Fig. 6b, c). In the absence of ABA, no significant differences between germination rates were observed (Fig. 6a). Subsequently, we examined the expression patterns of several ABA signaling pathway-related genes, including OsLEA3, OsP5Cs, OsProT, and NAC5, in the leaves and roots of transgenic and WT plants (Fig. 7). In the absence of ABA treatment, no significant differences were observed in the expression levels of these genes between the transgenic and WT plants. However, when exposed to 100 mM NaCl, the transgenic lines exhibited significantly higher expression levels than the WT plants. These findings suggest that the molecular function of OsSIRH2-3 is associated with the ABA-dependent signaling pathway in rice.
Abscisic acid (ABA) sensitivity. Seeds of OsSIRH2-3-overexpressing and WT plants were grown on each half of MS agar mediums containing a 0, b 3, or c 5 µM ABA. Experiments were performed with three biological replicates (each sample number = 15). Orange triangle for WT, green circle for OE-10, and blue square for OE-17, respectively. D: day. Student’s t-test: ns, non-significant **P < 0.01, or ***P < 0.001
Analysis of relative expression patterns of ABA-dependent pathway-related genes in leaves (Top) and roots (Bottom) of OsSIRH2-3-overexpressing and WT plants with (100 mM) or without NaCl. The samples were harvested at 10 d without NaCl treatment or after 100 mM NaCl treatment, respectively. a, e OsLEA3, b, f OsProT, c, g OsP5Cs, (d, h) OsNAC5. q-PCR was performed with three technical and biological replicates (each sample number = 15). Each of the gene expression levels was normalized to that of OseEF-1a. Student’s t-test: ns, non-significant **P < 0.01 or ***P < 0.001
Na+ and K+ Accumulation in OsSIRH2-3-Overexpressing Plants Under Salt stress
Under saline conditions, the accumulation of Na+ induces K+ deficiency, leading to an ionic imbalance in cells and the accumulation of Na+ which ultimately causes cellular damage in plants. Under salt stress, homeostasis of K+ plays an important role in maintaining the cell membrane turgor pressure, enzyme activity, and plant development (Rahman et al. 2017).To evaluate the Na+ and K+ concentrations in the leaves, roots, and xylem sap of OsSIRH2-3-overexpressing and WT plants, we conducted CoroNa-green (Na+-specific indication) staining and ICP analysis with (100 mM) and without NaCl.
To visualize the accumulation of Na+ in root cells, we conducted CoroNa-green staining of OsSIRH2-3-overexpressing and WT plants. Without NaCl treatment, no differences in the green signal and fluorescence intensity were observed between the root tips of OsSIRH2-3-overexpressing and WT plants (Fig. 8a, c). However, the green signal and fluorescence intensity of OsSIRH2-3-overexpressing plants were lower than those of WT plants under 100 mM NaCl stress (Fig. 8b, c).
Coro-Na green staining and ICP analysis of OsSIRH2-3-overexpressing and WT plants. For ICP analysis, ten-day-old rice seedlings of plants were treated with 0 or 100 mM for 7 d. Coro-Na green staining of root tip tissues in OsSIRH2-3-overexpressing and WT plants without or with 100 mM NaCl. Ten-day-old rice seedlings were treated without or with 100 mM NaCl for 24 h. All experiments were performed with three biological replicates (each sample number = 6). Scale bar = 50 μm. a, b Images of the green signal of root tips in three-genotype plants under 0 or 100 mM NaCl. c Green fluorescence intensity in three-genotype plants in response to treatment with 0 and 100 mM NaCl. For ICP analysis, the dried leaf and root samples were used to determine the concentration of Na+ and K+ via ICP-OES with three biological replicates (each sample number = 10). The xylem sap was collected from ten-day-old seedlings of the plants without or with (100 mM) NaCl for 12 h. Collected samples were measured via ICP-OES with three biological replicates (each sample number = 10). a, d, j Na+, b, e, k K+, and c, f, l Na+/K+ ratio. The asterisks indicate a significant difference according to the student’s t-test; ns, non-significant ***P < 0.001
Without NaCl, OsSIRH2-3-overexpressing and WT plants showed no significant differences in Na+ and K+ concentrations in the leaves, roots, and leaf xylem sap (Fig. 8d, e, g, h, j, k). In contrast, under 100 mM NaCl, OsSIRH2-3-overexpressing plants exhibited lower Na+ and higher K+ levels in the leaves and roots than the WT plants (Fig. 8d, e, g, h, j, k). In addition, the Na+/K+ ratios in the leaves, roots, and xylem sap were not significantly different than without NaCl. However, in 100 mM NaCl, OsSIRH2-3-overexpressing plants maintained a lower Na+/K+ ratio than WT plants in all tissues tested (Fig. 8f, i, l).
Expression of Na+/K+ Transporter Genes
To gain further insight into the mechanism of salt tolerance in OsSIRH2-3-overexpressing transgenic plants, we quantified the expression of 10 genes encoding Na+ and K+ transporters using qRT-PCR in both the leaves and roots of transgenic and WT plants in the presence (100 mM) or absence of NaCl for 0, 3, 6, 12, and 24 h. The genes included OsHKT1;4, OsHKT1;5, OsHKT2;1, OsAKT1, OsNHX1, OsSOS1, OsHAK2, OsHAK7, OsGORK and OsSKOR, and were selected based on previous reports (Platten et al. 2006; Horie et al. 2007; Martínez-Atienza et al. 2007; Yao et al. 2010; Hamamoto et al. 2015; Shen et al. 2015; Suzuki et al. 2016; Kobayashi et al. 2017; Li et al. 2018; Adem et al. 2020; Yan et al. 2021).
Under normal conditions, no significant differences in gene expression were observed between the leaves and roots of transgenic and WT plants. However, under salt stress, all of the genes showed significant differences in transcript levels (Fig. 9).
Relative expression patterns of Na+/K+ transporter genes in the OsSIRH2-3-overexpressing and WT plants. The ten-day-old rice seedlings were treated without or with (100 mM) NaCl for 0, 3, 6, 12, and 24 h, respectively. The leaves (top) and roots (bottom) expressions were represented. a, k OsHKT1;4 b, l OsHKT1;5, c, m OsHKT2;1, d, n OsAKT1, e, o NHX1, f, p OsSOS1, g, q OsHAK2, h, r OsHAK7, i, s OsGORK, and j, t OsSKOR. qRT-PCR was performed with three biologicals and three technical replicates. OseEF-1a was used as an internal control. The asterisks indicate a significant difference according to the Student’s t-test: ns, non-significant *P < 0.05, **P < 0.01, or ***P < 0.001
The expression levels of Na+ transporter genes OsHKT1;4 and OsHKT1;5 were lower in OsSIRH2-3-overexpressing plants than in WT plants from 1 to 24 h (Fig. 9a, b, k, l). Another Na+ transporter gene, OsHKT2;1, was significantly down-regulated in OsSIRH2-3-overexpressing plants compared to WT plants (Fig. 9c, m). OsAKT1 showed decreased expression in the leaves and roots of both plants. However, a consistently lower expression was observed in transgenic plants, unlike in WT plants under salt stress (Fig. 9d, n). The expression level of OsNHX1, a Na+ antiporter gene, reached maximum at 3 h, then decreased up to 24 h in the leaves and roots of both plants. Its expression was also higher in transgenic plants than in WT plants (Fig. 9e, o). The Na+/H+ antiporter gene OsSOS1 was highly expressed in OsSIRH2-3-overexpressing plants compared to that in WT plants under salt stress (Fig. 9f, p).
In contrast, the K+ transporter genes OsHAK2 and OsHAK7 were highly expressed in OsSIRH2-3-overexpressing plants compared to WT plants under salt stress (Fig. 9g, h, m, n). The potassium efflux channel gene OsGORK showed lower expression in the leaves and roots of transgenic lines than in WT plants (Fig. 9i, s). The expression level of the potassium channel gene OsSKOR was also higher in both tissues of transgenic plants than in those of WT plants (Fig. 9j, t). Taken together, these results suggested that OsSIRH2-3 facilitates salt tolerance by modulating the expression of Na+/K+ transporter genes and thereby maintaining Na+/K+ homeostasis.
Discussion
High-salinity soils pose a major environmental challenge, contributing to 20–50% reductions in global crop yields. Soil salinity has been increasing in many countries due to global warming. Plants have evolved various mechanisms to increase their tolerance to salt stress (Shrivastava and Kumar 2015). The ubiquitin–proteasome system (UPS) mediated by RING E3 ubiquitin ligases is a regulator of salt stress tolerance (Al-Saharin et al. 2022). For example, E3 ubiquitin ligase RING finger protein OsSIRH2-14 regulates levels of the Na+ transporter OsHKT2;1 via the 26S proteasome. OsSIRH2-14-overexpressing plants also show increased salt tolerance (Park et al. 2019). Here, we found increased salt tolerance in OsSIRH2-3-overexpressing rice. Hence, OsSIRH2-3 E3 ubiquitin ligase is also a regulator of salt tolerance in rice.
Maintaining K+ and Na+ homeostasis is crucial to achieving salt tolerance (Noreen et al. 2010; Abbasi et al. 2016; Almeida et al. 2017). OsSIRH2-3 overexpression under salt stress decreased the Na+/K+ ratio. The expression levels of Na+ and/or K+ transporters were significantly affected by OsSIRH2-3 overexpression. Hence, OsSIRH2-3 also maintains Na+/K+ homeostasis by modulating the expression of Na+ and/or K+ transporters.
The HKT family of proteins in plants includes high-affinity K+-uptake and Na+-selective transporters (Hamamoto et al. 2015). HKT proteins are categorized into two classes: HKT1 and HKT2 HKT1 is involved in Na+-selective and Na+/K+ transport (Platten et al. 2006). Previous reports have demonstrated that overexpression of an HKT1 Na+ transporter, OsHKT1;4, was found to result in weaker resistance to salt stress (Suzuki et al. 2016; Oda et al. 2018). Similarly, we found that OsHKT1;4 expression was lower in OsSIRH2-3-overexpressing plants than in WT plants under salt stress. Therefore, low OsHKT1;4 expression in OsSIRH2-3-overexpressing plants contributes to improved salt tolerance by regulating Na+ transport. Another Na+ transporter, OsHKT1;5, prevents the transfer of Na+ to shoots in rice (Kobayashi et al. 2017). In OsSIRH2-3-overexpressing plants, the low expression level of OsHKT1;5 may also lead to low Na+ content in the xylem under salt stress. OsHKT2;1 of the HKT2 family is directly involved in Na+ influx in the root, with reduced expression levels under salt stress (Horie et al. 2007; Zhang et al. 2018). The low expression of OsHKT2;1 in OsSIRH2-3-overexpressing plants under high salinity is consistent with previous reports. This also suggests that OsSIRH2-3 overexpression inhibits Na+ entry into the root epidermis, and thereby improves salt tolerance.
SOS1 and NHX1 are Na+/H+ exchangers located in the plasma membrane and tonoplast, respectively. These antiporters are involved in the export and sequestration of Na+ under salt stress (Zhang et al. 2018). OsNHX1, which performs the selective transport of Na+ into vacuoles depending on the salt concentration in the cell, was highly expressed in the leaves and roots of OsSIRH2-3-overexpressing plants under salt stress. Also, OsSOS1 expression in leaves and roots was significantly upregulated in transgenic plants. High levels of expression of these antiporters in OsSIRH2-3-overexpressing plants under salt stress led to the efflux of Na+ from the cytosol and improve salt tolerance.
In plants, K+ transporters are vital for maintaining intercellular K+ homeostasis, which is also closely associated with salt tolerance (Shen et al. 2015). This gene family encodes various types of transporters, including shaker-like and KUP/HAK/KT transporters (Mäser et al. 2001; Véry and Sentenac. 2003). In rice, the HAK family of K+ transporters plays a significant role in regulating the K+ uptake in response to salt stress (Li et al. 2018). Understanding the functions of HAK proteins in rice may provide valuable insights into plant salt tolerance mechanisms. OsAKT1, which belongs to the Shaker family, plays a vital role in K+ uptake from the soil (Li et al. 2014; Wu et al. 2019). In rice, OsAKT1 is also downregulated in salt-tolerant variants (Golldack et al. 2003; Fuchs et al. 2005; Ahmad et al. 2016). Similarly, we found a significantly lower expression of OsAKT1 under salt stress in OsSIRH2–3-overexpressing rice. Thus, OsSIRH2–3 overexpression is also associated with the OsAKT1 expression, potentially affecting K+ uptake in the roots and influencing the Na+/K+ balance under salt stress conditions.
In rice, OsHAK transporter genes are involved in K+ uptake and thus tolerance to salt stress through changes in gene expression patterns (Okada et al. 2008). OsHAK2 was also previously found to be involved in the transport of both Na+ and K+, and its K+ transport activity was inhibited by high Na+ concentrations (Horie et al. 2011). Our finding of high levels of OsHAK2 expression in OsSIRH2-3-overexpressing plants under salt stress is consistent with previous findings. Another K+ transporter gene, OsHAK7, showed higher expression in OsSIRH2-3-overexpressing than in the WT plants. This is also consistent with the increased accumulation of K+ in OsSIRH2-3-overexpressing plants.
Shaker-like K+ channels such as OsGORK and OsSKOR are key regulators of K+ homeostasis in plants (Hedrich 2012). OsGORK, an outward-rectifying potassium efflux channel, plays a central role in stress-induced K+ efflux from the cytosol (Adem et al. 2020). We observed decreased expression of OsGORK in both the roots and leaves of transgenic plants under salt stress, which may explain high K+ levels in transgenic plants. OsSKOR functions in root stellar tissues and is responsible for loading K+ into the xylem (Kim et al. 2015). We observed higher expression of OsSKOR in both the leaves and roots of transgenic plants than in WT plants under salt stress. Upregulation of OsSKOR may contribute to enhanced K+ loading into the xylem, thereby maintaining K+ homeostasis in transgenic plants. Another hypothesis proposed to explain the increased K+ retention is based on the reduction in ROS-activated non-selective cation channels due to the low levels of ROS in the transgenic lines under salt stress (Demidchik et al. 2010). Decreased ROS levels in transgenic plants may inhibit non-selective cation channels, resulting in better K+ retention under salt stress. Taken together, the differential expression patterns of OsGORK and OsSKOR may lead to the modulation of ROS-activated non-selective cation channels and contribute to the regulation of K+ content in the transgenic plants under salt stress.
ROS are often generated under extreme conditions, and their accumulation has detrimental effects on plant growth, leading to growth inhibition and cell death (Das and Roychoudhury 2014). Plants have evolved scavenging systems to prevent ROS accumulation (Pang and Wang 2008). We found that OsSIRH2-3-overexpressing plants exhibit higher activities of antioxidant enzymes, including SOD, POD, and CAT, than WT plants. The increased activities of SOD, POD, and CAT, along with the decreased H2O2 content in OsSIRH2-3-overexpressing plants, may lead to an improved ability to scavenge ROS, thus preventing their accumulation. Despite the overexpression of OsSIRH2-3, no significant changes in antioxidant activity were observed for several factors under unstressed conditions; thus, OSIRH2-3 might not directly regulate antioxidant enzyme activity or production. Alternatively, the basal antioxidant capacity of the plant may be sufficiently robust under unstressed conditions, resulting in no changes in OsSIRH2-3 overexpression. Further studies are required to investigate specific antioxidant pathways and their regulation by OsSIRH2-3.
The accumulation of compatible solutes, such as proline and soluble sugars, also confer tolerance to salt (Hayat et al. 2012). Increased accumulation of proline and soluble sugars in OsSIRH2-3-overexpressing plants under salt stress is consistent with previous reports.
Another indicator of plant stress response is lipid peroxidation through MDA production under salt stress (Liang et al. 2018). Low MDA content is an indicator of salt-tolerance (Xiong et al. 2014; Tang et al. 2019). We also found lower MDA levels in OsSIRH2-3-overexpressing plants than in WT plants. These results suggest that the overexpression of OsSIRH2-3 contributes to a reduction in lipid peroxidation and membrane damage, thereby enhancing salt tolerance.
ABA plays a crucial role in the response to salt stress and accumulates rapidly under extreme stress conditions. ABA regulates various salt tolerance mechanisms. Increased ABA levels through the regulation of ABA biosynthesis genes have a positive effect on salt tolerance (Zhang et al. 2006; Sah et al. 2016). ABA sensitivity is also closely associated with salt tolerance in plants (Xu et al. 2011; Zhang et al. 2020). Our findings regarding the high ABA sensitivity during the germination of OsSIRH2-3-overexpressing plants, and the high expression levels of ABA signaling pathway-related genes, suggest that the overexpression of OsSIRH2-3 may improve salt tolerance through ABA-dependent pathways in rice. The unchanged ABA sensitivity and expression patterns of ABA-dependent genes in OsSIRH2-3 overexpressing lines under unstressed conditions indicated that OsSIRH2-3 may not modulate ABA signaling pathways or downstream gene expression without exogenous stressors, suggesting that the role of OsSIRH2-3 in stress responses may be context-dependent, with its effects on ABA signaling being more pronounced under stress conditions.
In conclusion, the findings of this study regarding the differential expression of stress-responsive genes, such as OsSIRH2-3 under various stress conditions have implications for breeding and engineering crops with enhanced resilience to climate change-induced stresses, contributing to the promotion of environmentally sustainable agricultural practices. We investigated the molecular mechanisms responsible for the role of OsSIRH2-3 in response to salt stress in transgenic rice plants. Overexpression of OsSIRH2-3 resulted in reduced Na+ accumulation in both leaves and roots, increased activity of ROS-scavenging enzymes, high germination rates, and increased sensitivity to ABA. Interestingly, we also observed significant changes in the expression of Na+/K+ transporter genes, suggesting that OsSIRH2-3 increases salt tolerance, and provides a potential strategy for enhancing crop productivity in high-salinity environments. Unfortunately, we were not able to detect direct evidence of the mechanism behind the regulation of expression of Na+/K+ transporter genes through the overexpression of OsSIRH2-3. An attempt to use yeast two-hybrid screening failed to identify the substrate(s) of OsSIRH2-3 (data not shown). Therefore, further investigations should focus on identifying protein (s) that interact with OsSIRH2-3 to understand the molecular mechanisms involved in RING E3 ubiquitin ligases.
Data Availability
All data generated or analyzed during this study are included in this published article. The data shown in this article are available from the corresponding authors upon a reasonable request.
Code Availability
Not Available.
References
Abbasi H, Jamil M, Haq A et al (2016) Salt stress manifestation on plants, mechanism of salt tolerance and potassium role in alleviating it: a review. Zemdirbyste-Agric 103:229–238. https://doi.org/10.13080/z-a.2016.103.030
Adem GD, Chen G, Shabala L et al (2020) GORK channel: a master switch of plant metabolism? Trends Plant Sci 25:434–445. https://doi.org/10.1016/j.tplants.2019.12.012
Ahmad I, Mian A, Maathuis FJM (2016) Overexpression of the rice AKT1 potassium channel affects potassium nutrition and rice drought tolerance. J Exp Bot 67:2689–2698. https://doi.org/10.1093/jxb/erw103
Ali Y, Aslam Z, Ashraf MY, Tahir GR (2004) Effect of salinity on chlorophyll concentration, leaf area, yield and yield components of rice genotypes grown under saline environment. Int J Environ Sci Technol 1:221–225. https://doi.org/10.1007/bf03325836
Almeida DM, Oliveira MM, Saibo NJM (2017) Regulation of Na+ and K+ homeostasis in plants: towards improved salt stress tolerance in crop plants. Genet Mol Biol 40:326–345. https://doi.org/10.1590/1678-4685-gmb-2016-0106
Al-Saharin R, Hellmann H, Mooney S (2022) Plant E3 ligases and their role in abiotic stress response. Cells 11:890. https://doi.org/10.3390/cells11050890
Ariyarathna HA, Ul-Haq T, Colmer TD, Francki MG (2014) Characterization of the multigene family TaHKT 2; 1 in bread wheat and the role of gene members in plant Na+ and K+ status. BMC Plant Biol 14:1–15. https://doi.org/10.1186/1471-2229-14-159
Bassil E, Tajima H, Liang Y-C, Ohto M-A, Ushijima K, Nakano R, Esumi T, Coku A, Belmonte M, Blumwald E (2011) The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell 23:3482–3497. https://doi.org/10.1105/tpc.111.089581
Carillo P, Gibon Y (2011) Protocol: extraction and determination of proline. PrometheusWiki 2011:1–5
Chapagain S, Park YC, Jang CS (2017) Functional diversity of RING E3 ligases of major cereal crops in response to abiotic stresses. J Crop Sci Biotechnol 20:351–357. https://doi.org/10.1007/s12892-017-0104-0
Chinnusamy V, Jagendorf A, Zhu JK (2005) Understanding and improving salt tolerance in plants. Crop Sci 45:437–448. https://doi.org/10.2135/CROPSCI2005.0437
Danquah A, De Zélicourt A, Colcombet J, Hirt H (2014) The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol Adv 32:40–52. https://doi.org/10.1016/j.biotechadv.2013.09.006
Das K, Roychoudhury A (2014) Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front Environ Sci 2:53. https://doi.org/10.3389/fenvs.2014.00053
Demidchik V, Tester M (2002) Sodium fluxes through nonselective cation channels in the plasma membrane of protoplasts from Arabidopsis roots. Plant Physiol 128:379–387. https://doi.org/10.1104/pp.010524
Demidchik V, Cuin TA, Svistunenko D, Smith SJ, Miller AJ, Shabala S, Sokolik A, Yurin V (2010) Arabidopsis root K+-efflux conductance activated by hydroxyl radicals: single-channel properties, genetic basis and involvement in stress-induced cell death. J Cell Sci 123:1468–1479. https://doi.org/10.1242/jcs.064352
Deshaies RJ, Joazeiro CAP (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78:399–434. https://doi.org/10.1146/annurev.biochem.78.101807.093809
Finkelstein RR, Gampala SSL, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14:S15–S45. https://doi.org/10.1105/tpc.010441
Froger A, Hall JE (2007) Transformation of plasmid DNA into E. coli using the heat shock method. J vis Exp. https://doi.org/10.3791/253
Fuchs I, Stölzle S, Ivashikina N, Hedrich R (2005) Rice K+ uptake channel OsAKT1 is sensitive to salt stress. Planta 221:212–221. https://doi.org/10.1007/s00425-004-1437-9
Fukase E, Martin W (2020) Economic growth, convergence, and world food demand and supply. World Dev 132:104954
Gay C, Gebicki JM (2000) A critical evaluation of the effect of sorbitol on the ferric–xylenol orange hydroperoxide assay. Anal Biochem 284:217–220. https://doi.org/10.1006/abio.2000.4696
Gerona MEB, Deocampo MP, Egdane JA, Ismali AM, Dionisio-sese ML (2019) Physiological responses of contrasting rice genotypes to salt stress at reproductive stage. Rice Sci 26:207–219. https://doi.org/10.1016/j.rsci.2019.05.001
Golldack D, Quigley F, Michalowski CB, Kamasani UR, Bohnert HJ (2003) Salinity stress-tolerant and-sensitive rice (Oryza sativa L.) regulate AKT1-type potassium channel transcripts differently. Plant Mol Biol 51:71–81. https://doi.org/10.1023/A:1020763218045
Hamamoto S, Horie T, Hauser F, Deinlein U, Schroeder JI, Uozumi N (2015) HKT transporters mediate salt stress resistance in plants: from structure and function to the field. Curr Opin Biotechnol 32:113–120. https://doi.org/10.1016/j.copbio.2014.11.025
Hayat S, Hayat Q, Nasser Alyemeni M, Wani AS, Pichtel J, Ahmad A (2012) Role of proline under changing environments: a review. Plant Signal Behav. https://doi.org/10.4161/psb.21949
Hedrich R (2012) Ion channels in plants. Physiol Rev 92:1777–1811. https://doi.org/10.1152/physrev.00038.2011
Horie T, Costa A, Kim TH, Han MJ, Horie R, Leung H-Y, Miyao A, Hirochika H, An G, Schroeder JI (2007) Rice OsHKT2; 1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO J 26:3003–3014. https://doi.org/10.1038/sj.emboj.7601732
Horie T, Sugawara M, Okada T, Taira K, Nakayama PK, Katsuhara M, Shinmyo A, Nakayama H (2011) Rice sodium-insensitive potassium transporter, OsHAK5, confers increased salt tolerance in tobacco BY2 cells. J Biosci Bioeng 111:346–356. https://doi.org/10.1016/j.jbiosc.2010.10.014
Hossain MS, Dietz KJ (2016) Tuning of redox regulatory mechanisms, reactive oxygen species and redox homeostasis under salinity stress. Front Plant Sci 7:548. https://doi.org/10.3389/fpls.2016.00548
Janz D, Polle A (2012) Harnessing salt for woody biomass production. Tree Physiol 32:1–3. https://doi.org/10.1093/treephys/tpr127
Ji H, Pardo JM, Batelli G, Oosten MV, Bressan RA, Li X (2013) The salt overly sensitive (SOS) pathway: established and emerging roles. Mol Plant 6:275–286. https://doi.org/10.1093/mp/sst017
Jia W, Wang Y, Zhang S, Zhang J (2002) Salt-stress-induced ABA accumulation is more sensitively triggered in roots than in shoots. J Exp Bot 53:2201–2206. https://doi.org/10.1093/jxb/erf079
Kim HY, Choi E-H, Min MK, Hwang H, Moon SJ, Yoon I, Byun MO, Kim BG (2015) Differential gene expression of two outward-rectifying shaker-like potassium channels OsSKOR and OsGORK in rice. J Plant Biol 58:230–235. https://doi.org/10.1007/s12374-015-0070-4
Kim JH, Lim SD, Jung K-H, Jang CS (2023) Overexpression of a C3HC4-type RING E3 ligase gene, OsRFPHC-13, improves salinity resistance in rice, Oryza sativa, by altering the expression of Na+/K+ transporter genes. Environ Exp Bot 207:105224. https://doi.org/10.1016/j.envexpbot.2023.105224
Kobayashi NI, Yamaji N, Yamamoto H et al (2017) OsHKT1; 5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J 91:657–670. https://doi.org/10.1111/tpj.13595
Li J, Long Y, Qi G-N, Li J, Xu ZJ, Wu WH, Wang Y (2014) The Os-AKT1 channel is critical for K+ uptake in rice roots and is modulated by the rice CBL1-CIPK23 complex. Plant Cell 26:3387–3402. https://doi.org/10.1105/tpc.114.123455
Li W, Xu G, Alli A, Yu L (2018) Plant HAK/KUP/KT K+ transporters: function and regulation. Seminars in cell & developmental biology. Elsevier, pp 133–141
Liang W, Ma X, Wan P, Liu L (2018) Plant salt-tolerance mechanism: a review. Biochem Biophys Res Commun 495:286–291. https://doi.org/10.1016/j.bbrc.2017.11.043
Lim SDON, Hwang J-G, Jung CGYO, Hwang S-G, Moon JC, Jang CS (2013) Comprehensive analysis of the rice RING E3 ligase family reveals their functional diversity in response to abiotic stress. DNA Res 20:299–314. https://doi.org/10.1093/dnares/dst011
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25:402–408
Martínez-Atienza J, Jiang X, Garciadeblas B, Mendoza I, Zhu JK, Pardo JM, Quintero FJ (2007) Conservation of the salt overly sensitive pathway in rice. Plant Physiol 143:1001–1012. https://doi.org/10.1104/pp.106.092635
Maser P, Thomine S, Schroeder JI et al (2001) Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol 126:1646–1667. https://doi.org/10.1104/pp.126.4.1646
Millar N (2001) Biology statistics made simple using Excel. Sch Sci Rev 83:23–34
Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7:405–410. https://doi.org/10.1016/s1360-1385(02)02312-9
Monsur MB, Ivy NA, Haque MM, Hasanuzzaman M, Sabagh AE, Rohman MM (2020) Oxidative stress tolerance mechanism in rice under salinity. Phyton (b Aires) 89:497
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681. https://doi.org/10.1146/annurev.arplant.59.032607.092911
Nemati I, Moradi F, Gholizadeh S, Esmaeili MA, Bihamta MR (2011) The effect of salinity stress on ions and soluble sugars distribution in leaves, leaf sheaths and roots of rice (Oryza sativa L.) seedlings. Plant Soil Environ 57:26–33
Ni Z, Kim E-D, Chen ZJ (2009) Chlorophyll and starch assays. https://doi.org/10.1038/nprot.2009.12
Noreen Z, Ashraf M, Akram NA (2010) Salt-induced regulation of some key antioxidant enzymes and physio-biochemical phenomena in five diverse cultivars of turnip (Brassica rapa L.). J Agron Crop Sci 196:273–285. https://doi.org/10.1111/j.1439-037x.2010.00420.x
Oda Y, Kobayashi NI, Tanoi K, Ma JF, Itou Y, Katsuhara M, Itou T, Horie T (2018) T-DNA tagging-based gain-of-function of OsHKT1; 4 reinforces Na exclusion from leaves and stems but triggers Na toxicity in roots of rice under salt stress. Int J Mol Sci 19:235. https://doi.org/10.3390/ijms19010235
Okada T, Nakayama H, Shinmyo A, Yoshida K (2008) Expression of OsHAK genes encoding potassium ion transporters in rice. Plant Biotechnol 25:241–245. https://doi.org/10.5511/plantbiotechnology.25.241
Pang C-H, Wang B-S (2008) Oxidative stress and salt tolerance in plants. Springer, pp 231–245. https://doi.org/10.1007/978-3-540-72954-9_9
Park YC, Lim SD, Moon J, Jang CS (2019) A rice really interesting new gene H 2-type E 3 ligase, OsSIRH2-14, enhances salinity tolerance via ubiquitin/26 S proteasome-mediated degradation of salt-related proteins. Plant Cell Environ 42:3061–3076. https://doi.org/10.1111/pce.13619
Platten JD, Cotsaftis O, Berthomieu P et al (2006) Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci 11:372–374. https://doi.org/10.1016/j.tplants.2006.06.001
Polle A, Chen S (2015) On the salty side of life: molecular, physiological and anatomical adaptation and acclimation of trees to extreme habitats. Plant Cell Environ 38:1794–1816. https://doi.org/10.1111/pce.12440
Rahman A, Nahar K, Mahmud JA, Hasanuzzaman M, Hossain MS, Fujita M (2017) In international rice, undefined salt stress tolerance in rice: emerging role of exogenous phytoprotectants. books.google.com
Sah SK, Reddy KR, Li J (2016) Abscisic acid and abiotic stress tolerance in crop plants. Front Plant Sci 7:571. https://doi.org/10.3389/fpls.2016.00571
Shen Y, Shen L, Shen Z, Jing W, Ge H, Zhao J, Zhang W (2015) The potassium transporter O s HAK 21 functions in the maintenance of ion homeostasis and tolerance to salt stress in rice. Plant Cell Environ 38:2766–2779. https://doi.org/10.1111/pce.12586
Shrivastava P, Kumar R (2015) Soil salinity: a serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J Biol Sci 22:123–131. https://doi.org/10.1016/j.sjbs.2014.12.001
Song SY, Chen Y, Chen J, Dai XY, Zhang WH (2011) Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 234:331–345. https://doi.org/10.1007/S00425-011-1403-2
Su Y, Luo W, Lin W, Ma L, Kabir MH (2015) Model of cation transportation mediated by high-affinity potassium transporters (HKTs) in higher plants. Biol Proced Online 17:1–13. https://doi.org/10.1186/s12575-014-0013-3
Sun L, Lei P, Wang Q, Ma J, Zhan Y, Jiang K, Xu Z, Xu H (2020) The endophyte Pantoea alhagi NX-11 alleviates salt stress damage to rice seedlings by secreting exopolysaccharides. Front Microbiol 10:3112. https://doi.org/10.3389/fmicb.2019.03112
Suzuki K, Yamaji N, Costa A, Okuma E, Kobayashi NI, Kashiwagi T, Katsuhara M, Wang C, Tanoi K, Murata Y, Schroeder JI, Ma JF, Horie T (2016) OsHKT1; 4-mediated Na+ transport in stems contributes to Na+ exclusion from leaf blades of rice at the reproductive growth stage upon salt stress. BMC Plant Biol 16:1–15. https://doi.org/10.1186/s12870-016-0709-4
Tang Y, Bao X, Zhi Y et al (2019) Overexpression of a MYB family gene, OsMYB6, increases drought and salinity stress tolerance in transgenic rice. Front Plant Sci 10:168. https://doi.org/10.3389/fpls.2019.00168
Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J 47:969–976
Véry A-A, Sentenac H (2003) Molecular mechanisms and regulation of K+ transport in higher plants. Annu Rev Plant Biol 54:575–603. https://doi.org/10.1146/annurev.arplant.54.031902.134831
Waters S, Gilliham M, Hrmova M (2013) Plant high-affinity potassium (HKT) transporters involved in salinity tolerance: structural insights to probe differences in ion selectivity. Int J Mol Sci 14:7660–7680. https://doi.org/10.3390/ijms14047660
Wei M, Zhuang Y, Li H, Li P, Huo H, Shu D, Huang W, Wang S (2020) The cloning and characterization of hypersensitive to salt stress mutant, affected in quinolinate synthase, highlights the involvement of NAD in stress-induced accumulation of ABA and proline. Plant J 102:85–98. https://doi.org/10.1111/tpj.14613
Wu L-B, Holtkamp F, Wairich A, Frei M (2019) Potassium ion channel gene OsAKT1 affects iron translocation in rice plants exposed to iron toxicity. Front Plant Sci 10:579. https://doi.org/10.3389/fpls.2019.00579
Xiong L, Schumaker KS, Zhu J-K (2002) Cell signaling during cold, drought, and salt stress. Plant Cell 14:S165–S183. https://doi.org/10.1105/tpc.000596
Xiong H, Li J, Liu P, Duan J, Zhao Y, Guo X, Li Y, Zhang H, Ali J, Li Z (2014) Overexpression of OsMYB48-1, a novel MYB-related transcription factor, enhances drought and salinity tolerance in rice. PLoS ONE 9:e92913. https://doi.org/10.1371/journal.pone.0092913
Xu G-Y, Rocha PSCF, Wang M-L, Xu ML, Cui YC, Li LY, Zhu YX, Xia X (2011) A novel rice calmodulin-like gene, OsMSR2, enhances drought and salt tolerance and increases ABA sensitivity in Arabidopsis. Planta 234:47–59. https://doi.org/10.1007/s00425-011-1386-z
Yan G, Fan X, Zheng W, Gao Z, Yin C, Li T, Liang Y (2021) Silicon alleviates salt stress-induced potassium deficiency by promoting potassium uptake and translocation in rice (Oryza sativa L.). J Plant Physiol 258:153379. https://doi.org/10.1016/j.jplph.2021.153379
Yao X, Horie T, Xue S, Leung HY, Katsuhara M, Brodsky DE, WuSchroeder Y (2010) Differential sodium and potassium transport selectivities of the rice OsHKT2; 1 and OsHKT2; 2 transporters in plant cells. Plant Physiol 152:341–355. https://doi.org/10.1104/pp.109.145722
Yoshida T, Mogami J, Yamaguchi-Shinozaki K (2014) ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr Opin Plant Biol 21:133–139. https://doi.org/10.1016/j.pbi.2014.07.009
Zhang J, Jia W, Yang J, Ismail AM (2006) Role of ABA in integrating plant responses to drought and salt stresses. Field Crops Res 97:111–119. https://doi.org/10.1016/j.fcr.2005.08.018
Zhang Y, Fang J, Wu X, Dong L (2018) Na + /K + balance and transport regulatory mechanisms in weedy and cultivated rice (Oryza sativa L.) under salt stress. BMC Plant Biol. https://doi.org/10.1186/S12870-018-1586-9
Zhang X, Long Y, Huang J, Xia J (2020) OsNAC45 is involved in ABA response and salt tolerance in rice. Rice 13:1–13. https://doi.org/10.1186/s12284-020-00440-1
Acknowledgements
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) and funded by the Ministry of Education, Science, and Technology (grant no. 2019R1A2C1009840).
Funding
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) and funded by the Ministry of Education, Science, and Technology (grant no. 2019R1A2C1009840).
Author information
Authors and Affiliations
Contributions
CSJ conceived the study. MSC and JHK conducted the experiments and analyzed the data. MSC and CSJ drafted the manuscript. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethical Approval
Not Available.
Consent to Participate
Not Available.
Consent for Publication
Not Available.
Supplementary Information
Below is the link to the electronic supplementary material.
12374_2024_9433_MOESM1_ESM.pptx
Figure S1. Relative expression levels of abiotic stress and hormone marker gene. The ten-day-old rice seedlings were treated with two abiotic stress (100 mM NaCl and 20 % PEG) and 50 µM ABA for 0, 1, 6, 12, and 24h. (a) The expression level of OsNAC10 under 100 mM NaCl. (b) The expression level of OsSalT under 20 % PEG. (c) The expression level of OsRAB16a 50 μM ABA. The OseEF-1a was used as an internal control. The expressional values are calculated with three biological replicates (each sample number =10). The OseEF-1a (LOC_Os03g08010) gene was used as an internal control. The asterisks indicate a significant difference according to the student’s t-test; ***P < 0.001. Figure S2. q-PCR analysis of six independent OsSIRH2-3-overexpressing lines. WT plants were used as controls. The expressional values are calculated with three biological replicates (each sample number =10). The OseEF-1a (LOC_Os03g08010) gene was used as an internal control. The asterisks indicate a significant difference according to the student’s t-test; ns: non-significant; or ***P < 0.001. Figure S3. Representative image of total chlorophyll extraction. Ten-day-old leaves of OsSIRH2-3-overexpressing and WT plants were used. The experiments were conducted on three biological replicates (each sample number =15). Supplementary file1 (PPTX 1367 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Choi, M.S., Kim, J.H. & Jang, C.S. Rice H2-Type RING E3 Ligase Gene, OsSIRH2-3, Positively Regulates Salt Tolerance by Maintaining Na+/K+ Homeostasis. J. Plant Biol. 67, 283–298 (2024). https://doi.org/10.1007/s12374-024-09433-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12374-024-09433-9