Abstract
The production of fertile inflorescences is essential to make crosses for the development of new varieties of sugarcane. Within the Saccharum species and commercial hybrids of sugarcane, there is a wide range in flowering times and some varieties rarely flower; this can make it impossible to achieve some desired crosses. While photoperiod houses can assist with inducing some varieties to flower or synchronise flowering, understanding the expression of genes within the flowering pathway could provide an explanation for such variation. To this end experiments were designed to assess the photoperiod conditions within a controlled environment facility and measure changes in expression of 18 genes associated with flowering during the initial four weeks of the floral induction period at three time points across the day-light cycle. Two sugarcane varieties were selected for their differences in flowering propensity, Q208 and Q183, flowering and non-flowering, respectively. The 18 genes examined were associated with photoperiod perception, the circadian clock, the floral pathway, and floral meristem identity. Of these, 16 were detected and showed differences in expression during the light cycle and over the 4 weeks under photoperiod induction in Q208. The assessment of a subset of those genes in Q183 showed higher expression levels of ShLHY, ShCO and ShTFL1 in Q183 than Q208. One of the genes, TFL1 is a known flowering inhibitor of the penultimate step prior to floral induction. These gene profiles build on the proposed sugarcane flowering pathway and highlight key genes for manipulation to promote and synchronise flowering to assist sugarcane breeding programmes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sugarcane is grown in the tropics and subtropics, contributing 86% of the world’s sugar production with the remainder coming from sugar beets (OCED-FAO 2019). Since the late nineteenth century sugarcane breeders have produced new cultivars with improved traits, with the first step being the production of sugarcane flowers to enable crosses. Crosses are limited to the varieties with overlapping flowering time; for example, varieties that flower early are not available to cross with varieties that flower late (Durai et al. 2014). Plants have several pathways that can induce the production of flowers, including photoperiod, gibberellins, vernalisation, and autonomous/endogenous signalling (Mouradov et al. 2002; Albani and Coupland 2010; Fornara et al. 2010; Song et al. 2013). In addition, it is known that some plants utilise more than one pathway to induce flowering (Srikanth and Schmid 2011). However, the photoperiod pathway is particularly prevalent with some plants responding to increasing day lengths (long day, LD plants), such as wheat and Arabidopsis, while other plants like rice and sorghum are classified as short-day (SD) plants as they respond to decreasing day lengths. Sugarcane responds to shortening days with a specific number of induction cycles, where 12–35 days are required during the induction period, not necessarily consecutive but within a set period of time (Arceneaux 1965; James and Miller 1971; Julien 1971, 1973; Glassop et al. 2014). This specific induction profile further classifies sugarcane as an intermediate-short day (ISD) plant; however, it is commonly referred to as a SD plant (Vijayasaradhy and Narasimhan 1953; Burr et al. 1957; Coleman 1968; Midmore 1980). While manipulating planting dates to synchronise flowering has been successful for some varieties, sugarcane breeders have had more success by taking advantage of the photoperiod induction of flowering through the use of photoperiodic chambers to synchronise flowering between varieties and achieve crosses that may not be possible as a result of natural flowering (Coleman 1962; El-Manhaly et al. 1980).
As well as having optimal moisture, nutrients and temperature sugarcane must reach a level of maturity, defined as possessing 2–4 mature internodes (12–16 weeks), before changes in photoperiod can induce flowering (Burr et al. 1957; Clements and Awada 1965; Coleman 1968; Julien 1973; Julien et al. 1974; Moore and Nuss 1987; Glassop et al. 2014). Floral induction in sugarcane requires a consistent and persistent increasing nyctiperiod; an increase of 30–60 s each 24 h period and for 6.5 to 14 weeks before the sugarcane panicle emerges (MacColl 1977; Glassop et al. 2014;). The importance of the nyctiperiod for sugarcane floral induction was illustrated through a series of experiments whereby the night cycle was interrupted, resulting in disrupted floral induction and no flowering (Julien 1973; Midmore 1980; Moore and Nuss 1987; Berding and Hurney 2005). Changes in conditions disrupting the induction and development of the panicle can result in reversion to vegetative growth (Clements and Awada 1965; Moore and Nuss 1987; Glassop et al. 2014). Indication of floral induction and return to vegetative growth may be observed by a section of stalk with small internodes forming a zig-zag pattern, a multi-stalked top (also known as a witch’s broom), a panicle structure covered with small stalk tops, or in final developmental stages of the floral organs pollen sterility (Clements and Awada 1965; James and Miller 1971; Moore 1971; Clements 1975; Glassop et al. 2014).
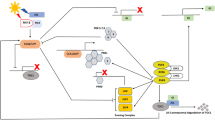
The induction of flowering is a multi-gene pathway. Initial changes in photoperiod are perceived by one group of genes and together with the internal biological clock genes cause a chain reaction of altered expression patterns that ultimately results in transition of the vegetative meristem to a floral meristem and the production of an inflorescence (Fig. 1). The changes in photoperiod are perceived in the spindle leaves of the sugarcane plant; with the spindle consisting of a tight whorl of 6–15 immature leaves (Moore 1974). Confirmation of the role of the spindle leaves was illustrated through experiments removing spindles leaves resulting in either delaying or inhibiting flowering (Panje and Raja Rao 1961; Coleman 1965; Panje et al. 1968; Moore and Berding 2014). Experiments conducted by Chu and Serapion (1972) showed that two sugarcane varieties, with profuse and medium flowering tendencies, required a persistent pressure for floral induction from spindle leaves, with plants grown in floral inducing conditions not producing inflorescences if spindle leaves were removed, even after 10 weeks of exposure. These experiments of removing young leaves, not only in sugarcane, have long illustrated that a signal, previously called florigen, originated in the leaves and transitioned to the meristem for floral induction. This florigen signal has further been identified as the mRNA and/or protein from the Flowering Locus T gene (FT) (King et al. 2006; Turck et al. 2008; Qin et al. 2017). It is the changes in timing/profiles and amplitude of expression of FT and the many other genes associated with the flowering pathway that result in the production of an inflorescence (Yanovsky and Kay 2003).
A simplified schematic of a potential gene pathway for control of flowering in sugarcane, adapted from published pathways in Arabidopsis and cereal crops (Blázquez 2000; Higgins et al. 2010; Izawa et al. 2003). Arrows indicate positive regulation and lines with closed circles indicate negative regulation, as based on published literature. Results of expression levels of genes enclosed in boxes are presented. Gene names: AGAMOUS LIKE 20 (AGL20), APETALA 1 (AP1), CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), CONSTANS (CO), CRYPTOCHROME CIRCADIAN REGULATOR 1 and 2 (CRY1, CRY2) EARLY HEADING DATE 1 (EHD1), FLOWERING LOCUS D (FD), FLOWERING LOCUS T (FT), FRUITFUL (FUL), GRAIN HEADING DATE 7/MATURITY GENE 6 (GHD7/Ma6), GIGANTEA (GI), LEAFY (LFY), LATE ELONGATED HYPOCOTYL (LHY), MATURITY GENE 1/PSUEDO-RESPONSE REGULATOR 37 (Ma1/PRR37), PHYTOCHROME A (PHYA), PHYTOCHROME B/MATURITY GENE 3 (PHYB/Ma3), PSEUDO-RESPONSE REGULATOR 1 (PRR1), PSEUDO-RESPONSE REGULATOR 73 (PRR73), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS (SOC1), SHORT VEGETATIVE PHASE (SVP), TERMINAL FLOWER 1 (TFL1), TIME OF CAB 1 (TOC1)and TWIN-SISTER OF FLOWERING TIME (TSFT)
The flowering pathway is very complex and studies in Arabidopsis have provided researchers a significant baseline to initiate studies in other plant species. There is also a high level of genetic conservation between species which has allowed identification of homologous genes. While monitoring changes in gene expression levels through vegetative to floral production in the model crop Arabidopsis can be done very quickly this type of experimental analysis in other plants is rare, mainly due to the time frame required and space to grow replicate plants. Hassankhah et al. (2020) were able to access 15-year-old Persian walnut trees and after three years of assessment of the timing of flower development were then able to complete a series of floral gene expression studies. Focussing on the genes involved with the last steps to floral meristem transition Hassankhah et al. (2020) were able to show the changes in expression over 12 months, reporting a combination of peaks and troughs for four of the five genes tested, Flowering Locus T (JrFT), Cauliflower/Apetala 1 (JrCAL/AP1), Leafy (JrLFY) and Terminal Flower 1 (JrTFL1), and a sustained peak for the remaining gene, Suppressor of Overexpression of Constans 1 (JrSOC1), over the induction, initiation and differentiation periods (Hassankhah et al. 2020). During winter the floral inducing genes had very low to nil expression levels and the JrTFL1 had its highest expression levels (Hassankhah et al. 2020). Koskela et al. (2012) also saw a decrease in FvTFL1 expression levels during the induction and development of flowers in SD strawberry plants. The decrease in FvTFL1 was countered with increases in FvAP1 and Fruitful 1 (FvFUL1) (Koskela et al. 2012). Interestingly the expression profile of FT did not match that seen in walnuts with a peak in FvFT at the start of the induction that reduced to very low levels within 6 weeks (Koskela et al. 2012). These expression profiles demonstrate the variability that exists across species and this prior knowledge from other species was used as a basis to examine the changes in sugarcane during photoperiod floral inducing conditions.
Despite the use of photoperiod chambers to alter flowering times in sugarcane some varieties remain unresponsive yet have traits that are desirable for the breeding programme. Understanding the changes in gene expression profiles may assist with identifying genetic roadblocks in sugarcane varieties that do not have a propensity to flower. Changes in the photoperiod are detected by a group of genes known as circadian clock regulator genes, including Pseudo-Response Regulator genes, Phytochrome genes and Chlorophyll A/B binding genes, which, in conjunction with the circadian clock genes, contribute to the initiation of the floral photoperiod induction pathway (Dutta et al. 2018). The circadian clock genes include CCA1, LHY, PRR1 and TOC1. A simplified pathway for floral induction then follows a chain reaction of GI, CO, FT, AP1 and LFY genes (Glassop and Rae 2019). In this study we measured the patterns of expression of 16 genes (2 of 18 not detected) in the critical first four weeks of induction during the light period. Then, we contrasted the expression of a subset of genes in a non-flowering variety to identify differences that may determine the response to photoperiod signals in sugarcane.
Materials and Methods
Plant Material
Planting material was treated for ratoon stunting disease (RSD) and reducing fungal infection, as per commercial practice. Single eye setts of Q208 and Q183 were treated for RSD by cold soaking for 40 h in water followed by long hot water treatment by incubating at 50 °C for 3 h were supplied by Sugar Research Australia (SRA, Meringa). Prior to planting in germination trays, setts were dipped in fungicide prepared as per manufacturer’s instructions (Yates 150 g Mancozeb Plus Garden Fungicide and Miticide, Australia). Plants were germinated in potting mix (Searles Peat 80 Plus Premium Potting mix, Australia) in a glasshouse (CSIRO, St Lucia, 27.5°S, 153°E) where temperatures were maintained between 22 and 32 °C. Once plants were approximately 15 cm tall, they were transplanted to individual 8 L pots and irrigated daily. For each treatment at each time point there were 4 replicates.
Controlled Environment Facility and Photoperiod Induction Settings
The Controlled Environment Facility (CEF, CSIRO, Queensland Bioscience Precinct, St Lucia, Qld) settings included: Day-light conditions of 29 °C and 60% humidity for 14 h 30 min (500 µmol photons m−2 s−1) and 24 °C and 90% humidity during the dark cycle. Photoperiod floral induction involved the reduction in day length to a starting point of 12 h 55 min and then a daily reduction of 1 min. These photoperiod floral induction conditions were previously shown to induce flowering in sugarcane (Rae et al. 2015).
Differences in Gene Expression Between Sugarcane Varieties That Have Different Propensity to Flower During Photoperiod Floral Induction
Q208 has a propensity to flower, readily flowering in field conditions, while Q183 is classified as a reluctant to flower variety in the QCanesSelect® database, based on field observations. Henceforth, Q208 and Q183 will be referred to as flowering and non-flowering, respectively. These two varieties were selected to examine differences in expression of genes associated with floral induction and initiation. Varieties Q208 and Q183 are covered by plant breeder’s rights (https://www.ipaustralia.gov.au/plant-breeders-rights). Following transplanting to 8 L pots the plants were moved to the CEF. Photoperiod floral induction was initiated when plants had been growing for 13 weeks in total. Samples of the spindle leaves (SL) were collected, approximately 2, 5 and 8 h after the start of the day-light cycle (T2, T5 and T8), weekly from 1 to 4 weeks after the initiation of the photoperiod induction (W1, W2, W3 and W4). The times of collection during the day-light cycle were selected based on the peak expression of most of the genes examined here over 24 h, as published by Glassop and Rae (2019). Examination of changes in gene expression over the first 4 weeks in photoperiod inducing conditions in spindle leaves covers the first two stages of floral development including the induction period and panicle initiation as described by Moore and Nuss (1987). The top of the plant was cut, approximately 10 cm below the youngest visible dewlap, and the section cut longitudinally in two to reveal the meristem. The meristem was then dissected away from the spindle leaves. The spindle leaf samples were quickly frozen in liquid nitrogen and stored at -80 °C before freeze drying. Samples collected at week 1, 2, 3 and 4 had been exposed to reducing photoperiod totalling 7, 14, 21 and 28 min, respectively.
Genes
Sugarcane genes, including some alleles, associated with photoperiod perception and floral induction were selected from previously published literature to assess expression levels in the samples collected (Glassop et al. 2014; Glassop and Rae 2019). Genes tested were AGAMOUS LIKE 20 (AGL20), APETALA 1 (AP1), CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), CONSTANS (CO), EARLY HEADING DATE 1 (EHD1), FLOWERING LOCUS T-A and- C (FT-A and FT-C), GRAIN HEADING DATE 7/MATURITY GENE 6 (GHD7/Ma6), GIGANTEA (GI), LEAFY (LFY), LATE ELONGATED HYPOCOTYL (LHY), MATURITY GENE 1/PSUEDO-RESPONSE REGULATOR 37 (Ma1/PRR37), PHYTOCHROME B/MATURITY GENE 3 (PHYB/Ma3), PSEUDO-RESPONSE REGULATOR 1/TIME OF CAB 1 (PRR1/TOC1), PSEUDO-RESPONSE REGULATOR 73 (PRR73), SHORT VEGETATIVE PHASE (SVP), TERMINAL FLOWER 1 (TFL1), and TWIN-SISTER OF FLOWERING TIME (TSFT). Previously published primers were used that had been designed to cross intron/exon boundaries or to produce a product that would exclude gDNA amplification by spanning an exon (Glassop and Rae 2019), primer details are included in Table S1. The housekeeping gene used to normalise the qPCR was Actin Depolymerising Factor (ADF) (Iskandar et al. 2004; Casu et al. 2015). Primer efficiency was tested on pooled samples and accepted if greater than 1.8, in accordance with criteria detailed by Nolan et al. (2006).
RNA Extraction, cDNA Synthesis and RT-PCR
Leaf samples were dried (Christ Alpha 1-4LSC freeze drier Sciteck Australia Pty Ltd, Australia) to a constant weight (until sample weight was stable for 24 h). Dried samples were stored in airtight containers with desiccator beads at room temperature until required; then ground to a fine powder with a ball mill (Retsch, MEP Instruments Pty. Ltd, Australia). RNA extraction followed the manufacturer’s instructions (QIAGEN Plant RNeasy Kit) with an additional 10 min incubation at room temperature after the addition of buffer RLT/βME. QIAGEN QuantiTect Reverse Transcription Kit was used to synthesise cDNA with anchored oligo dT primers. Triplicate real-time PCR reactions contained approximately 32 ng cDNA, 2.4–3 µM primers (0.3 µM for ADF gene, due to its high level of expression) and SYBR® Green Master Mix (ThermoFisher Scientific, Australia). Reactions were run on the Applied Biosystem ViiA7™ Real-Time PCR System (ThermoFisher Scientific, Australia). Results were only processed when technical replicates were within 0.5 Ct value of each other and the no-template controls showed no amplification (Nolan et al. 2006). The double delta threshold cycle (Ct) method was used to process qPCR results (Nolan et al. 2006).
Statistical Analysis
GenStat (v 19.1.0.21390, VSN International Ltd) was used to perform ANOVA, PCA analysis and Fisher’s protected least significant difference tests. Significant differences were accepted for P < 0.05.
Results
Changes in Gene Expression During the Early Stages of Floral Induction in Q208 a Flowering Variety
To capture expression changes, we measured gene expression weekly during the first four weeks (W1, W2, W3, W4) of exposure to photoperiod inducing conditions, sampling at three times over the day cycle at, two, five and eight h after the onset of the day cycle (T2, T5 and T8). Eighteen genes were assessed in the spindle leaf tissue of the flowering variety, Q208. The spindle leaves are the youngest leaves and are connected to the meristematic region of the sugarcane stalk apex; hence changes in photoperiod can quickly be perceived and gene expression in the leaves and apical meristem altered accordingly.
Of the 18 genes tested in Q208, ShEHD1 and ShTSFT had very low expression levels; where qPCR results did not meet experimental standards for any of the samples tested, with large variation within the technical replicates (results not shown). Of the remaining 16 genes tested, four are associated with photoperiod perception (ShPHYB/Ma3, ShMa1/PRR37, ShGHD7/Ma6, ShPRR73), three with circadian clock regulation (ShLHY, ShTOC1, ShCCA1), seven with floral induction (ShGI, ShCO, ShFT-A, ShFT-C, AGL20, SVP, TFL1), and two with meristem identity (ShAP1, ShLFY).
Over the first two weeks, ShPHYB/Ma3 expression was highest at the beginning and end of the light cycle and decreased in the middle of the light cycle at T5. By W3 there was low expression at both T2 and T5, and by W4 there were no significant differences between the times sampled (Fig. 2). Peak expression of ShMa1/PRR37 was observed soon after the start of the light cycle at T2 for all four weeks, with less expression for the remainder of the light cycle (Fig. 2). ShGHD7/Ma6 had a similar expression profile to that of ShPHYB/Ma3 with lowest expression at T5 for the first three weeks, though maximum expression was seen at the beginning of the light cycle for the first two weeks (Fig. 2). Expression of ShPRR73 was not significantly different for any time points (Fig. 2).
Heat map of changes in gene expression over sampling period in spindle leaf of sugarcane varieties Q208 and Q183. Values used to produce heat map are the average relative expression of each gene. The variation in the heat map is normalised within each gene over all time points (W and T), with changes from dark to light shades representing the highest and lowest relative expression. Sampling Time is 2, 5 and 8 h after the start of the light cycle for 4 weeks. Sampling Times with an asterisk have significantly different expression levels across the 4 weeks. Asterisks above the gene heat map indicates weeks where there is significant difference in expression between the sampled times within that week. (P < 0.05). Individual gene expression graphs are shown in Supplementary Figs. 1– 4
Changes in expression were measured in variety Q208 for four genes associated with circadian clock regulation: ShLHY, ShTOC1, ShCCA1 and ShGI. The expression of ShCCA1 had a large significant peak at T2 at the beginning of the light cycle and was very low at the other time points (Fig. 2). This pattern continued over the four weeks, although the magnitude of expression at T2 reduced over the four weeks. Profiles for both ShLHY and ShTOC1 expression were similar. Both showed significantly different peaks at T8 and low expression during the earlier part of the light cycle (Fig. 2). Expression levels of ShTOC1 in Q208 decreased significantly for all three sampling times, T, over the four weeks, W, (Fig. 2). There were also significant decreases in ShLHY expression over the four weeks at T5 and T8, (Fig. 2). Expression of ShGI was significantly different over the course of the day cycle for W1 and W2, only (Fig. 2). ShGI expression decreased over the four weeks for T5 and T8, with no change in expression at T2 (Fig. 2).
The internal clock genes and the genes associated with perceiving changes in photoperiod influence the expression of genes involved in floral induction further down the cascade. Expression of ShAGL20 showed no consistent pattern over four weeks and time of day (Fig. 2). Significant differences in ShAGL20 expression over times T2 – T8 were only observed in W4 (Fig. 2). Over the period of the light cycle, ShSVP showed a significant trough at T5 for the first two weeks (Fig. 2). Two alleles of ShFT were tested (ShFT-A and ShFT-C) and showed different expression profiles (Fig. 2). For ShFT-A, significant differences in expression during the light cycle were only detected at W4 when expression was highest at T8 at the end of the light cycle (Fig. 2). Expression of ShFT-C showed a significant decline at T8 across the four weeks, with the highest expression at W1 (Fig. 2).
Expression of ShCO peaked at T8, significantly so for W2 and W4 (Fig. 2). One of the genes associated with negative regulation of flowering, TFL1, was also assessed. ShTFL1 expression peaked significantly in the middle of the light cycle during W1 (Fig. 2). Expression of ShTFL1 then decreased at all three time points, T2, T5 and T8, over the four weeks (Fig. 2). The later stages of the flowering pathway involve genes that influence meristem identity and control the transition from vegetative to floral meristem, including ShAP1 and ShLFY. In variety Q208, ShAP1 had the highest expression at T8 for the first three weeks, and this did not show any significant changes over the 4-week period (Fig. 2). Differences in expression of ShLFY across the light cycle were not significant, though at T5 there was a significant change in expression over the four weeks (Fig. 2).
Changes in Gene Expression During the Early Stages of Floral Induction in Q183 a Non-Flowering Variety
A subset of the genes tested in variety Q208 was then selected for analysis in the non-flowering variety Q183. Two genes involved in circadian clock regulation (ShLHY and ShTOC1), four genes involved in floral induction (ShGI, ShCO, ShFT-A and TFL1) and one gene associated with meristem identity (ShLFY) were measured in spindle leaves, as above. Samples were collected weekly during the first four weeks (W1, W2, W3, W4) of exposure to photoperiod inducing conditions, sampling at three time points, two, five and eight hours after the onset of the day cycle (T2, T5 and T8).
In Q183 genes ShLHY and ShTOC1, associated with the internal clock, had similar expression patterns, with a significantly different peak of expression at T8 and low expression during the earlier part of the light cycle (Fig. 2). Over the four weeks, there were significant decreases in ShLHY expression at T8 and in ShTOC1 expression at T5 (Fig. 2).
In variety Q183, the floral induction genes, ShGI and ShCO and one of the negative regulators, ShTFL1 were measured. For both ShGI and ShCO, expression across the day cycle peaked at T8 (Fig. 2). Over the four weeks, ShGI expression decreased at times T5 and T8, though at T2 a slight increase in expression was observed at W2 and W4 (Fig. 2). For ShCO, decreasing expression was also observed over the four weeks at times T5 and T8 (Fig. 2). ShTFL1 expression was only significantly different over the day cycle in W3 where the highest expression was at T8 (Fig. 2). At both T5 and T8 the expression levels of ShTFL1 decreased significantly over the four weeks of photoperiod induction (Fig. 2).
Expression of ShFT-A was only significantly different over the day cycle for W1 and W3, with the highest expression in T5–T8 and T8, respectively (Fig. 2). Over the course of the four weeks there were significant changes in ShFT-A expression at T2, that showed a peak at W4, and T5, that decreased over the four weeks (Fig. 2).
The meristem identity gene, LFY was measured in variety Q183. There was no significant difference in expression of ShLFY across the three time points in the light cycle and this continued for all four weeks (Fig. 2). Interestingly there was a significant increase in expression at T2 in W4 (Fig. 2).
Differences in Gene Expression Between Flowering and Non-Flowering Varieties
For many components of the flowering pathway, key signals are perceived through changes to diurnal patterns or relative ratios of gene expression over time. We generated comparative heat maps as a method to display the patterns of expression, independent of quantity, and used these to compare the subset of seven genes that had been assessed in both Q208 and Q183 (Fig. 2). For each gene dataset in a variety, the average relative expression values were normalised and then values for each sampling point were assigned into five 20-percentile categories from low to high.
In both varieties, genes ShLHY and ShTOC1, associated with the internal clock, had similar expression patterns, with low expression during the earlier part of the light cycle and a significantly different peak at T8 (Fig. 2). The significant decrease in ShLHY expression over the four weeks for Q208 was matched by a decrease in expression in Q183. ShTOC1 expression also declined over the four weeks in both varieties; this was significant at all time points for Q208 but only at T5 for Q183 (Fig. 2).
Similar expression profiles were also observed for the floral induction genes ShGI and ShCO; in both varieties expression peaked at T8, significantly so for some weeks (Fig. 2). The only difference in expression of ShGI between Q208 and Q183 was in W1 where the expression trend of Q208 at T5 was higher than at T8, and the reverse for Q183 (Fig. 2). Peak expression of ShCO was at T8 for both varieties. Decreasing expression of ShCO over the four weeks also seemed to be a consistent trend, as it was observed for all sampling time points for Q208 and times T5 and T8 for Q183 (Fig. 2).
The pattern of ShFT-A expression from W1 to W3 was similar between Q208 and Q183, with lowest expression at T2 (Fig. 2). Differences were measured in W4 where Q183 had a peak at T2 while in Q208 it was greater at T8.
In contrast, some differences were observed between the two varieties in the expression of ShTFL1, the sugarcane homologue of the TFL1 gene which is one of the genes that negatively regulate the flowering pathway. While ShTFL1 expression peaked at T5 in the middle of the light cycle in both varieties at the start of the experiment and then generally decreased over the following weeks, expression at T8 at the end of the day remained higher in Q183 relative to the other time points, with a significant difference measured at W3 (Fig. 2).
ShLFY also showed some differences in expression profiles between varieties, in the relative expression at different timepoints through the day. In Q208 there was a significant decrease in expression of ShLFY over the 4 weeks in the middle of the day (T5), which was not mirrored in Q183 (Fig. 2). Q183 saw a significant increase in expression at T2 (Fig. 2). Expression at T8 the end of the day had slightly higher expression levels in Q183 compared to Q208 (Fig. 2).
Principal Component Analysis
Principal component analysis (PCA) assesses data using multivariate techniques, identifying patterns of similarity of the variables within complex pathways. In this case identifying correlations between the expression profile of seven genes collected over three time points and four weeks in two sugarcane varieties. PCA was used to identify any key factors that may illuminate the differences between the two varieties. Principal component 1 and 2 accounted for 65 and 13% of the variation, respectively (Fig. S5). When the PCA graph was coloured according to variety there was a greater proportion of Q183 samples spreading out to the right of the cluster than Q208 (Fig. S5). Changing the colouring of the PCA graph to reflect the time of day of sampling the scattered samples divide into T5 and T8 groups, with the T5 samples in the top half and the T8 samples in the lower half (Fig. S5). The scattered samples mainly came from W1 to W3, when the PCA graph was coloured according to week of sampling (Fig. S5). Within principal component 1 expression of ShTFL and ShCO are the strongest components separating samples negatively and positively, respectively; while principal component 2 has separation of samples due to effects of ShLHY, negatively, and ShTFL, positively.
Expression of LHY was greater at T8 for both varieties, though only significantly so for Q183 W1 – W4 (Fig. 3). Further downstream from LHY expression in the circadian clock is the expression of CO, which had a similar profile at T8 for both varieties, while at T2 the expression in Q183 increased at W4 compared to Q208 (Fig. 3). CO is proposed to positively regulate several genes that initiate or inhibit flowering, including the floral inhibitor TFL1 (Fig. 1). While the expression of TFL1 in Q183 decreased over the course of the four weeks it was significantly greater than expression in Q208 at various times over the day cycle (Fig. 3).
Average relative expression of genes TFL1, CO and LHY in spindle leaves of sugarcane varieties Q183 and Q208 sampled at 2, 5 and 8 h after the beginning of the light cycle. The legend applies to all graphs. N = 4. Error bars have not been included to simplify the graphs. The tables to the right of each graph details Fisher’s Protected Least Significant Difference (P < 0.05) between varieties and time (T) at each week, and only pertain to comparison between sampling times for each individual week, not between weeks. The y-axis varies between genes. Values, standard errors of means and all LSD’s are supplied in Table S4
Discussion
The production of fertile flowers is critical for sugarcane breeding, but there is a wide window of floral production encompassing the smaller flowering windows of individual varieties. The crossing of some varieties is prohibited due to timing of flowers and varieties that are unresponsiveness to floral inducing conditions. Some sugarcane varieties will produce inflorescences within 11–12 weeks from the beginning of the photoperiod induction, while other varieties will flower later, as much as 20–24 weeks (Paliatseas 1971). Various lengths of the nyctiperiod have been tested, ranging from 11 to 14 h, with some tests at constant cycles while others increased the dark cycle by 30, 45 or 60 s day−1 (Coleman 1962; Daniels 1962; Paliatseas 1962, 1974; Daniels et al. 1965; Clements 1972; Nuss 1977; Berding and Moore 2001; Diaz Romero and Cuenya 2002; Rizk et al. 2002, 2004, 2007; Berding and Hurney 2005; Miah and Paul 2006; Srivastava et al. 2006; Berding et al. 2007, 2010; Singh et al. 2009). The photoperiod conditions utilised here followed the regime reported by Berding and Hurney (2005), with a starting time of 12 h and 55 min; however, due to limitations of the CEF lighting controls the dark cycle was increased by 60 s day−1 instead of 45 s day−1. This regime was demonstrated to initiate flowering (Rae et al. 2015).
The genes controlling floral induction and inflorescence production have been extensively studied in other species like the model plant Arabidopsis and crops including rice, sorghum and Brachypodium (Bäurle and Dean 2006; Imaizumi and Kay 2006; Colasanti and Coneva 2009; Greenup et al. 2009; Higgins et al. 2010; Murphy et al. 2011). However, the identification of homologous genes in sugarcane has only recently started and detailing the pathway for floral induction is still preliminary (Coelho et al. 2013; Hotta et al. 2013; Glassop et al. 2014; Glassop and Rae 2019). While confirmation of the roles of each gene within the flowering pathway is best done by the production of transgenic plants or complementing mutant lines, the analysis of gene expression in sugarcane plants grown in floral inducing conditions will provide useful information to build on this foundation.
The sugarcane genome is complex with each gene having as many as eight to ten copies and more than one allele (Grivet and Arruda 2001; Coelho et al. 2013; Glassop and Rae 2019). As part of our experimental plan primers were designed to regions of homology between alleles, where sequence information was available, but may not represent all potential alleles; the cumulative allele expression profiles was measured. Fifteen different genes and one gene with two alleles associated with photoperiod perception and floral induction pathways displayed significant changes in expression over time in one or both varieties tested in spindle tissue (Fig. 2).
Of the 16 genes examined over the three time points during the light cycle for four weeks, all had their highest expression levels in weeks 1 and/or 2 (Fig. 2). Those genes that had their highest expression peak 2 h after the initiation of the light cycle in Q208 include ShMa1/PRR37 and ShCCA1 (Fig. 2). Only one gene, ShTFL1, peaked during the middle of the light cycle and four genes in the later part of the light cycle including, ShAP1, ShLHY, ShTOC1 and ShCO in Q208 (Fig. 2). The expression profile of genes assessed in both varieties were similar for ShLHY, ShTOC1, and ShCO; with profiles of ShFT-A, ShGI, ShLFY and ShTFL1 unique to each variety (Fig. 2).
Photoperiod Perception Genes
One of the first genes to perceive changes in photoperiod is PHYB/Ma3. In Q208 ShPHYB/Ma3 expression was significantly different over the course of the light cycle and changed with increasing weeks of floral induction (Fig. 2). ShPHYB/Ma3 expression in Q208 was different to that reported in Q174 (Glassop and Rae 2019). In Q174 there was no significant difference in ShPHYB/Ma3 expression over 24 h; however, in variety RB855453 there was a strong peak during the middle of the light cycle, though this difference may not be associated with the floral induction but other growth condition differences (Hotta et al. 2013; Glassop and Rae 2019). The expression of ShPHYB/Ma3 in sugarcane shows varietal differences and requires further research, including multiple varieties with differences in flowering propensity grown under the same conditions, to confirm its role in the sugarcane flowering pathway. The expression of ShPHYB/Ma3 then influences the expression of Ma1/PRR37, GHD7/Ma6 and PRR37, via transcription factors (Fig. 1).
While the gene ShPRR73 was identified by Casu et al. (2007) in Q117 the expression profile has not been previously reported in sugarcane. In Q208, the expression of ShPRR73 showed a peak at the beginning and the later part of the light cycle. Further analysis is required to determine expression during the dark cycle to confirm optimal sampling time to identify changes in expression during photoperiod induction and its role as one of the multiple pseudo-response regulators within the sugarcane flowering pathway (Fig. 2). When expression profiles of ShMa1/PRR37 and ShGHD7/Ma6 were examined in sugarcane variety Q174 over 24 h in non-floral inductive conditions, over the course of the light cycle both genes had their highest expression at the start, coinciding with results observed in Q208 (Fig. 2) (Glassop and Rae 2019). The role of GHD7/Ma6 has been associated with timing of floral induction, promoting or delaying flowering under SD or LD conditions, respectively, in sorghum (Yang et al. 2014a). In rice and sorghum, OsGHD7 and SbGHD7 have two peaks of expression, the first occurring in the morning and the second during the night when grown under LD non-flowering conditions, and only one peak in the morning in SD flowering conditions (Hori et al. 2013; Yang et al. 2014a). The expression peaks of ShGHD7/Ma6 in sugarcane do not precisely match those reported in rice and sorghum, occurring later in sugarcane during the day cycle. The change in timing of the peak expression of ShGHD7/Ma6 may be due to floral induction but requires further analysis over both day and night to confirm its role in the sugarcane flowering pathway. Sorghum had similar expression profiles for SbGHD7 and SbMa1/PRR37, two peaks over 24 h, in long- and short-day growth conditions (Yang et al. 2014a); this is in contrast to the ShMa1/PRR37 expression that shows a single peak during the dark cycle in non-flower inductive conditions in variety Q174 (Glassop and Rae 2019). Focussing on the light cycle only, the expression of ShMa1/PRR37 in Q208 did match that reported in Q174 with expression at the start of the light cycle being greater than the remainder of the light period (Fig. 2) (Glassop and Rae 2019). As the expression pattern of ShMa1/PRR37 did not match that reported in other short-day floral inductive plants, further research is required to understand the role it may play in the sugarcane floral pathway. The expression profile of ShPHYB/Ma3 is like that of ShGHD7/Ma6 and ShPRR73 and may indicate synchronised expression resulting from the perceived photoperiod by ShPHYB/Ma3; which along with the decreasing morning expression of ShMa1/PRR37 during SD floral induction conditions start the transition to flowering.
Circadian Clock Associated Genes
The expression pattern of the circadian clock genes, ShCCA1 and ShTOC1, in Q208 matched that reported in Q174 with peaks at the beginning and end of the light cycle, respectively (Fig. 2)(Glassop and Rae 2019). The expression of Q208 ShCCA1 also matched that reported in sugarcane variety RB855453 and maize with a peak at the beginning of the light cycle decreasing over the light period (Fig. 2) (Wang et al. 2011; Hotta et al. 2013). While expression profiles of ShTOC1 in both Q208 and Q183 matched those reported in Q174, they did not match the profile reported by Hotta et al. (2013) in variety RB855453 which was measured under continuous light conditions (Fig. 2); this may be due to different alleles between the sugarcane varieties and/or differences in the plant growth conditions. However, the expression profiles did align with rice, bamboo, and tobacco PRR1/TOC1 genes with peaks in the later part of the light cycle transitioning into the dark cycle (Ogiso et al. 2010; Yon et al. 2012; Dutta et al. 2018). The expression pattern of bamboo BtTOC1 maintained the same expression pattern in young leaves in both flowering and non-flowering plants; however, the amplitude of expression in flowering plants was half that reported in non-flowering plants (Dutta et al. 2018). In sugarcane variety IACSP96-7569, Manechini et al. (2021) reported differential expression of ScTOC1 between non-inductive and inductive photoperiod treatments, with decreased expression in the floral inductive treatment. This decreased expression in BtTOC1 and ScTOC1 between flowering and non-flowering plants was also seen between Q208 and Q183 with a higher level of expression of ShTOC1 in the non-flowering Q183 variety (Fig. 2). There was also greater expression of ShLHY in Q183 than Q208, though when the profile was compared to BtLHY the peaks occurred at opposite ends of the light cycle, with ShLHY peaking in the later part of the cycle and BtLHY at the beginning of the light cycle (Fig. 2) (Dutta et al. 2018). The difference in expression between flowering and non-flowering varieties Q208 and Q183, respectively, was opposite of that reported in sugarcane variety IACSP96-7569 where the expression of ScLHY was upregulated under floral inductive conditions compared to non-inductive conditions (Manechini et al. 2021). This difference may reflect that expression levels of LHY is specific to varieties and changes in expression within that variety affect the transition to flowering but cannot be compared between varieties.
The expression of the circadian clock genes has been extensively studied in Arabidopsis with gene peaks occurring in sequential order. In Arabidopsis the circadian clock associated genes follow that pattern of LHY/CCA1 ➔ PRR1/TOC1, over the course of 24 h; this was also observed in sugarcane variety Q174 but in Q208 the peaks were CCA1 ➔ LHY/TOC1 and in Q183 no CCA1 was measured but the LHY/TOC1 peaks also coincide (Fig. 2) (Glassop and Rae 2019). The difference in peak order between Q174 and the varieties tested here may be due to the growth conditions, inductive/non-inductive floral conditions, though the similarity between Q208 and Q183 may indicate that these genes are not directly associated with the propensity to flower phenotype.
Floral Pathway and Meristem Identity Genes
The remaining genes examined are associated with the transition from vegetative to floral induction pathway. Expression of ShGI in Q174 grown under non-inductive conditions showed no significant difference in expression over the course of the light cycle, while in Q208 and Q183 the significantly different expression profile may be attributed to the floral inductive growth conditions (Fig. 2)(Glassop and Rae 2019). In Q183 ShGI peak expression was in the latter half of the light cycle like that reported by Hotta et al. (2013). In other short-day receptive plants, such as rice and soybean, GI expression levels transitioned with the light to dark period and in rye grass the peak occurred earlier in the day cycle under floral inductive conditions (Filichkin et al. 2011; Hori et al. 2013; Li et al. 2013; Gagic et al. 2015). In bamboo the BtGI peak profile matched that of rice and soybean and when profiles were compared between flowering and non-flowering bamboo plants there was no difference (Dutta et al. 2018). Manechini et al. (2021) also reported no difference in ScGI expression between flowering and non-flowering plants. This suggests that the role of GI, in sugarcane and bamboo, in flowering may not be critical or is linked with changes in other flowering pathway genes. With expression profiles of ShGI not precisely matching that seen in other short-day plants, further investigation is required to confirm its role in the sugarcane flowering pathway and if the difference in peak times between Q208 and Q183 alters flowering ability (Fig. 2).
Within the proposed sugarcane flowering pathway, expression levels of GI would have an activating effect on CO (Glassop and Rae 2019). Expression of ShCO has not previously been assessed over time, though Coelho et al. (2013) had identified 10 different ShCO alleles that had unique in silico expression profiles in different tissues sampled; with the CoContig2 allele detected in eight of the 17 sugarcane libraries assessed, including four of the seven inflorescence libraries. Expression of ShCO in spindle leaves had its highest expression level at the end of the light cycle in both Q208 and Q183 (Fig. 2). This expression profile matched that in non-flowering bamboo BtCO allele A, but not allele B, as BtCO-B had a peak at the beginning of the light cycle; though samples from flowering bamboo had an additional small peak at the start of the light cycle (Dutta et al. 2018). Constans plays a critical role in Arabidopsis flowering under long day conditions, in particular the presence of CO proteins at the end of the light cycle, that would not normally be present, to interact with FT mRNA to induce flowering (Lagercrantz 2009). Constans may play a similar role in the sugarcane flowering pathway due to its presence at the end of the light cycle, but this requires further investigation. The expression of CO has been shown to positively regulate EHD1 which in turn positively regulates the expression of FT (Higgins et al. 2010). Two of the known negative regulators of EHD1 are Ma1/PRR37 and GHD7/Ma6; which similarly down-regulate the expression of FT. In sorghum, SbEHD1 expression is inhibited by SbMa1/PRR37 and SbGHD7/Ma6, but SbEHD1 activates expression of FT (Yang et al. 2014a). The two sugarcane genes, ShEHD1 and ShTSFT, previously amplified from other sugarcane samples (data not shown) and aligned to homologous sorghum sequences, were not amplified in the samples collected for analysis reported here. Expression of ShEHD1 and ShTSFT may be very low at the time points sampled or not expressed in the spindle leaf and further experimental design would need to be completed to confirm their role in the sugarcane flowering pathway.
The expression of ShAGL20 during the light cycle was not similar between Q208 (Fig. 2) and Q174 (Glassop and Rae 2019). Expression of ShAGL20 in Q174 slowly increased over the course of the light cycle but in Q208 expression was varied, peaking in the middle for week 1 and in the later part of the day for the other weeks (Fig. 2). The different expression profiles of ShAGL20 between the two varieties may be due to the non-inductive (Q174) /inductive (Q208) floral growth conditions (Glassop and Rae 2019). Expression of Q208 ShAGL20 did have a similar expression pattern to soybean GmGAL1 (homologue of SOC1/AGL20) when soybeans were grown under short-day floral inducing conditions, where the peak occurring during the light cycle in long day conditions now occurs at the transition to the dark cycle (Zhong et al. 2012). This is an example of the complexity of the flowering induction pathway where the expression of genes/proteins now coincide due to shifts in expression under floral inductive conditions that allow interactions with other genes/proteins that would not occur during non-floral growth conditions (Lagercrantz 2009; Song et al. 2013; Yang et al. 2014b).
The expression of AGL20 has been shown to be negatively regulated by SVP (Blázquez 2000; Izawa et al. 2003; Higgins et al. 2010). Diurnal expression analysis of ShSVP in Q174 showed no variation over the 24 h but in the floral inductive growth conditions of Q208 ShSVP had significant peaks in expression at the beginning and end of the day cycle during the first two weeks of induction, though in weeks three and four this was significantly reduced (Fig. 2) (Glassop and Rae 2019). The decrease in ShSVP expression may be indicative of the transition from vegetative to floral production with reduced repression on SOC1/AGL20 and FT as proposed in Arabidopsis flowering pathways (Higgins et al. 2010).
The expression profiles of Q208 ShFT-A, ShFT-C and Q183 ShFT-A were not similar to each other, with none of the profiles matching that reported in Q174 which had a ShFT-A peak at the beginning of the light cycle and no differences for ShFT-C (Fig. 2)(Glassop and Rae 2019). A difference in FT expression was reported in bamboo between flowering and non-flowering plants. Four FT alleles in bamboo were divided into two different expression profile groups, with BtFT-1 and -2 expression peaking during the dark cycle on flowering plants and no peaks in non-flowering plants, and BtFT-3 and -4 expression with a small peak occurring in the middle or the end of the light cycle in non-flowering and flowering samples, respectively (Dutta et al. 2018). Coelho et al. (2013) had identified eight FT alleles from contigs and singlet reads, with no in silico expression detected in the seven sugarcane inflorescence libraries. In sugarcane variety IACSP96-7569 there was no differential expression of ScFT between flowering and non-flowering treatments (Manechini et al. 2021). Confirming the detection and measurement of the appropriate FT allele associated with sugarcane floral induction will be essential to build on the sugarcane flowering pathway and may explain the reduced flowering capacity of Q183.
When expression levels of FT are increased during floral induction of Arabidopsis, expression of AP1 is activated (Abe et al. 2005). The Q208 ShAP1 peak at the end of the light cycle is similar to that reported in Q174 (Fig. 2) (Glassop and Rae 2019). The transition of the vegetative meristem to floral meristem requires both AP1 and LFY. The expression of ShLFY over 24 h showed a significant increase in expression at the end of the light cycle in Q174 (Glassop and Rae 2019); this profile was similar to that observed in Q208 and Q183 (Fig. 2). Interestingly the expression of Q208 ShLFY was lower in W2-4, while remaining high in Q183 through W1-3 (Fig. 2). Expression profiles of ShAP1 and ShLFY in the meristem may elicit their role within the flowering pathway and the difference in flowering propensity between Q208 and Q183 that is not evident in the expression profiles within the spindle leaves within the first four weeks of floral inducing conditions.
Within the flowering pathway there are both positive and negative regulation of genes that would promote or prohibit flowering. One negative interaction is the influence of TFL1 on expression of LFY (Higgins et al. 2010). Expression of ShTFL1 in spindle leaves of plants grown in non-inductive conditions was detected in sugarcane variety Q174, where there were no significant changes in expression levels over 24 h, which generally matched that observed in Q208 and Q183 (Fig. 2) (Glassop and Rae 2019). ShTFL1 expression in Q208 and Q183 spindle leaves and FvTFL1 in strawberry primary shoot apices had similar profiles with the highest expression detected at the start of the floral induction period, which quickly decreased with time (Fig. 2) (Koskela et al. 2012). While the expression of FvTFL1 was detected at all sampled time points the low expression levels did not inhibit floral production in strawberries, suggesting that a threshold of TFL1 expression levels is required in order to achieve inhibition (Koskela et al. 2012). Despite the similar expression profile of ShTFL1 over the 4 weeks between Q208 and Q183, the expression levels in Q183 were approximately 10–80 times higher than in Q208 and may result in the repression of flowering in Q183 (Fig. 2).
Moore and Nuss (1987) detailed that over the first 4 weeks of floral photoperiod inducing conditions sugarcane would transition from vegetative meristem to floral meristem. The intense focus on the first phases of floral induction and panicle initiation in this study was done with the primary aim to discern any differences between the flowering variety Q208 and the non-flowering variety Q183. Seven genes were examined including ShLHY and ShTOC1, associated with the internal clock, ShGI, ShCO and ShFT-A, associated with the floral pathway, ShTFL1, involved with inhibition of floral induction, and ShLFY a floral meristem identity gene. PCA analysis showed some separation between the two varieties, particularly for several Q183 samples at T5 and T8 during W1—3 (Fig. S5). The expression profiles of ShFT-A, ShLFY, ShGI, and ShPRR1/TOC1 did not contribute to any differences between the flowering and non-flowering varieties Q208 and Q183. The separation of the samples was due to the expression levels of ShTFL1, ShCO and ShLHY. If the sugarcane flowering pathway is like that reported in rice, then expression levels of ShCO are affected by expression levels of the circadian clock genes, including ShLHY, and light perceiving genes ( Higgins et al. 2010; Glassop and Rae 2019). Any variation in how sugarcane varieties react to changes in light perception would in turn affect flowering genes further down the flowering pathway and may explain the differences in ShCO expression between Q208 and Q183 (Fig. 3). Higher levels of ShTFL1 were detected in Q183 than Q208 (Fig. 3), which may be the key factor to Q183 being reluctant to flower; however, further investigation into the light perceiving and circadian clock genes expression levels in Q183 may identify further roadblocks to flowering. A different subset of flowering pathway genes were used to assess the differences between flowering and non-flowering bamboo and found levels of BtLHY, BtZTL, BtCO-A and -B and, BtFT-1, -2, -3 and -4 expression was significantly greater in flowering leaf samples than non-flowering samples (Dutta et al. 2018). While sugarcane homologues to the bamboo genes were not responsible for the discrimination in the sugarcane PCA analysis, expression levels of ShFT-A, ShLHY and ShCO were similar between Q208 and Q183, but Q183 did have higher levels of ShTFL a gene associated with inhibition of floral induction.
Long-term Changes Resulting in the Production of an Inflorescence
Within the Saccharum cultivars there is a wide range of times for the emergence of inflorescences from the beginning of the photoperiod induction; reportedly ranging from 11 to 24 weeks (Paliatseas 1971). This variation is also seen in commercial sugarcane varieties and some varieties are classified as reluctant to flower (Clements 1972; Rizk et al. 2004, 2007). Interestingly it was described that variation in flowering time between varieties may be due to differences in the rate of developmental stages, i.e. both varieties are induced at the same time but it takes one variety twice as long to produce an inflorescence compared to another variety (Mangelsdorg 1957; Paliatseas 1962; Arceneaux 1965). It is unclear if in sugarcane the floral induction signals are perceived at the same time followed by different developmental rates or if the floral induction signals need to be cued at each development stage specific to each variety. The assessment of genes in the flowering pathway over the duration of the floral induction period may assist with eliciting answers to these questions.
Conclusion
Flowering is a complex network requiring multiple cues to be met through different pathways to transition from vegetative growth to the production of an inflorescence. The use of mutant and transgenic lines has been invaluable in identifying the genes involved and their role, particularly in Arabidopsis, rice and Brachypodium (Blázquez 2000; Greenup et al. 2009; Higgins et al. 2010). While there are a few unique genes only occurring in SD and LD photoperiod flowering pathways, the conservative pathway provides a strong foundation to start in unresearched species (Higgins et al. 2010). The result of these floral gene pathway expression studies has shown that there is a unique pattern of gene expression peaks specific to the flowering variety Q208 and reluctant to flower variety Q183. Whereby, higher expression levels of ShLHY, ShCO and ShTFL1 in Q183 may be the cause of reduced/no flowering. Results presented here have provided a baseline for future analysis of the genes associated with the sugarcane flowering pathway, whereby future experiments can be designed to strengthen correlations identified and detect variations between varieties. Understanding the genetic component of the sugarcane flowering pathway is essential to identify differences between varieties and their propensity to produce flowers. This knowledge may be used to make genetic manipulations that could induce flowering and synchronise flowering times, a necessity for sugarcane breeders wishing to make crosses between varieties that currently do not flower at the same time.
References
Abe, M., Y. Kobayashi, S. Yamamoto, Y. Daimon, A. Yamaguchi, Y. Ikeda, H. Ichinoki, M. Notaguchi, K. Goto, and T. Araki. 2005. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056. https://doi.org/10.1126/science.1115983.
Albani, M.C., and G. Coupland. 2010. Comparative analysis of flowering in annual and perennial plants. In Current topics in developmental biology, ed. C.P.T. Marja, 323–348. Waltham, MA: Academic Press. https://doi.org/10.1016/S0070-2153(10)91011-9.
Arceneaux, G. 1965. Flowering of sugarcane. Proceedings of International Society of Sugar Cane Technologists 12: 780–784.
Bäurle, I., and C. Dean. 2006. The timing of developmental transitions in plants. Cell 125: 655–664. https://doi.org/10.1016/j.cell.2006.05.005.
Berding, N., and A.P. Hurney. 2005. Flowering and lodging, physiological-based traits affecting cane and sugar yield: What do we know of their control mechanisms and how do we manage them? Field Crops Research 92: 261–275. https://doi.org/10.1016/j.fcr.2005.01.015.
Berding, N., and P.H. Moore 2001. Advancing from opportunistic sexual recombination in sugarcane: Lessons from tropical photoperiodic research. Processdings of the international Society of Sugar Cane Technologists, 24: 482–487.
Berding, N., R.S. Pendrigh, and V. Dunne. 2007. Can flowering in sugarcane be optimised by use of differential declinations for the initiation and development phases? Proceedings of the International Society of Sugar Cane Technologists, 32: 699–711.
Berding, N., R.S. Pendrigh, and V. Dunne. 2010. Pursuing higher efficacy for managed photoperiodic initiation of sugarcane flowering in the tropics. Sugar Cane International 32: 234–250.
Blázquez, M.A. 2000. Flower development pathways. Journal of Cell Science 113: 3547–3548. https://doi.org/10.1242/jcs.113.20.3547.
Burr, G., C. Hartt, H. Brodie, T. Tanimoto, H. Kortschak, D. Takahashi, F. Ashton, and R. Coleman. 1957. The sugarcane plant. Annual Review of Plant Physiology 8: 275–308. https://doi.org/10.1146/annurev.pp.08.060157.001423.
Casu, R.E., J.M. Nielsen, G.D. Bonnett, and J.M. Manners. 2007. Identification of transcripts associated with cell wall metabolism and development in the stem of sugarcane by Affymetrix GeneChip Sugarcane Genome Array expression profiling. Functional and Integrative Genomics 7: 153–167. https://doi.org/10.1007/s10142-006-0038-z.
Casu, R.E., A.L. Rae, J.M. Nielsen, J.M. Perroux, G.D. Bonnett, and J.M. Manners. 2015. Tissue-specific transcriptome analysis within the maturing sugarcane stalk reveals spatial regulation in the expression of cellulose synthase and sucrose transporter gene families. Plant Molecular Biology 89: 607–628. https://doi.org/10.1007/s11103-015-0388-9.
Chu, T., and J. Serapion. 1972. The role of leaves in production of flowering stimulus in sugarcane. Proceedings of the International Society of Sugar Cane Technologist 14: 365–371.
Clements, H.F. 1972. Flower induction of Saccharum species and hybrid clones. Proceedings of the International Society of Sugar Cane Technologists 14: 317–322.
Clements, H.F. 1975. Flowering of sugarcane: Mechanics and control. Honolulu, Hawaii: Hawaii Agricultural Experiment Station-Technical Bulleting no.92.
Clements, H.F., and M. Awada. 1965. Experiments of the artificial induction of flowering in sugarcane. Proceedings of the International Congress of Sugar Cane Technologists 12: 795–812.
Coelho, C.P., A.P.C. Netto, J. Colasanti, and A. Chalfun. 2013. A proposed model for the flowering signaling pathway of sugarcane under photoperiodic control. Genetics and Molecular Research 12: 1347–1359. https://doi.org/10.4238/2013.April.25.6.
Colasanti, J., and V. Coneva. 2009. Mechanisms of floral induction in grasses: Something borrowed, something new. Plant Physiology 149: 56–62. https://doi.org/10.1104/pp.108.130500.
Coleman, R.E. 1962. Control of flowering and the use of pollen storage as techniques in a sugar cane breeding programme. Proceedings of the International Society of Sugar Cane Technologists 11: 533–540.
Coleman, R.E. 1965. Some aspects of flowering stimulus production in sugarcane. Proceedings of the International Society of Sugar Cane Technologists 12: 813–818.
Coleman, R.E. 1968. Physiology of flowering in sugarcane. Proceedings of the International Society of Sugarcane Technologists 13: 992–1000.
Daniels, J. 1962. Experimental control of flowering in Saccharum spontaneum L. Proceedings of the International Society of Sugar Cane Technologists 11: 527–533.
Daniels, J., K.T. Glasziou, and T.A. Bull. 1965. Flowering in Saccharum spontaneum. Proceedings of the International Society of Sugar Cane Technologists 12: 1027–1032.
Diaz Romero, C., and M.I. Cuenya. 2002. Implementation of new photoperiodic treatments for induction of flowering in selected lines of sugarcane. Avance Agroindustrial 23: 10–13.
Durai, A.A., P. Govindaraj, and A.S. Pazhany. 2014. Flowering behaviour of sugarcane genotypes from different agro climatic zones of India. Sugar Tech 16: 157–163. https://doi.org/10.1007/s12355-013-0258-1.
Dutta, S., P. Biswas, S. Chakraborty, D. Mitra, A. Pal, and M. Das. 2018. Identification, characterization and gene expression analyses of important flowering genes related to photoperiodic pathway in bamboo. BMC Genomics 19: 190. https://doi.org/10.1186/s12864-018-4571-7.
El-Manhaly, M.A., A.H. Nour, N.M. El-Talkwawy, and Y.M. Tawfik. 1980. The effect of various planting dates on flowering of sugarcane varieties. Proceedings of the International Society of Sugar Cane Technologists. 2: 1336–1340.
Filichkin, S.A., G. Breton, H.D. Priest, P. Dharmawardhana, P. Jaiswal, S.E. Fox, T.P. Michael, J. Chory, S.A. Kay, and T.C. Mockler. 2011. Global profiling of rice and poplar transcriptomes highlights key conserved circadian-controlled pathways and cis-regulatory modules. PLoS ONE 6: e16907. https://doi.org/10.1371/journal.pone.0016907.
Fornara, F., A. de Montaigu, and G. Coupland. 2010. SnapShot: Control of flowering in Arabidopsis. Cell 141: 550-550.e552. https://doi.org/10.1016/j.cell.2010.04.024.
Gagic, M., M. Faville, I. Kardailsky, and J. Putterill. 2015. Comparative genomics and functional characterisation of the GIGANTEA gene from the temperate forage perennial ryegrass Lolium perenne. Plant Molecular Biology Reporter 33: 1098–1106. https://doi.org/10.1007/s11105-014-0820-3.
Glassop, D., A. Rae, and G. Bonnett. 2014. Sugarcane flowering genes and pathways in relation to vegetative regression. Sugar Tech 16: 235–240. https://doi.org/10.1007/s12355-013-0284-z.
Glassop, D., and A.L. Rae. 2019. Expression of sugarcane genes associated with perception of photoperiod and floral induction reveals cycling over a 24-hour period. Functional Plant Biology 46: 314–327. https://doi.org/10.1071/FP18136.
Greenup, A., W.J. Peacock, E.S. Dennis, and B. Trevaskis. 2009. The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals. Annals of Botany 103: 1165–1172. https://doi.org/10.1093/aob/mcp063.
Grivet, L., and P. Arruda. 2001. Sugarcane genomics: Depicting the complex genome of an important tropical crop. Current Opinion in Plant Biology 5: 122–127. https://doi.org/10.1016/s1369-5266(02)00234-0.
Hassankhah, A., M. Rahemi, H. Ramshini, S. Sarikhani, and K. Vahdati. 2020. Flowering in Persian walnut: Patterns of gene expression during flower development. BMC Plant Biology 20: 136. https://doi.org/10.1186/s12870-020-02372-w.
Higgins, J.A., P.C. Bailey, and D.A. Laurie. 2010. Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. PLoS ONE 5: e10065. https://doi.org/10.1371/journal.pone.0010065.
Hori, K., E. Ogiso-Tanaka, K. Matsubara, U. Yamanouchi, K. Ebana, and M. Yano. 2013. Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response. The Plant Journal 76: 36–46. https://doi.org/10.1111/tpj.12268.
Hotta, C.T., M.Y. Nishiyama Jr., and G.M. Souza. 2013. Circadian rhythms of sense and antisense transcription in sugarcane, a highly polyploid crop. PLoS ONE 8: e71847. https://doi.org/10.1371/journal.pone.0071847.
Imaizumi, T., and S. Kay. 2006. Photoperiodic control of flowering: Not only by coincidence. Trends Plant Science 11: 550–558. https://doi.org/10.1016/j.tplants.2006.09.004.
Iskandar, H.M., R.S. Simpson, R.E. Casu, G.D. Bonnett, D.J. Maclean, and J.M. Manners. 2004. Comparison of reference genes for quantitative real-time polymerase chain reaction analysis of gene expression in sugarcane. Plant Molecular Biology Reporter 22: 325–337. https://doi.org/10.1007/BF02772676.
Izawa, T., Y. Takahashi, and M. Yano. 2003. Comparative biology comes into bloom: Genomic and genetic comparison of flowering pathways in rice and Arabidopsis. Current Opinion in Plant Biology 6: 113–120. https://doi.org/10.1016/S1369-5266(03)00014-1.
James, N. I., and J. D. Miller. 1971. Shoot apex development in early-, mid- and late-season flowering sugarcane clones. Proceedings of the 14th International Congress of Sugar Cane Technologists 14: 334–340.
Julien, M. H. R. 1971. The photoperiodic control of flowering in Saccharum. Proceedings of the 14th International Congress of Sugar Cane Technologists 14: 323–333.
Julien, M.H.R. 1973. Physiology of flowering in Saccharum. Journal of Experimental Botany 24: 549–557. https://doi.org/10.1093/jxb/24.3.549.
Julien, R., G.C. Soopramanien, and D. Lorence. 1974. Juvenility, senility, climate and flowering in Saccharum. Proceedings of the International Socienty of Sugar Cane Technologists 15: 984–990.
King, R.W., T. Moritz, L.T. Evans, J. Martin, C.H. Andersen, C. Blundell, I. Kardailsky, and P.M. Chandler. 2006. Regulation of flowering in the long-day grass Lolium temulentum by gibberellins and the FLOWERING LOCUS T gene. Plant Physiology 141: 498–507. https://doi.org/10.1104/pp.106.076760.
Koskela, E.A., K. Mouhu, M.C. Albani, T. Kurokura, M. Rantanen, D.J. Sargent, N.H. Battey, G. Coupland, P. Elomaa, and T. Hytönen. 2012. Mutation in TERMINAL FLOWER1 reverses the photoperiodic requirement for flowering in the wild strawberry Fragaria vesca. Plant Physiology 159: 1043–1054. https://doi.org/10.1104/pp.112.196659.
Lagercrantz, U. 2009. At the end of the day: A common molecular mechanism for photoperiod responses in plants? Journal of Experimental Botany 60: 2501–2515. https://doi.org/10.1093/jxb/erp139.
Li, F., X. Zhang, R. Hu, F. Wu, J. Ma, Y. Meng, and Y. Fu. 2013. Identification and molecular characterization of FKF1 and GI homologous genes in soybean. PLoS ONE 8: e79036. https://doi.org/10.1371/journal.pone.0079036.
MacColl, D. 1977. Some aspects of the flowering of sugar cane in Barbados and its control in a breeding programme. Annals of Botany 41: 191–201. https://doi.org/10.1093/oxfordjournals.aob.a085267.
Manechini, J.R.V., P.H.dS. Santos, E. Romanel, M.dS. Brito, M.S. Scarpari, S. Jackson, L.R. Pinto, and R. Vicentini. 2021. Transcriptomic analysis of changes in gene expression during flowering induction in sugarcane under controlled photoperiodic conditions. Frontiers in Plant Science 12: article 635784. https://doi.org/10.3389/fpls.2021.635784.
Mangelsdorg, A.J. 1957. Sugar cane breeding—In retrospect and in prospect. Proceedings of the International Society of Sugar Cane Technologists 9: 560–575.
Miah, M.A., and N.K. Paul. 2006. Effect of photoperiod treatment on pollen viability and seed setting in sugarcane. Indian Sugar 56: 39–44.
Midmore, D.J. 1980. Effects of photoperiod on flowering and fertility of sugarcane (Saccharum spp.). Field Crops Research 3: 65–81. https://doi.org/10.1016/0378-4290(80)90008-8.
Moore, P. 1974. Investigations on the flowering of Saccharum II. Number of spindle leaves and date of induction. Proceedings of the International Society of Sugar Cane Technologists 15: 7–16.
Moore, P.H. 1971. Investigations of the flowering of Saccharum. I. Ontogeny of the inflorescence. Canadian Journal of Botany 49: 677–682. https://doi.org/10.1139/b71-104.
Moore, P.H., and N. Berding. 2014. Flowering. In Sugarcane: Physiology, biochemistry, and functional biology, ed. P.H. Moore and F.C. Botha, 379–410. Milton: Wiley. https://doi.org/10.1002/9781118771280.ch15.
Moore, P.H., and K.J. Nuss. 1987. Flowering and flower synchronization. In Developments in crop science II sugarcane improvement through breeding, ed. J.H. Don, 273–311. New York: Elsevier. https://doi.org/10.1016/B978-0-444-42769-4.50012-6.
Mouradov, A., F. Cremer, and G. Coupland. 2002. Control of flowering time: Interacting pathways as a basis for diversity. The Plant Cell 14: S111–S130. https://doi.org/10.1105/tpc.001362.
Murphy, R.L., R.R. Klein, D.T. Morishige, J.A. Brady, W.L. Rooney, F.R. Miller, D.V. Dugas, P.E. Klein, and J.E. Mullet. 2011. Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proceedings of the National Academy of Sciences 108: 16469–16474. https://doi.org/10.1073/pnas.1106212108.
Nolan, T., R.E. Hands, and S.A. Bustin. 2006. Quantification of mRNA using real-time RT-PCR. Nature Protocols 1: 1559–1582. https://doi.org/10.1073/pnas.1106212108.
Nuss, K.J. 1977. Synchronization of flowering to implement a proven cross breeding system in sugarcane. Proceedings of the International Society of Sugar Cane Technologists 16: 111–119.
OCED-FAO 2019. Sugar, in OCED-FAO Agricultural Outlook. http://www.agri-outlook.org/commodities/Sugar.pdf. Accessed 25 October 2021.
Ogiso, E., Y. Takahashi, T. Sasaki, M. Yano, and T. Izawa. 2010. The role of casein kinase II in flowering time regulation has diversified during evolution. Plant Physiology 152: 808–820. https://doi.org/10.1104/pp.109.148908.
Paliatseas, E.D. 1962. Further studies on flowering of sugar cane in Louisiana. Proceedings of the International Society of Sugar Cane Technologists 11: 504–515.
Paliatseas, E.D. 1971. Flowering of sugarcane with reference to induction and inhibition. Proceedings of the International Society of Sugar Cane Technologists 14: 354–364.
Paliatseas, E.D. 1974. Flowering of sugarcane in Louisiana as related to interspecific hybridization. Proceedings of the International Society of Sugar Cane Technologists 15: 46–54.
Panje, R.R., and T. Raja Rao. 1961. Prevention of flowering in sugarcane. Current Science 30: 211–212.
Panje, R.R., T. Raja Rao, and K.K. Srivastava. 1968. Studies on the prevention of flowering in sugarcane I. Effect of suppression of flowering by defoliation on the yield and juice-quality of cane. Proceedings of the International Society of Sugar Cane Technologists 13: 468–475.
Qin, Z., J. Wu, S. Geng, N. Feng, F. Chen, X. Kong, G. Song, and K, Chen, A, Li, L. Mao, and L. Wu. 2017. Regulation of FT splicing by an endogenous cue in temperate grasses. Nature Communications 8: 14320. https://doi.org/10.1038/ncomms14320.
Rae, A. L., D. Glassop, and G. D. Bonnett. 2015. Faster flowering - new opportunities for genetic improvement: final report 2012/024, Sugar Research Australia. p 48.
Rizk, T.Y., H.A. Khalil, and H.M. Nosaer. 2002. Photoperiodic response of five locally developed sugar cane varieties. Arab Universities Journal of Agricultural Sciences 10: 619–627.
Rizk, T.Y., H.A. Khalil, and H.M. Nosaer. 2004. Flower initiation of certain groups of sugar cane varieties to photoperiod regimes and growth regulators. Egyptian Journal of Agronomy 26: 9–19.
Rizk, T.Y., H.A. Khalil, and H.A. Nosaer. 2007. Effect of two-photoperiod regimes on flower induction of five non flowering sugar cane varieties. African Crop Science Conference Proceedings 8: 155–159.
Singh, S.K., J. Dilip, S. Sharma, R.P. Singh, and S.B. Singh. 2009. Induction of flowering in sugarcane by photoperiod manipulation under sub-tropical conditions. Indian Sugar 58: 33–38.
Song, Y.H., S. Ito, and T. Imaizumi. 2013. Flowering time regulation: Photoperiod- and temperature-sensing in leaves. Trends in Plant Science 18: 575–583. https://doi.org/10.1016/j.tplants.2013.05.003.
Srikanth, A., and M. Schmid. 2011. Regulation of flowering time: All roads lead to Rome. Cellular and Molecular Life Sciences 68: 2013–2037. https://doi.org/10.1007/s00018-011-0673-y.
Srivastava, R., S. Singh, and P. Singh. 2006. Artificial induction of flowering in sugarcane under sub-tropical conditions—A successful approach. Sugar Tech 8: 184–186. https://doi.org/10.1007/BF02943658.
Turck, F., F. Fornara, and G. Coupland. 2008. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annual Review of Plant Biology 59: 573–594. https://doi.org/10.1146/annurev.arplant.59.032607.092755.
Vijayasaradhy, N., and R. Narasimhan. 1953. Control of flowering in sugarcane. Proceedings of the International Society of Sugar Cane Technologists 8: 371–401.
Wang, X., L. Wu, S. Zhang, L. Wu, L. Ku, X. Wei, L. Xie, and Y. Chen. 2011. Robust expression and association of ZmCCA1 with circadian rhythms in maize. Plant Cell Reports 30: 1261–1272. https://doi.org/10.1007/s00299-011-1036-8.
Yang, S., R.L. Murphy, D.T. Morishige, P.E. Klein, W.L. Rooney, and J.E. Mullet. 2014a. Sorghum Phytochrome B inhibits flowering in long days by activating expression of SbPRR37 and SbGHD7, repressors of SbEHD1, SbCN8 and SbCN12. PLoS ONE 9: e105352. https://doi.org/10.1371/journal.pone.0105352.
Yang, S.S., B.D. Weers, D.T. Morishige, and J.E. Mullet. 2014b. CONSTANS is a photoperiod regulated activator of flowering in sorghum. BMC Plant Biology 14: 148. https://doi.org/10.1186/1471-2229-14-148.
Yanovsky, M.J., and S.A. Kay. 2003. Living by the calendar: How plants know when to flower. Nature Reviews Molecular Cell Biology 4: 265–276. https://doi.org/10.1186/1471-2229-14-148.
Yon, F., P.-J. Seo, J.Y. Ryu, C.-M. Park, I.T. Baldwin, and S.-G. Kim. 2012. Identification and characterization of circadian clock genes in a native tobacco Nicotiana Attenuata. BMC Plant Bioliogy 12: 172. https://doi.org/10.1186/1471-2229-12-172.
Zhong, X., X. Dai, J. Xv, H. Wu, B. Liu, and H. Li. 2012. Cloning and expression analysis of GmGAL1, SOC1 homolog gene in soybean. Molecular Biology Reports 39: 6967–6974. https://doi.org/10.1007/s11033-012-1524-0.
Acknowledgements
The authors would like to thank Terry Grant for his fastidious attention to the photoperiod controls and plant maintenance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Glassop, D., Bonnett, G.D. & Rae, A.L. Changes in Floral Induction Genes During Photoperiod Induction in Flowering and Reluctant-to-Flowering Sugarcane Varieties. Sugar Tech 25, 751–765 (2023). https://doi.org/10.1007/s12355-022-01233-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12355-022-01233-0