Abstract
Insect wounding activates a large number of signals that function coordinately to modulate gene expression and elicit defense responses. How each signal influences gene expression in absence of wounding is also important since it can shed light on changes occurring during the shift to wound response. Using simulated Helicoverpa armigera herbivory on chickpea, we had identified at least 14 WRKY genes that showed 5–50 fold increase in expression within 5–20 min of wounding. Our studies show that contrary to their collective effects upon wounding, individual chemical cues show distinct and often opposite effects in absence of wounding. In particular, jasmonic acid, a key early defense hormone, reduced transcripts of most WRKY genes by > 50% upon treatment of unwounded chickpea leaves as did salicylic acid. Neomycin (a JA biosynthesis inhibitor) delayed and also reduced early wound expression. H2O2 transiently activated several genes within 5–20 min by 5–8 fold while ethylene activated only a few WRKY genes by 2–5 fold. The summation of the individual effects of these chemical cues does not explain the strong increase in transcript levels upon wounding. Detailed studies of a 931 nt region of the CaWRKY41 promoter, show strong wound-responsive GUS expression in Arabidopsis even in presence of neomycin. Surprisingly its expression was lost in the coi1, ein2 and myc2myc3myc4 mutant backgrounds suggesting the requirement of intact ethylene and JA signaling pathways (dependent on MYCs) for wound-responsive expression. The studies highlight the complexity of gene regulation by different chemical cues in the presence and absence of wounding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
During growth, a plant is constantly exposed to various microbes and organisms that depend on it for survival. While most biotic interactions with the plant may be benign or beneficial, some predatory interactions can be detrimental to its growth or even survival. Insect herbivory by chewing and sucking pests is one of the most common predatory interactions that a plant has to protect itself against. In response to mechanical wounding and insect damage, a large number of changes occur at the site of wounding and its surrounding tissue. These include activation of early and late wound responses that address the immediate damage caused and subsequent infections. The early signaling steps include depolarization of the plasma transmembrane potential (Vm), rise in cytosolic Ca++, production of reactive oxygen species (ROS), glutamic acid and mitogen-activated protein kinase (MAPK) activity (Erb and Reymond 2019; Miller et al. 2009; Kumari et al., 2019). Glutamate activates GLUTAMATE RECEPTOR-LIKE proteins, which increase intracellular Ca++ levels and help in the propagation of long distance signaling through slow wave potential (Toyota et al. 2018; Shao et al., 2020). Early wound responses also bring about a rapid change in the signaling of different phytohormones such as JA, ethylene, auxin, ABA, SA and GA (Diezel et al. 2009; Erb et al. 2012; Pandey et al. 2017). These, in turn, modulate downstream signaling and activate different pathways which collectively contribute to the diversity of responses. The specific responses to biotic stress factors are often mediated through the action of transcription factors (TFs) of certain families. Important among these are WRKY, MYC and ERF families that regulate responses to various types of organisms. To what extent the individual signal molecules can independently activate their specific targets (such as TFs) on their own, and whether they also simultaneously require other cues to be in an activated state for their full action, is still not clear.
WRKY transcription factors belong to a large plant-specific family of TFs and function either as activators or suppressors of gene expression (Eulgem and Somssich 2007; Pandey and Somssich 2009; Adachi et al. 2015). They have been identified as being important in the development, defense as well as abiotic stress responses. The family is characterized by the presence of a conserved 60 amino acid WRKY domain with the sequence WRKYGQK at its N- terminus followed by one or more zinc finger motifs (Rushton et al. 2010). The WRKY domain binds the W box (TTGACC) in the promoters of target genes to regulate their transcription (Rushton et al. 1996; Ciolkowski et al. 2008).
Many WRKY family members are activated upon pathogen attack and may mediate responses by regulating hormone interactions (Eulgem 2005; Ryu et al. 2006). NaWRKY3 and NaWRKY6 from N. attenuata mediate herbivory-induced defense responses by modulating JA and JA-Ile/-Leu levels (Skibbe et al. 2008) while AtWRKY70 mediates R gene resistance by altering the balance between SA and JA-dependent defense pathways (Li et al. 2006; Knoth et al. 2007). Similarly, AtWRKY33 expression enhances resistance towards the necrotrophic fungi, Alternaria brassicicola and Botrytis cinerea (Zheng et al. 2006), while AtWRKY53 and AtWRKY70 regulate systemic acquired resistance (Wang et al. 2006). Transgenic tobacco lines expressing Capsicum annum CaWRKY27 show enhanced resistance to R. solanacearum due to modulation of SA, JA and ethylene pathways (Dang et al. 2014) while CaWRKY6 promotes resistance against R. solanacearum resistance by activating CaWRKY40 (Cai et al. 2015). Lines over-expressing OsWRKY89 show greater resistance to fungal blast and Sogatella furicifera by modulating wax deposition on the leaf surface (Wang et al. 2007). OsWRKY53 acts as a suppressor for herbivore-induced responses (Hu et al. 2015). Other WRKYs such as SlWRKY70, SlWRKY72a and 72b, TaWRKY53 (from wheat), mediate defenses against aphids (Atamian et al. 2012; Bhattarai et al. 2010; Van Eck et al. 2010).
Cicer arietinum L. or chickpea is the primary legume in India (with India as the largest producer) but faces losses in productivity due to prevailing biotic and abiotic stresses (Dhaliwal et al. 2010). The pod borer, Helicoverpa armigera, is the major pest of chickpea (Sequeira et al. 2001; Dhaliwal et al. 2010) and alone accounts for losses of up to 30% or more (Dinesh, et al. 2017). Hence, a detailed understanding of the earliest wound responses mounted by chickpea against Helicoverpa is necessary so as to develop strategies to deter these as soon as they begin feeding on leaves/pods. In pursuit of these, we had performed simulated herbivory on chickpea leaves using H. armigera oral secretions and carried out a comparative analysis of transcriptomes of unwounded leaves with insect saliva-pretreated/mechanically wounded chickpea leaves at 20 min. The analysis revealed differential expression of about 8.4% of the chickpea transcriptome within 20 min of wounding (Pandey et al. 2017). Interestingly, a large number of genes of the WRKY family were identified amongst the earliest wound-inducible genes in the transcriptomic data. In this study, we have tried to dissect the early wound response of these genes to study how various factors contributing to the wound response can individually influence the expression of these genes in absence of wounding. Our studies show that each factor contributes differently to the regulation of each gene and that the wound response is not a summation of individual effects of the major chemical cues.
Materials and methods
Plant material
Chickpea seeds (Cicer arietinum, variety Pusa-362) were grown in pots in a growth chamber at 22 ± 2 ºC with a 16 h light/8 h dark period at a light intensity of 100 µEm−2 s−1 and 75% relative humidity. For neomycin treatment, chickpea seeds were grown in the field of NBRI from November to March.
Treatments
JA, SA and H2O2 treatment Eight-week old chickpea plants were treated either with 100 µM jasmonic acid (JA, dissolved in ethanol), 2 mM salicylic acid (SA, dissolved in ethanol), or 5 mM H2O2 by lightly and uniformly spraying on to the surface of leaves of individual plants using a hand sprinkler with constant flow. Samples were collected at intervals of 0, 20, 60, and 120 min after treatment. A set of plants sprayed with water containing the same amount of ethanol (used in the hormone sprays), and kept for the same time intervals as those of JA/SA treatments, served as a negative control. Samples were frozen in liquid N2 and stored at -70 ºC until further use.
Ethylene treatment For ethylene treatment, chickpea plants were kept in a desiccator and ethylene (10 µl L−1) was injected in the desiccator with a needle. Samples were collected at the time points of 0, 20, 60 and 120 min.
Neomycin treatment Eight-week-old field-grown chickpea plants were pre-treated with 100 µM neomycin solution for 1 h by spraying. Thereafter, the leaves were exposed to oral secretions of Helicoverpa armigera (10 µl oral secretions spread over the leaf with a soft brush) and then rapidly wounded with a pair of forceps by repeated pricking about 9–10 times (18–20 punctures) within a span of 10 s. The leaf tissue was collected at time points of 0, 5, 20, 60 and 120 min after simulated herbivory of neomycin-pretreated leaves and qRT-PCR was carried out and compared with wounded, neomycin-untreated leaves.
RNA isolation and real-time PCR analysis
Total RNA was extracted from the frozen chickpea leaves using a plant total RNA isolation kit (Sigma, India) according to the manufacturer’s instructions. The cDNAs were synthesized from RNA samples using the REVERTAID MMLV kit (Fermentas). Gene-specific real-time PCR primers are listed in Supplementary Table S1. Real-time PCR was performed in 10 μl for a set of selected WRKY genes using SYBR Green PCR Master Mix (ABI, USA) using the following cycle conditions: 94 °C for 2 min, followed by 30 cycles of 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s, and a final 5 min extension at 72 °C. EF1α and HSP90 were used as internal controls for the normalization of relative mRNA in different RNA samples. Reactions were carried out on an ABI StepOnePlus real-time PCR system with three biological replicates, each with three technical replicates. Relative gene expression was calculated using the 2−ΔCT method for all comparisons except neomycin treatment where 2−ΔΔCT was used (Livak and Schmittgen, 2001).
In silico analysis of promoter sequences
To identify putative cis regulatory elements in the promoter region of different wound-inducible WRKYs, a region of 2 kbp upstream from the translational initiation codon was extracted from genomic regions using the desi chickpea genome sequence ICC4958 available in the NCBI database and analyzed using the web-based online program PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/), a database of plant cis-acting regulatory DNA elements. The accession numbers of the WRKY gene sequences are provided in Supplementary Table S2.
Promoter studies in wild type and mutant Arabidopsis and histochemical GUS assays
The promoter region of one of the WRKY genes, WRKY41, was chosen for detailed studies and validation. A region of 931 nt was isolated from genomic DNA of chickpea using gene-specific primers (Table S1) based on the sequence in the chickpea genome database available in NCBI database and cloned first in pTZ57R/T (Fermentas) and then introduced in pBI101.2 at SalI. Primers were designed such that the initiation codon of the gene was in translational fusion with the GUS gene. The construct was introduced into the Agrobacterium strain GV3101. Promoter studies were carried out in stable transgenic Arabidopsis thaliana, ecotype Columbia (Col-0) using the floral dip method as described (Clough and Bent, 1998). Phytohormone treatments for promoter analysis were carried out by treating four week-old transgenic Arabidopsis plants expressing the promoter with either JA (100 μM), ethylene (10 µL L−1), SA (2 mM) or mock (water or 0.1% ethanol) for 2 h before color development.
To identify the hormones and factors regulating the function of CaWRKY41 promoter, the WRKY41pro construct was introduced into various Arabidopsis mutant backgrounds such as ein2-1 (defective in ethylene responses), coi1-1 (defective in JA responses) and myc2myc3myc4 (defective in multiple MYC functions affecting specific JA responses). All the plants were grown on soilrite in a culture room maintained at 22 ± 2 °C under a 16 h light period (light intensity 100 μE m−2 s−1, relative humidity 78 ± 4% at 25 °C).
Histochemical GUS staining was carried out as described (Gattolin et al. 2006). Tissue samples were incubated in 1 mg ml−1 X-gluc solution containing 50 mM sodium phosphate buffer pH 7.2, 0.5 mM K3Fe(CN)6 and 0.5 mM K4Fe(CN)6 for 16–24 h at 37 °C. After incubation, tissues were destained in 70% ethanol at 37 oC and examined. Light microscopy was performed on a Leica Wild M3Z microscope (Leica Germany).
Statistical analysis
Significant differences between control and treated plants were analyzed using ANOVA: single factor with the help of Analysis tool pack (Data Analysis) in Microsoft Excel 2010 where * indicates P < 0.05, ** indicates P < 0.01, *** indicates P < 0.001.
Results
A large number of WRKY genes are activated early in chickpea in response to simulated herbivory by Helicoverpa armigera.
A comparative transcriptome sequencing of simulated herbivory on chickpea leaves was previously carried out in our lab using RNA from unwounded and 20 min mechanically wounded chickpea leaves (pre-exposed to H. armigera saliva) to assess the early transcriptional responses of chickpea towards wounding (Pandey et al. 2017). One of the most prominent groups to be differentially regulated upon simulated herbivory was the WRKY family, with 32 genes being up-regulated within 20 min of wounding.
In order to validate the transcriptomic data, 14 out of 32 up-regulated WRKY genes showing greater than five-fold change in actual transcript levels (Log2FC ≥ 2) were selected for real time PCR validation. These included WRKY11, WRKY17, WRKY33A, WRKY33B, WRKY33C, WRKY40A, WRKY40B, WRKY40C, WRKY41, WRKY46, WRKY53, WRKY70A, WRKY70B and WRKY72. Of the 14 genes chosen, WRKY33C, WRKY40A, WRKY41 and WRKY53 were most strongly up-regulated with 30–45 fold higher expression compared to the unwounded control while the others showed 5–16 fold higher expression within 5–20 min of wounding (Fig. 1). The qRT-PCR expression profiles of the selected WRKY genes were consistent with the patterns obtained by RNA-Seq analyses, indicating that changes in expression determined by transcriptome sequencing were true to nature and that the WRKYs responded strongly and rapidly to wound signals.
Real-time PCR validation of expression of the wound-inducible chickpea WRKY genes identified from the transcriptome. Total RNA was isolated from chickpea leaves subjected to simulated herbivory by H. armigera saliva and mechanical wounding for 0, 5 and 20 min. The data was normalized against reference genes EF1α and HSP-90. Expression analysis was performed in technical triplicates on three biological replicates. Error bars show the standard error ± SE of three biological control. * on the bar indicate significant differences at *P < 0.05, **P < 0.01, ***P < 0.001 with respect to the controls
JA strongly suppresses the expression of most wound-inducible WRKY genes in absence of wounding
Like most plants, chickpea responds to simulated herbivory and mechanical wounding by activating and remodeling several phytohormone pathways (Pandey et al. 2017). Jasmonic acid is the primary defense-related hormone involved in mediating short term and long term responses to herbivory. The JA pathway is also rapidly up-regulated upon simulated herbivory in chickpea (Pandey et al. 2017). To check whether the strong up-regulation observed upon simulated herbivory in chickpea was primarily induced by JA, the expression of the WRKY genes was tested in response to JA treatment under unwounded conditions.
Quite against expectations of what was observed upon wounding, the expression profiles of the majority of the wound-inducible WRKY genes namely WRKY11, WRKY17, WRKY33A, WRKY40B, WRKY40C, WRKY41, WRKY53 and WRKY70A showed a rapid reduction in transcript levels upon JA treatment to less than 50% of the control values in 20 min (Fig. 2). Three of these, WRKY33A, WRKY41 and WRKY53, were reduced to just 20% within 20 min of JA treatment. WRKY33B also showed a JA-dependent reduction in transcript levels but these reduced to below 50% after 1 h. Transcript levels of WRKY33C and WRKY46 did not show much change while WRKY72 and WRKY40A were induced upon JA treatment in 20 min. WRKY70B was unusual in showing a slight induction at 20 min after JA treatment, but a strong reduction to less than 20% after 60 min followed again by a rise in transcript levels at 2 h after JA treatment. The results showed that JA strongly inhibited transcript accumulation of most wound-inducible genes in absence of wounding and suggested that it may either not be involved in the early wound response or may only partly contribute to it or that it may have a role reversal depending on the conditions.
Real-time expression profiles of WRKY genes in response to jasmonic acid. Leaves of eight-week-old unwounded chickpea plants were treated with 100 µM jasmonic acid. Samples were collected at 0, 20, 60 and 120 min. Reactions were run and analyzed as described in Fig. 1
Suppression of JA biosynthesis by neomycin treatment only partially suppresses the wound-induced expression of WRKY genes
Although most of the wound up-regulated WRKYs in chickpea showed a strong reduction in transcript levels upon treatment with JA in absence of wounding, the possibility existed that these genes may still be activated by JA but only in combination with some accessory factor(s) that are activated upon wounding. To test this, the wound-inducible expression of the WRKY genes, following simulated herbivory, was studied after pretreatment of leaves with neomycin. Neomycin, a poly-cationic aminoglycoside antibiotic, blocks the accumulation of oral secretion-induced Ca++ elevation and the conversion of JA to its bioactive form JA-Ile (Vadassery, et al. 2014). Thus, pre-treatment with neomycin would prevent formation of active JA-Ile and thereby block the activation of JA-dependent wound-inducible genes but not JA-independent wound-inducible genes.
Interestingly, pre-treatment of chickpea leaves with neomycin, one hour prior to wounding, reduced the transcript levels of nine WRKY genes namely, WRKYs 11, 17, 33A, 33B, 33C, 40A, 41, 46 and 70A at 5 min as compared to mechanically wounded chickpea leaves that were pre-treated with H. armigera oral secretions but not with neomycin (Fig. 3). The reduction ranged from ~ 50–80% at 5 min compared to the neomycin-untreated samples. For WRKY72, the reduction was seen at 20 min. In most cases, however, wound-induced transcription in neomycin pretreated leaves increased by 20 min and was close to the high levels observed upon wounding (in absence of neomycin treatment) suggesting that the transcript increase upon neomycin pre-treatment was only delayed compared to neomycin-untreated wounding. WRKY33A was an exception where pre-treatment with neomycin completely blocked wound-induced increase in transcription suggesting complete dependence on JA. On the other hand, WRKYs 40B, 40C and 70B did not undergo any change upon pre-treatment with neomycin and showed the same increase in transcription as seen upon wounding (in absence of neomycin) suggesting that their expression was independent of JA. The results suggested that JA-dependent as well as JA-independent factors contributed to the increase in transcript levels of the WRKYs.
Comparative real-time PCR analysis of WRKY genes upon simulated herbivory in presence and absence of neomycin. Chickpea plants were pretreated with 100 µM neomycin for an hour prior to simulated herbivory and expression studied at 5, 20, 60 and 120 min after simulated herbivory and compared to 0, 5 and 20 min wound-induced expression of simulated herbivory (neomycin-untreated; Fig. 1) with 0 min being the unwounded, neomycin untreated control in both sets of experiments. Reactions were run and analysed as described in Fig. 1
Ethylene induces several WRKY genes while SA suppresses most WRKYs in absence of wounding
Since the ethylene pathway, representing another important defense hormone, was also substantially up-regulated upon simulated herbivory in chickpea within 20 min (Pandey et al. 2017), it was investigated for its contribution to controlling WRKY gene expression. Ethylene, unlike JA, regulated the WRKY genes differently in absence of wounding with several genes being up-regulated. Five of the genes (WRKYs 40A, 40B, 70A, 70B and 72) were strongly induced by ethylene by 3–10 folds within 20 min of treatment (Fig. 4). For WRKY70B and WRKY72, the scale of induction in response to ethylene treatment and simulated herbivory was similar, suggesting that wound-induction of these genes may be wholly dependent on ethylene while for the other three (WRKY40A, WRKY40B and WRKY70A) the scale of induction was much less compared to that observed upon simulated herbivory suggesting other factors being responsible for their induction. Three genes, WRKY33A, WRKY33B and WRKY40C showed only a slight increase (~ three fold) at 60 min while a fourth, WRKY46, increased slightly at 20 min. Four genes, WRKY11, 17, 33C and 41, were not affected by ethylene treatment. WRKY53 was the only gene that was negatively regulated by ethylene with a transient decrease in transcript levels to less than 50% between 20–60 min.
Expression profiles of WRKY genes in response to ethylene. Leaves of eight-week-old unwounded chickpea plants were treated with 10 µl L−1ethylene. Samples were collected at 0, 20, 60 and 120 min. Reactions were run and analyzed as described in Fig. 1
Salicylic acid signaling, which is activated by biotrophic organisms, antagonizes JA responses in wounding but is known to be activated upon feeding by some insects, thereby enabling their survival (Diezel et al. 2009; Thaler et al. 2012; Rajendran et al. 2014; Schäfer et al. 2011). Treatment with SA, like that of JA, led to a rapid reduction in transcript levels of majority of the WRKY genes namely WRKY11, WRKY17, WRKY33A, WRKY33B, WRKY40C and WRKY53 (Fig. 5) with most being reduced to less than 50% of untreated samples in 20 min while WRKY33A was reduced to less than 10% within 20 min. WRKY40B showed a strong but transient reduction in transcript levels to less than 10% within 20 min of SA treatment before reaching background (unwounded) levels. In contrast, some of the WRKYs, namely WRKY33C, 46, 70A and 70B, were up-regulated by SA. Of these, the level of up-regulation for WRKY70A and WRKY70B (4.9 and 4.2 fold, respectively), was very close to their induction in response to simulated herbivory. Three other WRKY genes, namely WRKY41, WRKY40A and WRKY72, showed only a transient induction at 20 min upon treatment.
Expression profiles of WRKY genes in response to salicylic SA. Leaves of eight-week-old unwounded chickpea plants were treated with 2 mM SA. Samples were collected at 0, 20, 60 and 120 min. Reactions were run and analyzed as described in Fig. 1
H2O2 treatment activates the expression of most WRKY genes even in absence of wounding
Reactive oxygen species like hydrogen peroxide and the superoxide radical are common components of the plant defense in response to pathogen and herbivore attacks and among the earliest signals produced within minutes after wounding. Since the rapid and strong increase of most WRKY genes in response to simulated herbivory could neither be explained by JA nor ethylene, we investigated the role of H2O2 as one of the possible inducers in their regulation.
Indeed, majority of the WRKY genes (12 out of 14) were rapidly and strongly induced by H2O2 treatment (Fig. 6 a) and peaked within 20 min of H2O2 exposure while two others, WRKY53 and WRKY70B, showed a peak at 60 min. While the majority showed a 2–3 fold change in response to H2O2 treatment, three genes namely, WRKY17, WRKY33B and WRKY41 showed a 5–10 fold increase at the 20 min time point that matched the scale of induction observed upon simulated herbivory. WRKY72 was an exception in that H2O2 treatment induced a rapid decrease in transcript levels to less than 50% of the control values within 20 min of exposure and remained low thereafter. Transcript levels of WRKY40C also decreased in response to H2O2 treatment but only after 1 h. For most genes (with the exception of WRKY70B) the effect of H2O2 treatment on transcript levels was transient and no longer visible after 2 h of treatment. For WRKY41, the dynamics of induction in response to H2O2 differed from simulated herbivory as its induction was not as high as wounding. These expression analyses suggested that early wound responses were probably only partly dependent on H2O2 (for WRKY33B, WRKY17 and WRKY11) and could not be explained solely through induction by H2O2.
Expression profiles of WRKY genes in response to H2O2. Leaves of eight-week-old unwounded chickpea plants were treated with 5 mM H2O2. Samples were collected at 0, 20, 60 and 120 min. Reactions were run and analyzed as described in Fig. 1
A heat map summarizing the effects of all treatments on WRKY gene expression is shown in Fig. 7.
In silico sequence analysis of WRKY promoters for identification of putative cis regulatory elements
For identifying the putative cis regulatory elements of WRKY genes of chickpea, a 2 kb region upstream of the translational initiation codon of all genes was extracted using the chickpea genome database of ICC4958. The cis acting elements in all these promoters were analyzed using the PlantCare database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al. 2002). Various elements (Fig. 8a; Supplementary Table S3) involved in responses to phytohormone stimuli (JA, SA, ABA, ethylene), light responses, defense responses, cell and tissue specificity and those functioning as binding sites for known transcription factors were identified within the 2 kb region on the positive as well as negative strands since some elements are likely to function as enhancers. The most prevalent elements identified in the WRKY gene promoters were the BOX4 site (all 14 promoters), MYB, MYC and ethylene response element (ERE) (present in 13 promoters each), the anaerobic response element (ARE) (12 promoters), the light responsive elements G-box and GT1 motif (10 promoters) followed by defense response elements such as W box, TC-rich elements, Wun element and WRE (present in 7–9 WRKY genes). Also abundant (found in 7–11 promoters) were JA responsive elements, ABREs (responsive to ABA) and TCA elements (involved in SA response).
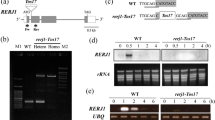
In silico analysis of CaWRKY promoters and functional validation of the CaWRKY41 promoter. a Schematic representation of the common cis elements present on the selected WRKY gene promoters. The various predicted cis regulatory elements with significant similarity to previously identified elements on the positive and negative strands are shaded with different color bars. Length of promoters is indicated with bars on scale that represent nucleotide number from the initiation codon. b–d Histochemical GUS assay in transgenic Arabidopsis plants expressing CaWRKY41pro under different conditions. GUS activity in transgenic Arabidopsis harboring WRKY41pro::GUS expression cassette after (b) mechanical wounding in the Arabidopsis Col-0 background, c upon treatment with JA, ethylene and SA (treatments as described in methods) and (d) in the ein2, coi1 and myc2myc3myc4 triple mutant backgrounds
The WRKY41 promoter shows wound-inducible and SA-responsive GUS expression
Most CaWRKY genes showed rapid wound–inducible expression and also possessed an array of cis elements responding to different cues including defense and defense hormones. To study this in vivo, the promoter of one of the wound-responsive WRKY genes, WRKY41 (labeled as WRKY50 by Kumar et al., 2016), was studied in detail. A region of 931 nt upstream of the translation initiation codon of WRKY41 (including 5’ UTR) was introduced into Arabidopsis in fusion with GUS (Supplementary Fig S1a and S1b). As shown (Fig. 8b), transgenic plants expressing WRKY41pro:GUS showed a strong and specific GUS expression only at the site of wounding but not in the absence of wounding (except where the leaf was excised in the unwounded plants). Interestingly, pre-treatment of transgenic plants with neomycin (which suppresses JA-Ile formation) did not seemingly affect the wound-induced expression of CaWRKY41pro.
Since, JA, SA and ethylene influence the transcription of defense-related genes during wound signaling, transgenic Arabidopsis plants expressing CaWRKY41pro::GUS were also treated with JA (100 μM), SA (2 mM) and ethylene (10 µl L−1) for 2 h (Fig. 8c). While the WRKY41 promoter region used for study lacked known JA and SA responsive elements (as per the PlantCare database) it did possess the ERE element and elements for MYB and MYC binding. Surprisingly, SA was able to activate the promoter as observed by blue color in unwounded SA-treated transgenic leaves. However, minimal change in GUS expression could be seen in unwounded transgenic leaves treated with JA and ethylene. The results suggested that as yet undiscovered SA responsive elements may drive the expression of the promoter upon exposure to SA.
To obtain further insight into the wound-responsive expression of the promoter, the WRKY41pro-GUS construct was introduced into different hormone response mutants namely coi1 (defective in JA responses), ein2 (defective in ethylene responses) and the myc2myc3myc4 triple mutant (defective in multiple MYC functions involved in JA signaling). Surprisingly, the wound-responsive expression was completely lost in all the three mutant backgrounds indicating that the wound-responsive GUS expression driven by WRKY41pro in Arabidopsis required a functional ethylene pathway as well as a functional JA pathway that depended on MYC2/3/4 (Fig. 8d).
Discussion
Wounding by chewing pests such as Helicoverpa (a major pest of chickpea) elicits a large number of defense responses in a plant, both early and late, for protection from further damage. Insect wound-induced chemical signals include Ca++ changes, glutamate, ROS, JA, ethylene, SA, etc. that function collectively to modulate gene expression within minutes to hours, leading to the final response that either deters the insect or allows it to feed (Erb et al. 2012). The contribution of individual chemical cues to the final expression of the target, and the extent of their crosstalk with different chemical cues, is still not very clear. How these signals influence gene expression in absence of wounding is also important since it can shed light on the changes in signal machinery that occurs as cells shift from an unwounded to a wound-responsive state. During our studies on the early events upon simulated H. armigera herbivory in chickpea, we identified the WRKY family as one of the most prominently up-regulated groups wherein at least 14 genes showed 5–50 fold increase in expression within 20 min of wounding (Pandey et al. 2017). Although these WRKY genes may be involved in different processes, their strong up-regulation upon simulated herbivory suggested commonalities in their regulation by wound-responsive cues. In order to unravel the complexities of the early wound response, this group was therefore chosen for a comparative study of the contribution of different wound response cues when provided individually (in absence of wounding) versus that when observed upon wounding.
Our results show that contrary to the strong induction observed upon wounding, different chemical cues, individually, show distinct and often opposite effects on gene expression in absence of wounding. Strikingly, jasmonic acid, known to be a key factor in early wound responses (Pieterse et al. 2012), reduces transcript levels in 10/14 genes by 50–75% within 20–60 min of treatment in absence of wounding and these remain low, while none of the rest shows a greater than two-fold increase. Although a decrease in some of the Group III chickpea WRKYs by JA had also been noted in a previous study (Kumar et al. 2016) the strong inhibition observed in the current context was counter-intuitive. It suggested that the early wound response of most of the WRKY genes was either independent of JA or, more likely, that the regulation by JA was altered depending on the presence or absence of wound-responsive factors. In absence of these factors under unwounded conditions, JA exerted an inhibitory effect. Wound responses are governed by both JA-dependent and JA-independent pathways (Titarenko et al. 1997). To identify the extent of contribution by JA, leaves were pretreated with neomycin prior to simulated herbivory. Neomycin inhibits the release of Ca++ upon wounding and blocks the conversion of JA to its active form, JA-Ile, thereby affecting JA-dependent processes across dicot and monocot plants (Vadassery et al. 2014, 2019). Any reduction in transcript levels in presence of neomycin would indicate the involvement of JA while any wound-induced up-regulation in its presence would indicate JA-independent regulation. Indeed, neomycin treatment substantially reduced the rapid wound-responsive up-regulation at 5 min by 50–70% in 9/14 WRKY genes with levels rising again by 20 min in most cases. For genes like WRKYs 11, 41, 53 and 72, peak wound-inducible expression post-neomycin treatment was only about 30–50% of that in absence of neomycin. Since the transcriptional response had slowed down and shifted (delayed) from 5 to 20 min, it indicated that JA biosynthesis contributed partly to substantially for the up-regulation of most WRKY genes upon wounding. The wound-induced increase of WRKY33A transcript levels appeared to be entirely dependent on JA since it was completely blocked by neomycin at all time points. In contrast, WRKYs 40B, 40C and 70B, showed no reduction in the transcript levels upon neomycin treatment compared to unwounded, neomycin-untreated samples indicating that their wound-responsive transcription was independent of JA. The results are interesting since they show that although JA can activate the transcript levels of the WRKY genes upon wounding, it reduces these under unwounded conditions. The reason why treatment with JA reduces transcript levels below basal levels under unwounded conditions will require further studies. The studies reveal a context-dependent role for JA wherein wounding may change the transcriptional dynamics of the WRKY genes and their responses towards JA in a way that facilitates their transcription upon wounding but a reduction in absence of wounding. Such context-dependent changes in regulation introduce an added dimension to the regulatory roles of hormones like JA. The transcriptional dynamics that undergo such a change (from the unwounded state to the wounded state) due to interaction of the JA pathway components with wound-responsive cues, within the first few seconds/minutes, begs further studies.
Treatment with SA also reduces transcript levels of several genes in absence of wounding. In fact, six of the ten genes that were suppressed by JA under unwounded conditions, namely WRKYs 11, 17, 33A, 33B, 40C, 53 and to some extent 41, also showed a strong reduction upon treatment with SA. Since JA contributes at least partially to the wound-induced up-regulation of most of the above WRKYs, their suppression by SA is not entirely surprising since JA and SA largely antagonize each other functionally (Thomma et al. 1998; Glazebrook 2005; Thaler et al. 2012). Herbivory usually leads to suppression of the SA pathway because of its negative effect on the JA pathway and insect defenses (Thaler et al. 2012). In fact, oral secretions of some insects like Schistocerca gregaria, Spodoptera littoralis and Spodoptera exigua (but not H. armigera) elicit SA responses in plants and these probably help in survival of the insects through suppression of the JA pathway (Diezel et al. 2009; Rajendran et al. 2014; Schäfer et al. 2011). SA is, however, an important player in biotrophic defenses, many of which are modulated through regulation of WRKY genes. In chickpea, WRKYs 33C, 70A and 70B were up-regulated 3–6 fold by SA even under unwounded conditions in a manner similar to the SA up-regulation of Grp III WRKYs (Kumar et al. 2016) suggesting involvement in SA-governed functions. Of these, chickpea WRKY70 (labeled as WRKY70A in this study) was recently shown to be activated by SA and reported to suppress multiple defense responses including ROS production thereby increasing susceptibility to Fusarium oxysporum (Chakraborty et al. 2020). The homologue of these genes in Arabidopsis, AtWRKY70, is also known to be induced by SA during pathogenesis (Li et al. 2004) and is a key TF that suppresses JA responses in Arabidopsis. SlWRKY70, a homologue of AtWRKY70, is also induced by SA and suppressed by JA and mediates resistance to aphids and the root knot nematode Meloidogyne javanica (Atamian et al. 2012). Since insect wounding exposes the plant to infection by opportunistic bacteria on the leaf surface as well as those in oral secretions, it is likely that the transcription of some of these WRKYs induced by JA under wounded conditions, and involved in SA responses, may be necessary to protect the plant from biotrophic infections caused by bacterial entry at the wound site.
Compared to JA, ethylene has more specific effects and is believed to function as a modulator of signaling by fine-tuning sensitivity to other cues (von Dahl et al. 2006; Diezel et al. 2009; Erb et al. 2012). Not surprisingly, it induced a moderate increase in 6/14 genes that was limited to 1.5–5 folds compared to the 5–50 fold increase observed upon simulated herbivory. The two exceptions were WRKY70B and WKRY72 where a 10–20 fold increase was observed that matched in scale with simulated herbivory. In most others, ethylene did not induce much of a change despite the presence of the ERE element in most promoters. With the exception of WRKY53, no other gene showed any reduction in transcript levels upon ethylene treatment. As in case of JA, it is likely that a combination of ethylene with wound-responsive factors may alter the regulation of the WRKY genes compared to that of ethylene alone under unwounded conditions.
Reactive oxygen species such as hydrogen peroxide are also produced in response to herbivory (Maffei et al. 2006) and are amongst the earliest cues that activate wound responses and gene expression. Different ROS can activate different pathways and possibly gene expression through changes in oxidation states of TFs (Desikan et al. 2001; Gadjev et al. 2006; Møller et al. 2007). Many ROS responsive genes possess distinct cis motifs (Petrov et al. 2012). Hydrogen peroxide is one of the most prominent ROS due to its stability and ability to move through membranes (Petrov and van Breusegem, 2012) and can strongly influence gene expression (Vandenabeele et al. 2003; He et al. 2018). Unlike the three hormones which largely failed to activate the genes much in absence of wounding, treatment with H2O2 strongly but transiently up-regulated 11/14 WRKY genes by 3–8 fold even under unwounded conditions within 20–60 min. For WRKYs 11, 17, 33B and 41, the increase appeared close to the levels observed upon simulated herbivory and early enough. Compared to any other hormone, the response to H2O2 in absence of wounding was stronger, suggesting that unlike JA, ethylene or SA, responses to H2O2 were probably not as dependent on other wound-responsive cues. Thus, early wound-induced transcription of at least some of the WRKY genes appeared to be partly dependent on and could be attributed to ROS like H2O2.
As is clear from these studies, the summation of the individual effects of the primary chemical cues under unwounded conditions does not explain the large wound-responsive increase in transcript levels since these (especially JA) actually reduce transcript levels under unwounded conditions. One inference is that although wounding may activate several different signals near simultaneously, their action on target genes is probably not independent of the other signals and not mutually exclusive. Instead, these may require the interaction of specific wound-responsive factors and other changes in the cell to alter the sensitivity of the genes to the chemical cues. A combinatorial action by different signals on the promoters of the wound-responsive WRKYs may synergistically alter the transcriptional state or the chromatin. How these factors enable activation of gene expression from an inhibited state to a highly activated state will require further studies at the chromatin level. Such detailed chromatin level studies have been performed for SA under biotrophic infections (Jin et al. 2018; Singh et al. 2015). Another possibility is that gene transcription and transcript stability may undergo dynamic changes depending on local hormone concentrations. Rising levels of JA, ethylene and H2O2 may activate the genes while their increase beyond a certain level may induce transcript instability. Hormones like auxin and SA are known to have concentration-dependent effects that can be inhibitory as well as activating (Kubeš et al. 2012; Caarls et al. 2015; Pasternak et al. 2019).
We have tried to validate the observations through the study of the WRKY41 promoter. Although studied in a heterologous system like Arabidopsis, the strong wound-responsive GUS expression driven by the 931 bp region (encompassing the promoter and 5’UTR of WRKY41) shows that wound-responsive cis elements on the CaWRKY41 promoter respond to and are recognized even in Arabidopsis suggesting conservation of the basic wound response machinery across families. The wound-responsive expression of CaWRKY41 is seen within minutes of wounding and is also apparent in presence of neomycin indicating that much of the WKRY41 expression may be JA-independent. Nevertheless, the promoter is neither activated in the coi1 mutant background nor in myc2myc3myc4 background. This indicates that transactivation of the promoter requires a functional JA pathway and is dependent on at least one of the MYC factors that are known to drive JA-dependent responses (Van Moerkercke et al. 2019). Indeed the CaWRKY41 promoter contains a site for MYC binding which may govern this expression. While the results seem contrary to those showing activation of the promoter in Arabidopsis even upon neomycin treatment, one inference could be that at least some of the wound-responsive changes in Arabidopsis may actually bypass JA biosynthesis and activate JA signaling downstream via COI1 and MYCs. The inability of CaWRKY41pro to get activated in the ein2 mutant background is also interesting and suggests requirement of a functional ethylene pathway for wound-responsive activation at least in Arabidopsis. Thus both the JA and ethylene pathways need to be functional for activation of the CaWRKY41 promoter in Arabidopsis. The promoter also shows SA responsiveness in Arabidopsis which is in tune with its activation by SA in chickpea. Despite these results, the 931 bp region does not show any known SA-responsive element indicating that novel SA-responsive cis elements within this region may drive its expression. The studies highlight the complexity of the interactions that lead to CaWRKY41 activation upon wounding. Like the strong, early wound-inducible RbPCD1 promoter that was recently described (Pandey et al. 2019), the CaWRKY41 promoter also possesses biotechnological value due to its early wound-responsive nature and is currently being characterized for further tests.
In conclusion, our studies show that individual wound-responsive chemical cues like JA, ethylene, SA, H2O2 have different and often opposite effects under unwounded conditions compared to their collective action under wounded conditions. Their action during wound responses is not a summation of the individual effects in isolation but probably requires interaction with several other wound-responsive compounds that function coordinately to induce gene expression. The study sheds light on the complexity of regulation of genes as they shift from the unwounded state to the wounded state.
References
Adachi H, Nakano T, Miyagawa N, Ishihama N, Yoshioka M, Katou Y, Yaeno T, Shirasu K, Yoshioka H (2015) WRKY transcription factors phosphorylated by MAPK regulate a plant immune NADPH oxidase in Nicotiana benthamiana. Plant Cell 27:2645–2663
Atamian HS, Eulgem T, Kaloshian I (2012) SlWRKY70 is required for Mi-1-mediated resistance to aphids and nematodes in tomato. Planta 235:299–309
Bhattarai KK, Atamian HS, Kaloshian I, Eulgem T (2010) WRKY72-type transcription factors contribute to basal immunity in tomato and Arabidopsis as well as gene-for-gene resistance mediated by the tomato R gene Mi-1. Plant J 63:229–240
Cai H, Yang S, Yan Y, Xiao Z, Cheng J, Wu J, Qiu A, Lai Y, Mou S, Guan D (2015) CaWRKY6 transcriptionally activates CaWRKY40, regulates Ralstonia solanacearum resistance, and confers high-temperature and high-humidity tolerance in pepper. J Exp Bot 66:3163–3174
Caarls L, Pieterse CMJ, Van Wees SCM (2015) How salicylic acid takes transcriptional control over jasmonic acid signaling. Front Plant Sci 6:170
Chakraborty J, Sen S, Ghosh P, Jain A, Das S (2020) Inhibition of multiple defense responsive pathways by CaWRKY70 transcription factor promotes susceptibility in chickpea under Fusarium oxysporum stress condition. BMC Plant Biol 20:1–23
Ciolkowski I, Wanke D, Birkenbihl RP, Somssich IE (2008) Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol Biol 68:81–92
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Dang F, Wang Y, She J, Lei Y, Liu Z, Eulgem T, Lai Y, Lin J, Yu L, Lei D (2014) Overexpression of CaWRKY27, a subgroup IIe WRKY transcription factor of Capsicum annuum, positively regulates tobacco resistance to Ralstonia solanacearum infection. Physiol Plant 150:397–411
Desikan R, A.-H.-Mackerness S, Hancock JT, Neill SJ (2001) Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol 127:159–172
Dhaliwal G, Jindal V, Dhawan A (2010) Insect pest problems and crop losses: changing trends. Indian J Ecol 37:1–7
Diezel C, von Dahl CC, Gaquerel E, Baldwin IT (2009) Different lepidopteran elicitors account for cross-talk in herbivory-induced phytohormone signaling. Plant Physiol 150:1576–1586
Dinesh K, Anusha S, Singh RB, Dangi N (2017) Estimation of avoidable yield losses caused by Helicoverpa armigera (Hubner) on chickpea. J Entomol Zool Stud 5:1476–1478
Erb M, Meldau S, Howe GA (2012) Role of phytohormones in insect-specific plant reactions. Trends Plant Sci 17:250–259
Erb M, Reymond P (2019) Molecular interactions between plants and insect herbivores. Annu Rev Plant Biol 70:527–557
Eulgem T (2005) Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci 10:71–78
Eulgem T, Somssich IE (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10:366–371
Gattolin S, Alandete-Saez M, Elliott K, Gonzalez-Carranza Z, Naomab E, Powell C, Roberts JA (2006) Spatial and temporal expression of the response regulators ARR22 and ARR24 in Arabidopsis thaliana. J Exp Bot 57:4225–4233
Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, Apel K, Inzé D, Mittler R, Van Breusegem F (2006) Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol 141:436–445
Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43:205–227
He H, Van Breusegem F, Mhamdi A (2018) Redox-dependent control of nuclear transcription in plants. J Exp Bot 69:3359–3372
Hu L, Ye M, Li R, Zhang T, Zhou G, Wang Q, Lu J, Lou Y (2015) The rice transcription factor WRKY53 suppresses herbivore-induced defenses by acting as a negative feedback modulator of mitogen-activated protein kinase activity. Plant Physiol 169:2907–2921
Jin H, Choi S-M, Kang M-J, Yun S-H, Kwon D-J, Noh Y-S, Noh B (2018) Salicylic acid-induced transcriptional reprogramming by the HAC–NPR1–TGA histone acetyltransferase complex in Arabidopsis. Nucleic Acids Res 46:11712–11725
Knoth C, Ringler J, Dangl JL, Eulgem T (2007) Arabidopsis WRKY70 is required for full RPP4-mediated disease resistance and basal defense against Hyaloperonospora parasitica. Mol Plant-Microbe Interact 20:120–128
Kubeš M, Yang H, Richter GL, Cheng Y, Młodzińska E, Wang X, Blakeslee JJ, Carraro N, Petrášek J, Zažímalová E (2012) The Arabidopsis concentration-dependent influx/efflux transporter ABCB4 regulates cellular auxin levels in the root epidermis. Plant J 69:640–654
Kumar K, Srivastava V, Purayannur S, Kaladhar VC, Cheruvu PJ, Verma PK (2016) WRKY domain-encoding genes of a crop legume chickpea (Cicer arietinum): comparative analysis with Medicago truncatula WRKY family and characterization of group-III gene (s). DNA Res 23:225–239
Kumari A, Chételat A, Nguyen CT, farmer EE, (2019) Arabidopsis H+-ATPase AHA1 controls slow wave potential duration and wound-response jasmonate pathway activation. Proc Natl Acad Sci USA 116:20226–20231
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Li J, Brader G, Kariola T, Tapio Palva E (2006) WRKY70 modulates the selection of signaling pathways in plant defense. Plant J 46:477–491
Li J, Brader G, Palva ET (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16:319–331
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25:402–408
Maffei ME, Mithöfer A, Arimura G-I, Uchtenhagen H, Bossi S, Bertea CM, Cucuzza LS, Novero M, Volpe V, Quadro S (2006) Effects of feeding Spodoptera littoralis on lima bean leaves. III. Membrane depolarization and involvement of hydrogen peroxide. Plant Physiol 140:1022–1035
Miller G, Schlauch K, Tam R, Cortes D, Torres MA, Shulaev V, Dangl JL, Mittler R (2009) The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci Signal 2:ra45–ra45
Møller IM, Jensen PE, Hansson A (2007) Oxidative modifications to cellular components in plants. Annu Rev Plant Biol 58:459–481
Pandey SP, Singh AP, Srivastava S, Chandrashekar K, Sane AP (2019) A strong early acting wound-inducible promoter, RbPCD1pro, activates cryIAc expression within minutes of wounding to impart efficient protection against insects. Plant Biotechnol J 17:1458–1470
Pandey SP, Somssich IE (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150:1648–1655
Pandey SP, Srivastava S, Goel R, Lakhwani D, Singh P, Asif MH, Sane AP (2017) Simulated herbivory in chickpea causes rapid changes in defense pathways and hormonal transcription networks of JA/ethylene/GA/auxin within minutes of wounding. Sci Rep 7:1–14
Pasternak T, Groot EP, Kazantsev FV, Teale W, Omelyanchuk N, Kovrizhnykh V, Palme K, Mironova VV (2019) Salicylic acid affects root meristem patterning via auxin distribution in a concentration-dependent manner. Plant Physiol 180:1725–1739
Pieterse CM, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SC (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28:489–521
Petrov V, Vermeirssen V, De Clercq I, Van Breusegem F, Minkov I, Vandepoele K, Gechev TS (2012) Identification of cis-regulatory elements specific for different types of reactive oxygen species in Arabidopsis thaliana. Gene 499:52–60
Petrov VD, Van Breusegem F (2012) Hydrogen peroxide a central hub for information flow in plant cells. AoB plants 2012
Rajendran S, Lin I-W, Chen M-J, Chen C-Y, Yeh K-W (2014) Differential activation of sporamin expression in response to abiotic mechanical wounding and biotic herbivore attack in the sweet potato. BMC Plant Biol 14:1–21
Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15:247–258
Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich I (1996) Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J 15:5690–5700
Ryu H-S, Han M, Lee S-K, Cho J-I, Ryoo N, Heu S, Lee Y-H, Bhoo SH, Wang G-L, Hahn T-R (2006) A comprehensive expression analysis of the WRKY gene superfamily in rice plants during defense response. Plant Cell Rep 25:836–847
Schäfer M, Fischer C, Baldwin IT, Meldau S (2011) Grasshopper oral secretions increase salicylic acid and abscisic acid levels in wounded leaves of Arabidopsis thaliana. Plant Signal Behav 6:1256–1258
Sequeira R, McDonald J, Moore A, Wright G, Wright L (2001) Host plant selection by Helicoverpa spp. in chickpea-companion cropping systems. Entomol Exp Appl 101:1–7
Shao Q, Gao Q, Lhamo D, Zhang H, Luan S (2020) Two glutamate- and pH-regulated Ca2+channels are required for systemic wound signaling in Arabidopsis. Sci Signal 13:eaba1453
Singh M, Bag SK, Bhardwaj A, Ranjan A, Mantri S, Nigam D, Sharma YK, Sawant SV (2015) Global nucleosome positioning regulates salicylic acid mediated transcription in Arabidopsis thaliana. BMC Plant Biol 15:1–21
Skibbe M, Qu N, Galis I, Baldwin IT (2008) Induced plant defenses in the natural environment: Nicotiana attenuata WRKY3 and WRKY6 coordinate responses to herbivory. Plant Cell 20:1984–2000
Thaler JS, Humphrey PT, Whiteman NK (2012) Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci 17:260–270
Thomma BP, Eggermont K, Penninckx IA, Mauch-Mani B, Vogelsang R, Cammue BP, Broekaert WF (1998) Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci 95:15107–15111
Titarenko E, Rojo E, Leon J, Sanchez-Serrano JJ (1997) Jasmonic acid-dependent and-independent signaling pathways control wound-induced gene activation in Arabidopsis thaliana. Plant Physiol 115:817–826
Toyota M, Spencer D, Sawai-Toyota S, Jiaqi W, Zhang T, Koo AJ, Howe GA, Gilroy S (2018) Glutamate triggers long-distance, calcium-based plant defense signaling. Science 361:1112–1115
Vadassery J, Ballhorn DJ, Fleming SR, Mazars C, Pandey SP, Schmidt A, Schuman MC, Yeh K-W, Yilamujiang A, Mithöfer A (2019) Neomycin: an effective inhibitor of jasmonate-induced reactions in plants. J Plant Growth Regul 38:713–722
Vadassery J, Reichelt M, Jimenez-Aleman GH, Boland W, Mithöfer A (2014) Neomycin inhibition of (+)-7-iso-jasmonoyl-L-isoleucine accumulation and signaling. J Chem Ecol 40:676–686
Vandenabeele S, Van Der Kelen K, Dat J, Gadjev I, Boonefaes T, Morsa S, Rottiers P, Slooten L, Van Montagu M, Zabeau M (2003) A comprehensive analysis of hydrogen peroxide-induced gene expression in tobacco. Proc Natl Acad Sci 100:16113–16118
Van Eck L, Schultz T, Leach JE, Scofield SR, Peairs FB, Botha AM, Lapitan NL (2010) Virus-induced gene silencing of WRKY53 and an inducible phenylalanine ammonia-lyase in wheat reduces aphid resistance. Plant Biotechnol J 8:1023–1032
Van Moerkercke A, Duncan O, Zander M, Šimura J, Broda M, Bossche RV, Lewsey MG, Lama S, Singh KB, Ljung K (2019) A MYC2/MYC3/MYC4-dependent transcription factor network regulates water spray-responsive gene expression and jasmonate levels. Proc Natl Acad Sci 116:23345–23356
Von Dahl CC, Hävecker M, Schlögl R, Baldwin IT (2006) Caterpillar-elicited methanol emission: A new signal in plant–herbivore interactions? Plant J 46:948–960
Wang D, Amornsiripanitch N, Dong X (2006) A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog 2:e123
Wang H, Hao J, Chen X, Hao Z, Wang X, Lou Y, Peng Y, Guo Z (2007) Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol Biol 65:799–815
Zheng Z, Qamar SA, Chen Z, Mengiste T (2006) Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J 48:592–605
Acknowledgements
We are grateful to Mr Ram Awadh for managing the glasshouse and field plants and Mr Rakesh Srivastava for rearing insects and providing oral secretions of H. armigera. We thank Dr RP Bhalerao, Umea Plant Science Centre, Sweden, for the Arabidopsis mutants. SS, SPP, LP and PS were supported by Senior Research Fellowships from the Council of Scientific and Industrial Research while APS was supported from funds from the CSIR projects NWP-03 and BSC107.
CSIR-NBRI Manuscript number: CSIR-NBRI_MS/2022/02/05
Funding
The work was supported from funds from the CSIR projects NWP-03 and BSC107.
Author information
Authors and Affiliations
Contributions
SS, SPP and APS designed the study, SS and SPP performed the treatments, SS carried out the real time experiments, SS, PS and LP carried out the promoter studies, SS, VP and APS analysed the data and SS and APS wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic Supplementary material.
Supplementary Table S1.
List of oligonucleotide primers specific for WRKY genes used in this study. Supplementary Table S2. Accession numbers of the WRKY genes identified in the study. Supplementary Table S3. Presence of different types of known cis regulatory elements in all 14 WRKY promoters. Supplementary Figure S1a. A schematic diagram showing the CaWRKY41 promoter-GUS fusion construct in pBI101.2. b The 931 bp CaWRKY41 promoter sequence used for study.
Rights and permissions
About this article
Cite this article
Srivastava, S., Pandey, S.P., Singh, P. et al. Early wound-responsive cues regulate the expression of WRKY family genes in chickpea differently under wounded and unwounded conditions. Physiol Mol Biol Plants 28, 719–735 (2022). https://doi.org/10.1007/s12298-022-01170-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-022-01170-y