Abstract
Molecular probes provide imaging signal and contrast for the visualization, characterization, and measurement of biological processes at the molecular level. These probes can be designed to target the cell or tissue of interest and must be retained at the imaging site until they can be detected by the appropriate imaging modality. In this article, we will discuss the basic design of molecular probes, differences among the various types of probes, and general strategies for their evaluation of cardiovascular disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Molecular imaging incorporates traditional imaging modalities with special probes that provide the imaging signal and contrast for the visualization, characterization, and measurement of biological processes at the cellular, subcellular, and even molecular levels. Akin to stains for histological analysis of tissue samples, these probes are injected into living subjects to image discrete biological and molecular events. Although these agents have many aliases, including but not limited to molecular imaging probes, molecular beacons, reporter probes, tracers, smart probes, activatable probes, nanoparticles, and contrast agents, they all enable serial in vivo visualization of biological processes. In this article, we will discuss the basic design of molecular imaging probes, differences in the properties of various probes, and how they are generally used to evaluate cardiovascular disease.
Basic Components of the Imaging Probe
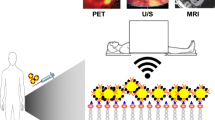
The molecular imaging probe is composed of three elements (Figure 1).1 The first element is an affinity component, which is a ligand that interacts with the target. Most targets are proteins, because they are relatively more abundant than other molecules present in the cell. Only 1 to 2 DNA and 10 to 1000 RNA, for example, are present in the cell, which is not sufficient to be adequately detected. In contrast, proteins, such as intracellular enzymes, cell surface receptors, membrane transporters, and extracellular enzymes, are relatively more abundant (from 0.01 to 1 million). Thus, affinity components, which are either antibodies or peptides, are typically designed to bind to protein targets. Nonspecific probes, however, have no affinity component.
Schematic of a molecular imaging probe designed to target a cell surface protein or extracellular target. A molecular imaging probe is composed of three parts: (1) Ligand: Antibodies and proteins that bind to the target cell; (2) Carrier: Carriers such as cells, viral particles, nanoparticles, etc., that bind to the ligand and the signal element; (3) Signal elements that generate signals including visible light, gamma rays, and x-rays that are detectable by an imaging system (i.e., MRI, PET/SPECT, or CT). MRI magnetic resonance imaging, PET positron emission tomography, SPECT single photon emission computed tomography, CT computed tomography
The second element is a signaling component that emits signals that can be detected by traditional imaging systems. In general, the signaling components for optical approaches, nuclear imaging systems, magnetic resonance imaging or ultrasound imaging systems are typically fluorochromes, radiotracers, iron oxide particles, or perofluorocarbon gas, respectively. The third component is a carrier that links the affinity component with the signaling component. Carriers are usually cells, liposomes, polymers, viral particles, nanoparticles, or microbubbles. Probes may also have other features designed to amplify signal and to increase imaging sensitivity, including trapping converted ligands inside the cell (i.e., phosphorylation of glucose by herpes simplex virus type I thymidine kinase, HSV1-TK) and changing their physical nature after target interaction (i.e., fluorescence quenching).
Ideal Molecular Imaging Probe
Clinical translation will require probes to meet efficacy as well as safety standards.1 To achieve adequate efficacy, a molecular imaging probe must be able to generate sufficient signal for detection (high sensitivity and contrast-noise ratio) and provide biochemical information that accurately reflects the biological process of interest (high specificity and target binding affinity). To ensure safe administration, the molecular probe and its metabolites must have minimal toxicity locally (i.e., target cells and tissues) and systemically. For economic feasibility, production costs must be low and high-throughput generation is required. Because not all molecular imaging probes have achieved these requirements, many are still under development, and only a few are used clinically, as discussed below.
Types of Molecular Imaging Probes
Molecular probes can be categorized based on answers to the following questions: (1) how they generate signal (i.e., optical vs radiolabeled probes)? (2) whether they continuously produce signal or only produce signal after target interaction (e.g., continuous vs activatable probes)? (3) whether the biologically process can be directly visualized by imaging a ligand probe as it binds to its receptor target or indirectly by expression of a reporter gene that measures the endogenous protein (direct vs indirect probes)? and (4) whether they bind specifically to a target or do not have a distinct set of targets (e.g., specific vs nonspecific probes)? In the following section, we will discuss different types of probes and their specific features, categorizing them based on how they generate signal.
Optical Probes
Optical probes (e.g., organic dyes) are composed of fluorescent molecules coupled to antibodies, peptides, non-peptide small molecules, and nanoparticles for targeting various biological processes. Signal is generated by excitation of the probe at a specific wavelength and is detected as emitted light at another wavelength using an optical imaging camera. Although these fluorescent molecules can emit light with wavelengths between 650 and 1350 nm (in the near-infrared range) where tissue penetration is the greatest, the most recent optical probes use wavelengths <900 nm to minimize light attenuation, which is directly proportional to wavelength. This strategy maximizes both sensitivity and specificity. Using standard fluorescent labels, however, remains challenging especially in multicolor experiments due to issues with signal intensity strength, background auto-fluorescence-limiting sensitivity, rapid photobleaching resulting in relatively short lifetimes, narrow excitation ranges, broad emission spectra causing high spectral overlap, and the limited number of agents available in the near-infrared range.
The advent of quantum dot technology has alleviated some of these limitations. Quantum dot nanoparticles are fluorescent semiconductor nanoparticles, which are composed of semiconductor material with diameters in the range of 2-10 nm and exhibit the following unique optical properties: (1) large molar extinction coefficient which increases probe brightness and improves imaging sensitivity to the single nanoparticle level; (2) narrow emission band enabling multicolor labeling; (3) broad excitation spectrum that allows simultaneous excitation of quantum dots for imaging and analyzing multiple molecular targets simultaneously; (4) large Stokes shifts (distance between the excitation and emission peaks) which can be exploited to further improve imaging sensitivity; and (5) high photostability enabling consistent image acquisition for longer periods. The major hurdle for clinical translation of quantum dot technology, however, remains the possibility of cytotoxicity.
Another alternative to standard fluorochromes is Raman spectroscopy.2 Raman spectroscopy measures scattering phenomenon instead of the absorption/emission, which is measured in fluorescence. Compared to fluorescence, Raman scattering has less photobleaching and exhibits narrow spectral features, enabling longer-term imaging and the conduct of multicolor experiments, respectively. One of the main challenges to Raman spectroscopy, however, is that the generated signal has very low intensity, limiting imaging sensitivity. With the advent of single wall carbon nanotubes, which enables the production of intense Raman peaks upon light exposure, Raman spectroscopy has emerged as a promising technique for molecular imaging. Their low cytotoxicity and rapid renal excretion further support their use in clinical molecular imaging.
Radiolabeled Probes
Radiolabeled probes including single photon emission computed tomography (SPECT) and positron emission tomography (PET) probes are perhaps the most common probes used for molecular imaging. They differ with respect to the radiotracer used and the mechanism by which they generate signal. SPECT probes (e.g., technetium 99 m, indium 111, iodine-123, and iodine 131) are produced by a generator and emit gamma rays that can be detected by a SPECT camera. In contrast, PET probes require an on-site cyclotron, emit positrons that collide with electrons to produce gamma rays that are 180° apart, and are detected by the PET scanner. Although these radiolabeled probes are readily available and highly sensitive, they have relatively low spatial resolution (clinical SPECT: 7-15 mm; clinical PET: 6-10 mm; microSPECT: 0.5-2 mm; and microPET: 1-2 mm), expose subjects to radiation, and lack anatomic information, which limit their clinical translation.
Ultrasound Probes
Ultrasound probes are composed of microbubbles that are directly conjugated to ligands or indirectly conjugated by attachment via a linker molecule (i.e., streptavidin).3 Microbubbles contain perfluorocarbon gas enclosed in spherical shells of lipid, protein, or biocompatible polymer. Because of their size (1-10 μm), they remain in the vasculature and, therefore, can only be used for imaging targets on the endothelium. Signal can be produced in one of two ways. Because of the differences in echogenicity between the gas in the microbubbles and soft tissue, microbubbles can be easily identified with ultrasound imaging. Alternatively, signal can be produced by destroying microbubbles using high acoustic powers, thereby, releasing the echogenic free gas. Although these probes are inexpensive and specific, microbubbles have low adhesion efficiency and cannot image extravascular targets, limiting their utility.
MR Probes
There are two major categories of magnetic resonance (MR) probes.4 The first are gadolinium probes that appear bright on T1-weighted images. To achieve adequate imaging sensitivity, multiple gadolinium particles must be linked together by a carrier such as a nanoparticle or protein. To image specific molecular targets, the carrier must be conjugated to a ligand such as an antibody or peptide. By incorporating multiple gadolinium chelates into nanoparticles or liposomes or micelles, signal can be further amplified. Alternatively, imaging sensitivity as well as specificity can be enhanced by means of novel gadolinium agents that oligomerize in response to target enzyme activation.
Iron oxide particles are the second class of MR probes. Unlike gadolinium, they provide negative contrast and appear as signal voids on T2-weighted images. There are three classes of iron oxide particles: (1) microparticles of iron oxide that have been used for targeting platelets (MPIOs: 0.9-8 μm); (2) short circulating iron oxide nanoparticles, otherwise known as superparamagnetic iron oxide particles (SPIOs: 60-150 nm) that have a thin and incomplete dextran coat; and (3) the long circulating, ultrasmall superparamagnetic iron oxide particles (USPIOs: 10 to 40 nm), which have a thick and complete dextran coat. Because iron oxide particles are nonspecifically taken up by cells, they have been used to image macrophages in the atheroma and stem cells after transplantation. Iron oxide particles can also be conjugated to specific epitopes like oxidized low-density lipoprotein, VCAM1, and other components to image atherosclerosis.
Reporter Gene and Reporter Probes
Reporter gene and reporter probes are used for indirect imaging, which can be more complex than direct molecular imaging, where the resultant image and image intensity are directly related to the target-probe interaction.5 The ideal reporter gene is absent from the cells used in the study or can be easily distinguishable from the native form of the gene, easily assayed, and has a broad emission spectra. The selected reporter gene is placed under the control of a promoter in a DNA construct and transfected into target cells. The transcribed reporter protein is usually an enzyme that catalyzes a reaction that produces a detectable signal or is transcellular membrane protein that enables entry of a molecular probe. Further detail on reporter gene imaging can be found under the section: “In vivo imaging strategies using molecular probes.”
Hybrid Probes for Multimodality Imaging
With the advent of multimodality imaging systems, a number of hybrid probes have been developed to provide simultaneous anatomic, functional, metabolic, or molecular information of selected targets. For preclinical imaging, for example, dual and triple reporter gene probes have been developed to perform initial in vivo imaging using radiolabeled and/or MR probes, followed by post-mortem imaging with optical probes.
Engineered nanoparticles also offer a unique opportunity to develop dual-, tri-, or multimodal imaging probes. For example, a single nanoparticle carrying a radiolabeled probe that is highly sensitive for even the most scant molecular targets can be coupled with an MR probe that provides high spatial resolution. Due to their low rate of extravascular penetration and long circulating times, however, nanoparticles are restricted to the visualization and treatment of processes within or near the vessel wall. Clinical translation has also been hampered by safety and regulatory concerns.
Optoacoustic or photoacoustic imaging is another promising multimodality approach, which is based on the discovery that sounds waves are formed after light is absorbed in tissue (e.g., “the photoacoustic effect”).6 The generated sound waves can then be detected by ultrasound machines. This approach takes advantage of the wide array of optical probes available for molecular imaging as well as the high spatial and temporal resolution and tissue penetration of ultrasound. Photoacoustic imaging has been used to differentiate components of the atherosclerotic plaque and label stem cells for tracking post transplantation.
In Vivo Imaging Strategies Using Molecular Probes
There are two basic ways to apply molecular probes to visualize biological processes in vivo. Using the first method, a probe targeting a surface receptor or intra/extracellular target on cells or tissue is injected into the living subject and imaged over time. Probes targeting an intracellular target face an additional challenge because they need a transport mechanism to bring them across the cell or organelle membrane for target to probe interaction. Nevertheless, probes targeting both intra and extracellular targets have been used to evaluate inflammation, thrombosis and apoptosis in the vulnerable plaque as well as myocardial viability, fibrosis and remodeling in heart failure.4 , 7 Using the second method, target cells are labeled with the probe and injected into the living subjects and tracked serially. This approach has been used to label white blood cells for evaluating inflammation and to label stem cells after transplantation.
Molecular Imaging Probes for Imaging Cell Surface, Extracellular Targets, or Intracellular Targets
Molecular imaging probes have been designed to evaluate many cardiovascular disease processes including proliferation, cell death, inflammation, thrombosis, angiogenesis, hypoxia, metabolism, and changes in receptor expression. First, key players or regulators participating in the disease process of interest are identified, and then probes are designed to bind these targets.8
Atherosclerosis, for example, is a disease process characterized by the formation of a plaque in arterial walls. Atherosclerotic plaque is composed of cholesterol, inflammatory cells, pro-thrombotic agents, and components of the extracellular matrix. Traditional imaging approaches like nuclear myocardial perfusion and x-ray angiography can only evaluate patients in the advanced stages of disease when extensive plaque causes impairment in blood flow or obstruction, respectively. Molecular imaging approaches, in contrast, can evaluate the morphology of the arterial wall as well as the composition of plaque. Because macrophages have been shown to play an important role in the initiation of plaque formation, various molecular probes have been developed to target these inflammatory mediators including superparamagnetic SPIOs and 18F-FDG, which are engulfed or trapped by macrophages in vivo, enabling imaging by MRI and PET, respectively. Other strategies to image early atherosclerosis include (1) activatable fluorescent probes that emit signal only after encountering cysteine proteases or metalloproteases, upregulation of which has been associated with unstable plaque: (2) 99MTc-labeled annexin V probes that target apoptosis in unstable human carotid plaque: and (3) 18F-labeled peptides or magnetic nanoparticles that bind adhesion molecules (e.g., VCAM-1, vascular adhesion molecule -1), which facilitate recruitment of leukocytes into the atherosclerotic wall.4 We refer the reader to other reviews for details of additional probes targeting atherosclerosis as well as probes targeting other cardiovascular disease processes.4 , 7 , 8
Molecular Imaging Probes to Label Cells
Imaging of the fate of labeled cells after delivery into the living subject can provide important insight into biological processes.4 , 5 , 9 Cells can be labeled directly by incubation with a specific molecular imaging probe (e.g., direct labeling) or indirectly by transfecting/transducing cells with a reporter gene (e.g., reporter gene labeling) (Figure 2(A)).
Schematic of cell labeling strategies. A Direct labeling (top): For direct labeling, cells are incubated with molecular probes (e.g., SPIOs, 99MTc, 111In-Oxime, and 18F-FDG) that are taken up by the cell through passive diffusion, transporters, or endocytosis. These probes emit specific signals that are detected by their respective imaging systems. SPIOs super paramagnetic iron oxide nanoparticles; 99M Tc 99M Technetium, 111 In-Oxime 111 Indium-Oxime, 18 F-FDG 18 F-Fluorodeoxyglucose. B Indirect labeling (bottom): For reporter gene labeling, a reporter gene and its promoter are introduced into the cell via a vector. The reporter gene integrates into the cellular genome. The reporter gene is then transcribed and translated by the host cell machinery to form its corresponding mRNA and a functional protein (e.g., enzyme such as firefly luciferase or herpes simplex virus type 1 kinase as shown here or transcellular membrane which is not shown). A signal is generated after introduction of the enzyme’s substrate (e.g., D-Luciferin or FHBG). Reporter genes can be introduced singly or fused to other reporter genes to make a triple construct that carries FLuc, HSV-TK, and EGFP for multimodality imaging using bioluminescence imaging, PET imaging, and histological analysis, respectively, as shown here. FLuc firefly luciferase, HSV-TK herpes simplex virus type 1 thymidine kinase, FHBG 9-(4-18F-fluoro-3-hydroxymethylbutyl)guanine), EGFP enhanced green fluorescent protein
For direct labeling, molecular probes can enter cells via passive diffusion (e.g., 111In-oxine molecules), endocytosis (e.g., quantum dots, SPIOs, gold nanorods, carbon nanotubes), or active transport (e.g., 18F-FDG). While some agents are trapped in the cell like glucose, which is rapidly phosphorylated preventing its egress, others may leak out of the cell over time, resulting in signal loss. Cell division can also lead to dilution of the contrast agent. Even more concerning is the loss of imaging specificity over time because dead cells engulfed by macrophages emit signals that cannot be differentiated from signals generated from viable cells of interest.
For reporter gene imaging, cells are transfected/transduced with vectors (i.e., viruses, plasmids, mini-circles, etc.) carrying a reporter gene, which integrates into the host genome (Figure 2(B)).4 , 5 Using the host’s cellular machinery, the reporter gene is transcribed into its messenger RNA and then translated into specific proteins such as (1) a membrane transporter that facilitates entry of exogenously delivered probes (e.g., sodium iodide symporter [NIS]), (2) an intracellular enzyme that interacts with an exogenously delivered probe (e.g., HSV1-TK); (3) an intracellular storage protein that accumulates a contrast agent (e.g., ferritin), and (4) a cell surface receptor that binds an exogenously delivered probe (e.g., dopamine type 2 receptor). Compared to direct labeling approaches, reporter gene techniques have higher specificity because only viable cells can produce signal, which does not dilute over time even if the cell divides.
Of the two labeling strategies, only direct labeling with radioisotope tracers has been used routinely in the clinic and in clinical trials. The indium white blood scan, for example, is a routine nuclear medicine procedure where white blood cells are removed from the patient, labeled directly with radioisotope 111In-oxine, and then injected intravenously into the patient. The labeled white blood cells then localize to the area of new inflammation and are imaged using a planar SPECT camera. Direct labeling has also been used to image the fate of bone marrow mononuclear after transplantation into patients with heart failure.
Clinical trials evaluating the fate of stem cells have also relied on direct labeling techniques. Although most reports have used 18F-FDG for PET imaging, the feasibility of 111In-oxine and 99mTc-HMPAO has also been demonstrated. These studies reported low cytotoxicity, but labeling efficiency was variable, ranging from <10% to >70% even with the same agent.4 The main limitation of using direct labeling with radioisotopes for tracking stem cell fate is that serial imaging is limited to the half-life of the tracer, which can be as short as 110 min for 18F-FDG. With a half-life of 2.8 days, however, studies using 111In-oxine have reported successful longitudinal follow-up for up to 3 days.
Although longer-term imaging can be achieved with reporter genes, their clinical translation continues to be hampered by the risk of immunogenicity associated with using nonhuman reporter genes and oncogenic transformation due to the potential for random integration of reporter genes. Strategies circumventing these obstacles, such as the development of a human mitochondrial thymidine kinase type 2 herpes simplex virus reporter gene as well as site-specific integration strategies (e.g., genome-editing methods using zinc-finger nucleases, transcription activator-like effector nucleases, and cluster regularly interspaced short palindromic repeats), will facilitate the clinical translation of reporter gene imaging.
Summary
In this article, we have reviewed the basic components of molecular probes, types of probes, and general strategies for implementation. While the clinical translation of these techniques are still in their infancy, we are confident that further advancement in probe design will enhance their efficacy and safety, and enable more widespread application of these techniques to improve our understanding of atherosclerosis, heart failure, and stem cell fate.
Abbreviations
- CT:
-
Computed tomography
- 18F-FDG:
-
18 F-Fluorodeoxyglucose
- HMPAO:
-
Hexamethylpropyleneamine oxime
- HSV-TK:
-
Herpes simplex virus type I thymidine kinase
- MR:
-
Magnetic resonance
- PET:
-
Positron emission tomography
- SPECT:
-
Single photon emission tomography
- SPIO:
-
Superparamagnetic iron oxide particles
- 99mTc:
-
Technetium 99 M
- VCAM-1:
-
Vascular adhesion molecule-1
References
Nolting DD, Nickels ML, Guo N, Pham W. Molecular imaging probe development: A chemistry perspective. Am J Nucl Med Mol Imaging 2012;2:273-306.
Jokerst JV, Pohling C, Gambhir SS. Molecular imaging with surface-enhanced Raman spectroscopy nanoparticle reporters. MRS Bull 2013;38:625-30.
Villanueva FS. Molecular imaging of cardiovascular disease using ultrasound. J Nucl Cardiol 2008;15:576-86.
Chen IY, Wu JC. Cardiovascular molecular imaging: Focus on clinical translation. Circulation 2011;123:425-43.
Nguyen PK, Riegler J, Wu JC. Stem cell imaging: From bench to bedside. Cell Stem Cell 2014;14:431-44.
Pu K, Shuhendler AJ, Jokerst JV, Mei J, Gambhir SS, Bao Z, et al. Semiconducting polymer nanoparticles as photoacoustic molecular imaging probes in living mice. Nat Nanotechnol 2014;9:233-9.
Osborn EA, Jaffer FA. The advancing clinical impact of molecular imaging in CVD. JACC Cardiovasc Imaging 2013;6:1327-41.
Shaw SY. Molecular imaging in cardiovascular disease: Targets and opportunities. Nat Rev Cardiol 2009;6:569-79.
Nguyen PK, Lan F, Wang Y, Wu JC. Imaging: Guiding the clinical translation of cardiac stem cell therapy. Circ Res 2011;109:962-79.
Acknowledgements
We would like to thank Ian Chen, MD, PhD and Joseph Wu, MD, PhD for their assistance in editing this manuscript. We would like to thank Amy Morris for her contribution to the figures. We would also like to thank the Stanford Cardiovascular Institute, American Heart Association, and the Veterans Affairs for their support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liang, G., Nguyen, P.K. Molecular probes for cardiovascular imaging. J. Nucl. Cardiol. 23, 783–789 (2016). https://doi.org/10.1007/s12350-016-0501-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12350-016-0501-8