Abstract
Owing to rapid global climate change, the occurrence of multiple abiotic stresses is known to influence the outburst of biotic stress factors which affects crop productivity. Therefore, it is essential to understand the molecular and cell biology of key genes associated with multiple stress responses in crop plants. SlHyPRP1 and DEA1, the members of eight-cysteine motif (8CM) family genes have been recently identified as putative regulators of multiple stress responses in tomato (Solanum lycopersicum L.). In order to gain deeper insight into cell and molecular biology of SlHyPRP1 and DEA1, we performed their expression analysis in three tomato cultivars and in vivo cell biological analysis. The semi-quantitative PCR and qRT-PCR results showed the higher expression of SlHyPRP1 and DEA1 in leaf, stem, flower and root tissues as compared to fruit and seed tissues in all three cultivars. The expression levels of SlHyPRP1 and DEA1 were found to be relatively higher in a wilt susceptible tomato cultivar (Arka Vikas) than a multiple disease resistant cultivar (Arka Abhed). In vivo cell biological analysis through Gateway cloning and Bi-FC assay revealed the predominant sub-cellular localization and strong protein–protein interaction of SlHyPRP1 and DEA1 at the cytoplasm and plasma membrane. Moreover, SlHyPRP1 showed in vivo interaction with stress responsive proteins WRKY3 and MST1. Our findings suggest that SlHyPRP1 with DEA1 are co-expressed with tissue specificity and might function together by association with WRKY3 and MST1 in plasma membrane for regulating multiple stress responses in the tomato plant.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In recent years the role of agriculture is crucial in mitigating and adapting to climate change (Gill et al. 2014). According to the Intergovernmental Panel on Climate Change (IPCC), almost one fourth of world anthropogenic greenhouse gas (GHG) emissions are due to agriculture, forestry and land use changes (Savvides et al. 2016; FAO 2019; Marwein et al. 2019). Climate change caused by extreme weather, drought, flooding and other disasters, affecting the livelihoods of millions of people across the world (FAO 2019). Plants being sessile, need to respond and face a wide range of abiotic and biotic stresses in the field lead to yield losses to a major extent (Cohen and Leach 2019). Most of the biotechnology and breeding efforts have been directed towards mitigating a single stress factor (Parmar et al. 2017). Generally, breeding for tolerance to single stress is not ideal as plants respond distinctively to different or concurrent stresses (Pandey et al. 2017). The occurrence of multiple abiotic stresses such as drought, temperature extremes and salinity are known to be the major factors that influence the outburst of a range of biotic stress factors (Chikkaputtaiah et al. 2017; Baruah et al. 2020). Together these multiple biotic and abiotic stresses severely affect overall plant growth, innate tolerance capacity, productivity and the yield (Debbarma et al. 2019). Hence, there is an urgent need to develop advanced crop varieties that are capable of tolerating or resisting multiple stresses (Lohani et al. 2020).
Tomato (Solanum lycopersicum L), is one of the widely consumed vegetables in the world with high nutritional values (Brooks et al. 2014). It is considered the ideal crop model for molecular genetics and cell biology studies because of its small diploid genome, short life cycle, known complete genome sequence and its stability for Agrobacterium mediated transformation (Schwarz et al. 2014). Moreover, many of the regulatory genes associated with various abiotic and biotic stress responses are known in tomatoes, which intern help implement crop improvement strategies through advanced biotechnological approaches in tomato crop. However, in recent years, tomato productivity in India and the world is affected largely due to different abiotic and biotic stress factors (Gerszberg et al. 2015).
Plant proline-rich proteins (PRPs) are cell wall structural proteins and based on their protein structure they are divided into three classes: hybrid proline-rich proteins (HyPRPs), PRPs with multiple copies of the POVEKPOVXK motif, and NHyPRP proteins (Huang et al. 2011; Yang et al. 2018). All three classes of PRPs possess two common motifs such as the proline rich motif at N-terminus and 8-cysteine motif (8CM) at C-terminus (Neto et al. 2013). HyPRPs present in seed plants belongs to the family of 8CM containing PRPs (Li et al. 2016). Apart from plant developmental processes, HyPRPs play important roles in response to different abiotic and biotic stresses (Yang et al. 2018; Kapoor et al. 2019; Saikia et al. 2020). The N-terminal domain of HyPRPs resembles the structural cell wall PRPs but lacks the characteristic amino acid motifs of PRPs (José-Estanyol et al. 2004; Dvořáková Lenka 2007). Several studies in plants have shown that HyPRPs such as CaHyPRP1, GbHyPRP1, NbHyPRP1, GhHyPRP4, CcHyPRP1, AZI1, OsPRP, and BnPRP play multiple functional roles during specific developmental stage and respond to biotic and abiotic stresses as positive and negative regulators (Goodwin et al. 1996; Gothandam and Flower 2010; Gengqing et al. 2011; Jung et al. 2012; Yeom et al. 2012; Li et al. 2016; Mellacheruvu et al. 2016; Saikia et al. 2020). Recently, the SlHyPRP1 (S. lycopersicum) and SpHyPRP1 (S. pennellii) genes of tomato were found to be negative regulators of different abiotic stresses such as drought, salinity, cold, oxidative stress, and phytohormone ABA (Li et al. 2016; Yang et al. 2018). Moreover, SlHyPRP1 has been reported to play a direct or indirect role in defense-related signaling pathways (Kapoor et al. 2019).
The Differentially expressed in response to arachidonic acid 1 gene (DEA1) is a circadian-regulated gene in tomato belonging to 8CM proline rich family proteins (Weyman et al. 2006a, b). The protein structure of DEA1 has a conserved domain shared by the 8CM superfamily of protease inhibitors, alpha amylase inhibitors, lipid transfer proteins and seed storage proteins (Marchler-Bauer et al. 2003; José-Estanyol et al. 2004). DEA1 shares sequence similarity to Arabidopsis EARL1 gene that confers cold tolerance (Weyman et al. 2006a). Over-expression of DEA1 conferred cold stress tolerance in yeast (Weyman et al. 2006b). GmDEA1 has been reported to be involved in the regulation of biotic stress responses in soybean (Klink et al. 2011). The transcript level of DEA1 was found to be altered by late blight caused by Phytophthora infestans in tomatoes (Weyman et al. 2006a). The promoter region of DEA1 was found to contain putative stress signaling elements (Weyman et al. 2006b). W-box motif and Alfin1-response motif are the stress-response motifs of the promoter region of the tomato DEA1 gene involved in biotic and abiotic stress responses. W-box motif is found to be involved in response to pathogen attack, wounding, and senescence (Eulgem et al. 2000) and Alfin1-response motif is involved in response to salinity stress (Winicov and Bastola 1999).
Thus, SlHyPRP1and DEA1, key genes of 8CM protein family might together possibly play important regulatory roles in multiple stress tolerance in tomato. Despite their potential significance as multi-stress responsive genes, the expression pattern in tomato cultivars and the cell biology of these two key proteins is not known. In the present study, we carried out the expression analysis of SlHyPRP1 and DEA1 in three tomato cultivars through semi-quantitative PCR and qRT-PCR. Further, we performed sub-cellular localization analysis of these proteins in tomato protoplasts and seedlings in vivo. Furthermore, in vivo protein–protein interaction analysis was carried out to validate the predictive in silico protein partners of SlHyPRP1 and DEA1 associated with stress responses through Bi-FC assay in tomato protoplasts. The present study provides knowledge on cell and molecular biology of two key multi-stress responsive genes in tomato crop model that can be explored to unravel their functional significance in multiple stress regulation in plants.
Materials and methods
Plant materials and growth condition
Three tomato (Solanum lycopersicum L.) cultivars have been used in this study namely, Micro tom, Arka Vikas and Arka Abhed. All three cultivars were procured from ICAR-IIHR Bangalore. The seeds were sterilized with 70% ethanol for 5 min, treated with 4% sodium hypochlorite (w/v) for 10 min followed by washing thrice with distilled water. The sterile germinated seeds were kept in ½ strength MS media, 3% sucrose and 0.3% phytagel (Sigma Aldrich) were cultured at dark for 3 days followed by plant growth chamber at 25 °C, 70% RH and 16/8 photoperiod at 150µE m−2 s−2. Early grown plants of all three cultivars were then transplanted in soilrite on pots and maintained in climate controlled greenhouse at standard growth conditions. Different tissue samples from each cultivar were collected at different growth stages and stored at −80 °C for gene expression studies. The leaves of 3-week old seedlings of cv. Arka Vikas were collected with a sterile scalpel for harvesting healthy tomato protoplasts and used for cell biological studies.
In silico analysis
With extensive literature mining, SlHyPRP1 and DEA1 genes responsive to abiotic and biotic stresses of cultivated tomato have been selected for this study. The gene sequences and amino acid sequences were retrieved from Sol Genomics Network (SGN) (https://solgenomics.net/), which is a clade-oriented database dedicated to the biology of the Solanaceae family. Similarity prediction of these genes with Solanaceae family was carried out using NCBI-BLAST search. The protein sequences of similar Solanaceous family were imported into DNAMAN software to get the best hit aligned amino acid sequences. The phylogenetic tree was constructed by the neighbor-joining method with 1000 bootstrap replicates. Benchling online informatics platform was used to predict the gene orientation and structure. Gene Structure Display Server (GSDS 2.0) tool was used to draw a structural domain. For in silico prediction of sub-cellular localization of SlHyPRP1 and DEA1 proteins, CELLO2GO (Yu et al. 2014) online software predictor was used with an E-value of 0.001 as the best predicted score. For in silico protein–protein interaction prediction, STRING software tool was used (Rahim et al. 2018; Szklarczyk et al. 2019).
RNA isolation, cDNA synthesis, RT-PCR and qRT-PCR
About 100 mg of leaf, root, stem, flower, fruit and seed tissues were collected from three different tomato cultivars (as mentioned above). Total RNA was extracted using RNeasy® Plant Mini Kit (Qiagen) as per manufacturer’s instructions and was quantified using nanodrop spectrophotometer (Eppendorf Bio spectrometer). The quality was also confirmed by running the total RNA on 2% w/v agarose gel. Total RNA (1 µg) from each tissue sample was used for the synthesis of cDNA. First strand cDNA was reverse transcribed with PrimeScript™ RT reagent kit with gDNA Eraser (DSS-TAKARA) as per the manufacturer’s instructions. For expression analysis of SlHyPRP1 and DEA1 genes, semi-quantitative PCR was performed using the DNA polymerase Green Emerald GT master mix. The elongation factor α 1 (EF1-α) was used as a house-keeping control gene. Thermal cycling conditions of 94 °C (5 min), 55 °C (45 s), 72 °C (15 s) for 35 cycles were used for PCR amplification. The respective products were run on 2% agarose gel and visualized on the GelDoc system. For expression quantification, quantitative real time PCR (qRT-PCR) was performed with SlHyPRP1 and DEA1 gene specific primers using SYBR green fluorophores (Applied Biosystems, USA). EF1-α was used as house-keeping internal control gene. The qRT-PCR program was set at 95 °C (7 min), 95 °C (10 s), 60 °C (30 s) for 40 cycles. The excitation of DNA fluorescence was measured relative to internal control (EF1-α). The expression levels of SlHyPRP1 and DEA1 in different tissues of each tomato cultivar was measured on a real time PCR detection system (Applied Biosystems, USA) following the previously described method (Livak and Schmittgen 2001). The statistical significance was calculated using the parametric two sample t-test variance among specific tissues of different cultivars using XLSTAT software (https://www.xlstat.com/en/). The experiment was performed with three independent biological replicates and each reaction was set up with three technical replicates. The experiment was repeated thrice. The primer sets used for the study are given in Table S1.
Gateway cloning for generating sub-cellular localization and split-YFP constructs
For generating gateway sub-cellular localization constructs, the ORF of SlHyPRP1 and DEA1genes were amplified using the primers with a stop codon (N-terminal YFP fusion) and without the stop codon (C-terminal YFP fusion). The primers were flanked by the gateway adaptor sites attB1 in forward and attB2 sites in reverse orientation. The PCR products were purified and cloned into gateway entry vector pDNR221 using BP Clonase II enzymes (Gateway ™ Technology, Invitrogen). The positive clones were confirmed using restriction analysis with appropriate enzymes. Sanger sequencing of each clone was performed to confirm the positive clone for correct orientation as well as for the presence of stop and without stop codons in the entry vector using M13 F and M13 R primers (Table S1). The gateway entry vector carrying the positive inserts were cloned into expression vectors, pENSG-YFP (N-terminal YFP fusion) and pEXSG-YFP (C-terminal YFP fusion) using LR Clonase II enzymes (Gateway ™ Technology, Invitrogen) and the positive clones were confirmed by restriction analysis. For generating gateway Split-YFP interaction constructs, entry clones of SlHyPRP1, DEA1 and predicted putative interacting partners namely WRKY3, MST1 and Snakin-2 were cloned into the gateway expression vectors pE-SPYNE and pE-SPYCE using LR Clonase II enzymes (Gateway ™ Technology, Invitrogen). The positive clones were confirmed by restriction analysis with appropriate enzymes. The primer sets used for generating gateway cloning constructs are given in Table S1. Vector maps of each construct were generated using VectorNTI software tool (Life Technologies, USA) and are given in Fig. S2.
Tomato protoplast isolation and transformation
Three-week-old tomato cv. Arka Vikas leaves were collected for healthy protoplast isolation. The protoplasts were isolated using the previously reported protocol (Ray et al. 2015) with slight modifications. DNA (1 µg) of each construct was transfected into 100 µl healthy protoplasts through standard Poly Ethylene Glycol (PEG) method. The transformed protoplasts were incubated overnight in dark at room temperature and visualized under confocal microscopy.
Particle bombardment (Gene delivery) in tomato seedlings
The tomato seedlings of cv. Arka Vikas were germinated on MS plates. Three weeks old seedlings were transformed with N and C terminal YFP constructs of SlHyPRP1 and DEA1 through particle delivery system (PDS1000, BioRad, USA). The protocol described by (Ueki et al. 2009) was used with minor modifications. Briefly, 5 µg of DNA coated with 30 mg (1 µm size) gold particles were bombarded on the seedlings at a helium pressure of 1100 PSI. The bombarded seedlings were incubated in dark for 24 h inside a petri dish with wet filter paper and visualized under confocal microscopy.
Confocal laser scanning microscopy
The sub-cellular localization and split-YFP constructs were visualized under a confocal laser-scanning microscope (Leica Microsystems, Wetzlar, Germany). EYFP PMT and DAPI detectors were used in this study. YFP fluorescence was detected at 488 nm and 514 nm excitation with an emission wavelength range of 505–530 nm. DAPI fluorescence was detected at 405 nm excitation and 507-530 nm emission. The chlorophyll auto-fluorescence was analyzed with 555 nm excitation and > 650 nm emission. Images were processed and exported in JPEG format. The true fluorescence intensity quantifications of positive signals were measured using Leica LAS-X software (.lif) in comparison to the negative controls (Fig. S3).
Results
Gene structure, multiple sequence alignments and phylogenetic analysis of SlHyPRP1 and DEA1
The extensive literature search and bioinformatics analysis showed, SlHyPRP1 and DEA1 as key proteins of 8CM family. The deduced structural arrangement of both the genes is represented diagrammatically in Fig. 1. The information on genomic DNA and CDS of SlHyPRP1 and DEA1 was obtained from NCBI. The NCBI accession number of SlHyPRP1 is AF308937 and DEA1 is NM_001246895. The SlHyPRP1 gene (1.4 kb) carries two exons interspaced by a single intron (Fig. 1A). The CDS region of SlHyPRP1 consists of N-terminal PRP region and C-terminal 8CM region (Fig. 1B). A signal peptide consisting of 24-30 bp was present in the upstream of N-terminal region of SlHyPRP1 (Fig. 1B). DEA1 is a single exonic gene of 416 bp flanked by 43 bp upstream and 269 bp downstream DNA sequence and the CDS contains C-terminal 8CM region (Fig. 1D). The multiple sequence alignments of SlHyPRP1 and DEA1 using DNAMAN software tool showed that the 8CM was conserved across Solanaceous species which is characteristic of both SlHyPRP1 and DEA1 proteins (Fig. S1). It is evident from the amino acid sequence alignment that proline is rich in N-terminal domain while unique conserved 8CM is present in C-terminal (Fig. S1). Therefore, the presence of these unique regions confirms that both SlHyPRP1 and DEA1 proteins belong to members of 8CM family proteins (Weyman et al. 2006b; Kapoor et al. 2019). The phylogenetic analysis of SlHyPRP1 showed 96% similarity between S. lycopersicum and S. pennellii (Fig. 1C). DEA1 of S. lycopersicum has shown 100% similarity with PRP of S. pennellii along with its nearest PRP of S. tuberosum (98% similarity). Furthermore, DEA1 showed a 91% similarity with EARL1 of Capsicum annuum and Capsicum chinense (Fig. 1E). The multiple sequence alignment and phylogenetic analysis of SlHyPRP1 and DEA1 indicates that 8CM of these two proteins was conserved between Solanaceous plants (Weyman et al. 2006a; Li et al. 2016). Interestingly, DEA1 also share 50–65% sequence similarity with Arabidopsis EARL1, a lipid transfer protein associated with abiotic stress tolerance in plants (Weyman et al. 2006b).
Schematic representation of gene structure, functional motifs and phylogenetic tree of SlHyPRP1 and DEA1. A. Genomic region (1.4 kb) of SlHyPRP1 gene with 2 exons interspaced by a single intron. B. CDS region (798 bp) of SlHyPRP1 depicting N-terminal proline rich region (green) with upstream signal peptide (24–30 bp)(red) and C-terminal 8CM (pink). C. Phylogenetic tree of tomato (S. lycopersicum) SlHyPRP1 with other Solanaceae plants. D. Single exonic (461 bp) gene structure of DEA1 flanked by upstream and downstream sequences. C terminal 8CM is shown in pink. E. Phylogenetic tree of tomato (S. lycopersicum) DEA1 with other Solanaceae plants
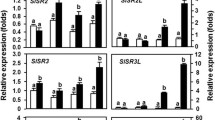
Tissue specific expression analysis of SlHyPRP1 and DEA1 in different tomato cultivars
SlHyPRP1 and DEA1 are known as putative regulators of different abiotic and biotic stress responses in plants (Li et al. 2016; Gujjar et al. 2018). Despite their significance as putative multi-stress responsive genes, very little information is available on their tissue specificity. Therefore, we performed a systematic expression analysis of SlHyPRP1 and DEA1 genes in three tomato cultivars namely, Micro tom, a model tomato cultivar (Shikata and Ezura 2016); a wilt susceptible Arka Vikas, and a multiple disease resistant Arka Abhed, the two most widely grown tomato cultivars in India (Upreti and Thomas 2015). Semi-quantitative PCR results have shown the predominant expression of SlHyPRP1 and DEA1 in leaf, stem, flower and root tissues while the least expression in fruit and seeds in all three cultivars (Fig. 2A). The internal control EF1-α showed uniform expression in all the tissues across all three cultivars (Fig. 2A). To further quantify the expression levels of SlHyPRP1 and DEA1 in different tissues, a systematic qRT-PCR was performed with the different tissues of three cultivars. The qRT-PCR results were found to be consistent with the semi-quantitative PCR results (Fig. 2B,C). The expression levels of SlHyPRP1 and DEA1 were found to be significantly higher in leaf, root, stem and flower tissues of cv. Arka Vikas and cv. Arka Abhed as compared to fruit and seed tissues (Fig. 2B, C). In cv. Micro tom, the gradual increase in expression levels of SlHyPRP1 was observed in root, stem, leaf and highest in flower tissue (Fig. 2C) while low expression was observed in seed and fruit tissues. DEA1 showed the highest expression in root whereas lowest in seeds and fruits while moderate expression in other tissues (Fig. 2C). The important observation from the qRT-PCR was that the expression levels of both SlHyPRP1 and DEA1 genes were found to be significantly higher in the cv. Arka Vikas, a bacterial wilt susceptible tomato cultivar compared to cv. Arka Abhed, a multiple disease resistant cultivar (Fig. 2B,C). The significance level of expression of qRT-PCR results was calculated using parametric two sample t-test variance among specific tissues of different cultivars relative to that in fruits. Each experiment was repeated thrice and the results were found to be consistent.
Expression analysis of SlHyPRP1 and DEA1 genes in different tissues of three tomato (S. lycopersicum) cultivars through semi-quantitative PCR and qRT-PCR. A. Semi-quantitative PCR analysis of SlHyPRP1 and DEA1. B and C. qRT-PCR analysis of SlHyPRP1 and DEA1. Values were normalized to Ef-1α as house-keeping gene and are calculated relative to that in fruit. The significant difference in expression level of SlHyPRP1 and DEA1 were evaluated by student’s t-test within each cultivar, taking each tissue separately with respect to the tissue having least expression. The bar represents mean ± SE from three technical replicates (*P < 0.05, **P < 0.01, ***P < 0.001)
Analysis of sub-cellular localization of SlHyPRP1 and DEA1
To study the subcellular localization of SlHyPRP1 and DEA1, we performed a transient cell biological assay in protoplasts and seedlings of tomato (cv. Arka Vikas). Prior to the cell biological assay, we carried out in silico localization predictions of SlHyPRP1 and DEA1 using the Cello2Go software tool. SlHyPRP1 showed localization at plasma membrane with the highest score of 26.7% (Fig. 3A) and DEA1 in extra cellular region with the highest score of 67.1% (Fig. 3B). For the in vivo sub-cellular localization study, we generated N and C terminal YFP fusion constructs of SlHyPRP1 and DEA1 using the Gateway cloning approach as described in material and methods (Fig. 3C). Molecular confirmation of constructs was carried out using restriction analysis (Fig. 3D–H, Fig. S2) and Sanger sequencing (Fig. 3I–L). Both YFP:SlHyPRP1 and SlHyPRP1:YFP showed predominant localization at the cytoplasm and plasma membrane (Fig. 4A, D). Similarly, YFP:DEA1 and DEA1:YFP also predominantly localized to plasma membrane and cytoplasm (Fig. 5A, D). The protoplasts transformed with empty vectors as negative controls did not show any YFP signals (Fig. 4B, E and 5B,E). In order to independently confirm their sub-cellular localization, we transformed both N and C terminal YFP fusions of SlHyPRP1 and DEA1 into tomato seedlings using the PDS1000 He system and observed under a confocal microscope. Both SlHyPRP1 and DEA1 showed strong localization at the plasma membrane and cytoplasm in both N-terminal and C-terminal fusions (Figs. 4C, F and 5C, F). In order to confirm the true YFP signals and eliminate the possibility of artefacts, we further quantified the YFP signals of each image using Leica XLS.lif software tool and found that the YFP signals were real in comparison to negative controls (Fig. S3 A–L). Each in vivo subcellular localization experiment was repeated thrice and the results were found to be consistent.
In silico localization predictions and Gateway cloning constructs of SlHyPRP1 and DEA1. In silico localization prediction of SlHyPRP1 A and DEA1 B using Cello2Go software tool (Yu et al., 2014). SlHyPRP1 showed predicted localization maxima at plasma membrane (26.7%) and DEA1 showed highest score of localization in extracellular regions (67.1%). C. Schematic representation of N-terminal A, B) and C-terminal (C, D) constructs of SlHyPRP1 and DEA1 in gateway expression vectors. D. PCR amplification of SlHyPRP1 and DEA1 genes from gDNA and cDNA of tomato (S. lycopersicum cv. Arka Vikas) with gateway adapter primers. Restriction digestion confirmation of SlHyPRP1 E and DEA1 F with stop codon and without stop codon in gateway pDONR entry vectors. Restriction digestion confirmation of SlHyPRP1 G and DEA1 H in gateway binary destination vectors pENSG-YFP and pEXSG-YFP. Sanger sequence alignment results of positive clones of SlHyPRP1with stop codon I, SlHyPRP1 without stop codon J, DEA1 with stop codon K) and DEA1 without stop codon L in pDONR vectors
Sub-cellular localization of N and C terminal YFP fusions of SlHyPRP1 in tomato protoplasts and seedlings. Tomato protoplasts transformed with N terminal (A, a–d), and C terminal (D, m–p) YFP fusion of SlHyPRP1. YFP:SlHyPRP1 was imaged with GFP channel. . Tomato seedlings transformed through PDS1000 system with N terminal (C, i–l), and C terminal (F, u–x) fusion of SlHyPRP1. Tomato protoplasts transformed with empty plasmid used a negative control for N terminal (B, e–h) and C terminal (E, q–t) YFP fusions did not show YFP signal. Each experiment was repeated thrice
Sub-cellular localization of N and C terminal YFP fusions of DEA1 in tomato protoplasts and seedlings. Tomato protoplasts transformed with N terminal (A, a–d), and C terminal (D, m–p) YFP fusion of DEA1. YFP:DEA1 was imaged with GFP channel . Tomato seedlings transformed through PDS1000 system with N terminal (C, i–l), and C terminal (F, u–x) YFP fusion of DEA1. Tomato protoplasts transformed with empty plasmid used a negative control for N terminal (B, e–h) and C terminal (E, q–t) YFP fusions did not show YFP signal. Each experiment was repeated thrice
Further, we made a comparative analysis of sub-cellular localization of SlHyPRP1 and DEA1 from our in vivo study, in silico predictions as well as in the literature findings in other plants (Table 1). As suggested by the expression analysis data carried out in this study, the tomato cv. Arka Vikas, a known stress susceptible cultivar (Upreti and Thomas 2015) was chosen for our cell biological study. Taken together, our results indicate that SlHyPRP1 and DEA1 are predominantly localized to plasma membrane and cytoplasm.
In vivo protein–protein interaction of SlHyPRP1 and DEA1 with stress associated proteins
Multiple stress responses in plants are highly complex which might be regulated by several host genes (Saikia et al. 2020). There could be a cross talk of plant-abiotic stress factors, plant-microbial interaction and plant defense pathway genes (Jones et al. 2019). Therefore, it is a prerequisite to identify the strong interacting partners of regulatory proteins that might be functioning together in multiple stress responses. In order to predict the putative protein partners of SlHyPRP1 and DEA1 in silico, we used the STRING interaction prediction tool which is an advanced search tool for retrieval of in silico interacting proteins (Szklarczyk et al. 2019). This search tool has the capacity to show the possible strong to weak interacting partner proteins (Szklarczyk et al. 2019). In silico analysis predicted a strong interaction of SlHyPRP1 with MST1 (Mercaptopyruvate sulphur-trasnferase-like protein) and WRKY3 transcription factor (Fig. 6A). DEA1 is predicted to be showing strong interaction with Snakin-2, an anti-microbial protein (Fig. 6B). The edges represent protein–protein association through text mining and co-expression (Fig. 6A, B).
In silico prediction of protein partners of SlHyPRP1 and DEA1 and molecular cloning and confirmation of gateway Split-YFP constructs. Predicted protein interaction network of SlHyPRP1 and DEA1 and their associated proteins using STRING software tool (Szklarczyk et al. 2019). A. SlHyPRP1, MST1 and WRKY3 showed interaction by the edges representing protein–protein association through text mining.B. DEA1 showed protein interaction network with Snakin-2 by edges representing text mining and co-expression . C. Schematic representation of split YFP constructs in gateway expression vectors pE-SPYNE and pE-SPYCE. D–F. PCR amplification of Snakin-2, WRKY3 and MST1 with gateway adapter primers from cDNA template of tomato (S. lycopersicum cv. Arka Vikas). G–I. Selection of positive clones of Snakin-2, WRKY3 and MST-1 in gateway expression vectors (pE-SPYNE and pE-SPYCE) through restriction analysis. J. Sanger sequence alignment results of positive clones of Snakin-2, K.WRKY3, and L.MST-1 in pDONR vector (pDONR221)
To validate the in silico protein partners of SlHyPRP1 and DEA1 in vivo, we performed a systematic Split-YFP (Bi-FC) assay (Gehl et al. 2009; Baruah et al. 2020) using the protoplasts of tomato (cv. Arka Vikas). The principle of in vivo Split-YFP assay is based on the formation of fluorescent complex (YFP/GFP) when two interacting partner proteins fused with two non-fluorescent segments of fluorescent protein (Kerppola 2010). We generated split-YFP constructs of SlHyPRP1 and DEA1, and predicted interacting partners in expression vectors pE-SPYNE and pE-SPYCE using Gateway cloning approach (Fig. 6C). Molecular confirmation was carried out using PCR and restriction analysis (Fig. 6D–I; Fig. S2) and Sanger sequencing of inserts in pDONR221 (Fig. 6 J–L). We co-transformed different combinations of split-YFP constructs into tomato protoplasts as represented in Table 1 and visualized their interaction pattern under confocal microscope. SlHyPRP1 showed a strong interaction with WRKY3 and the interaction was observed at the nucleus (Fig. 7A). SlHyPRP1 also showed interaction with MST1 and the interaction was observed at cytoplasm (Fig. 7B). However, DEA1 did not show interaction with Snakin-2 as we could not detect any YFP signal despite their notable in silico interactions (Fig. 7C). Interestingly, we observed a strong interaction between SlHyPRP1 and DEA1 where the interaction was taking place at cytoplasm and plasma membrane (Fig. 7D), although there was no in silico interaction prediction between them. In order to exclude the possibility of artifacts or false positives, the combination of empty split YFP vectors (pE-SPYNE and pE-SPYCE) co-transformed into tomato protoplasts used as standard negative controls did not show any YFP signals (Fig. 7E). The strong YFP signals observed for positive interactors were quantified using Leica XLS.lif software tool and found that the YFP signals were real in comparison to the negative controls (Fig. S3 m-q). Each split-YFP experiment was repeated thrice and the results were found to be consistent.
In vivo protein–protein interaction of SlHyPRP1 and DEA1 with stress associated proteins through split-YFP approach. A. Tomato protoplasts co-transformed with split YFP-constructs SlHyPRP1pE−SPYNE and WRKY3pE−SPYCE showed strong interaction in the nucleus (a–d). B. SlHyPRP1pE−SPYNE and MST1pE−SPYCE showed strong interaction in the cytoplasm and cytoplasmic organelles (e–h). C. DEA1pE−SPYCE and Snakin-2pE−SPYNE showed no interaction between them (i–l). D. SlHyPRP1pE−SPYNE and DEA1pE−SPYCE showed strong interaction in the cytoplasm and plasma membrane (m–p). E. Tomato protoplasts co-transformed with empty split-YFP plasmids pE-SPYNE and pE-SPYCE were used as negative controls which did not show any YFP signal (q–t). Each experiment was repeated thrice
We made a comparative analysis of in silico predictions, in vivo findings, interaction site of protein partners and localization of individual proteins along with literature findings as represented in Table 1. Taken together, our in vivo results of positive interactions of SlHyPRP1 with WRKY3 and MST1 were in accordance with in silico predictions but contradictory to DEA1 interaction with Snakin-2. The new finding from our in vivo interaction assay is the strong interaction between SlHyPRP1 and DEA1.
Discussion
SlHyPRP1 and DEA1 expressed tissue specifically
The role of HyPRPs and 8CM family proteins in response to multiple abiotic and biotic stresses have been marked primarily from their expression studies (Kapoor et al. 2019). All the HyPRPs reported so far are known to be up-regulated by abiotic stresses and down-regulated in response to various biotic stresses (Yang et al. 2018). Temporal and tissue-specific patterns of gene expression are the key factors in determining the functionality of a biological system (Lu et al. 2012). Our results have shown that the expression of SlHyPRP1 was highest in flower followed by leaf, stem and root in all three tomato cultivars. Previous studies revealed the expression of soybean SbPRP and cotton GhHyPRP4 was higher in leaves (He et al. 2002; Huang et al. 2011) while, the expression of CaHyPRP1 in Capsicum was highest in roots, flowers and immature green fruits than in leaves (Yeom et al. 2012). Our tissue specific expression studies revealed that even though the stem tissue showed a lesser expression of SlHyPRP1 in cv. Arka Abhed, a clear progression of expression in both cv. Micro tom and cv. Arka Vikas was observed from root to flower. However, it was not the same in fruit and seed tissue.
Our study showed the highest expression of DEA1 in stem and root tissues of cv. Arka Vikas and cv. Arka Abhed. Another finding of our study was the relatively higher expression levels of both SlHyPRP1 and DEA1 genes in stress susceptible tomato cv. Arka Vikas compared to multiple disease resistant cv. Arka Abhed. This suggests that these genes might play a negative regulatory function in imparting multiple stress tolerance in tomato (Weyman et al. 2006a; Li et al. 2016). We also observed the variation in expression among the three cultivars which was understandable due to the genotypic variations among cultivars. This is also because, the expression level and specificity of gene expression in tissue depends upon gene architecture of a higher organism which ultimately shapes the phenotype (Das and Bansal 2019). Taken together, our results suggest that SlHyPRP1 and DEA1 genes are predominantly expressed in leaf, stem, flower and root tissues, and expressed at a lower level in fruit and seed tissues of different tomato cultivars. Since SlHyPRP1is known as negative regulator of stress (Li et al 2016), its expression was found to be higher in cv. Arka Vikas, a bacterial wilt susceptible tomato cultivar compared to cv. Arka Abhed, a multiple disease resistant cultivar. Interestingly, constitutive expression of DEA1 was also found to be higher in susceptible tomato cultivar. As the expression profile and tissue specificity of SlHyPRP1 and DEA1 between multiple tomato cultivars were found to be similar, it is likely that they are co-expressed.
SlHyPRP1 and DEA1 are predominantly localized to plasma membrane and cytoplasm
SlHyPRP1 localizes to cytosol and cytosolic organelles in wild tomato S. pennellii (Li et al. 2016). GbHyPRP1is localized to the cell periphery in cotton (Yang et al. 2018). HyPRP1 is also known to be involved in sulfite metabolism, which occurs in leaf cells such as mesophyll cells and chloroplasts which are cytoplasmic organelles involved in photosynthesis (Takahashi 2010; Li et al. 2016). We can infer from our localization results that, the HyPRPs are putative cell wall structural proteins (Li et al. 2016). In order to perform the osmo-protectant activity during stress (Gujjar et al. 2018) as well as for cell wall thickening and ROS accumulation in response to pathogen attack (Yang et al. 2018), they might be localized at plasma membrane and cytoplasm. DEA1 is an 8CM family protein primarily expressed in taproot and stem involved in biotic and abiotic stress responses (Eulgem et al. 2000). DEA1 was localized to the cell periphery and plasma membrane in leaf protoplasts of Nicotiana benthamiana (Weyman et al. 2006a). It is evident from our results that the presence of secretory signal sequence along with the hydrophobic nature of the protein might supports the localization of DEA1 predominantly in the plasma membrane (Weyman et al. 2006b). As we also observed its prominent localization in the cytoplasm, even though the previous study supports its localization only towards the plasma membrane but it was not clear whether they are integral membrane proteins or not. There can be a possibility of transient overexpression differences in cellular localization during normal growth and development or in response to external stresses (Weyman et al. 2006a). Overall our in vivo localization results of SlHyPRP1 and DEA1 are in accordance with in silico predictions.
SlHyPRP1 showed in vivo localized protein interaction with DEA1, WRKY3 and MST1
The putative interactors observed from the in silico and in vivo studies are either directly or indirectly associated with abiotic and abiotic stress responses. SlHyPRP1 showed strong interaction with WRKY3 and MST1 as evident from in silico and in vivo observations. WRKY transcription factors are important regulators of abiotic stresses and are also involved in plant defense regulation (Aamir et al. 2017; Hichri et al. 2017). SlWRKY3 is a nuclear localized protein known to be involved in salt tolerance in tomato (Hichri et al. 2017). SlHyPRP1 is a cell wall structural protein localized to cytoplasm and plasma membrane. However, the interaction between SlHyPRP1 and WRKY3 in the nucleus suggests that SlHyPRP1 might re-localize to the nucleus during stress response function with WRKY3. This signifies their functional similarities during stress signaling pathways. MST1 is a mitochondrion and chloroplast localized protein that catalyzes sulphur ions from mercaptopyruvate to cyanide ions for sulfite metabolism (Nakamura et al. 2000; Brychkova et al. 2013; Höfler et al. 2016). SlHyPRP1 showed strong interaction with MST1 in cytoplasm. SlHyPRP1 is also known to be involved in sulfite metabolism in the conversion of toxic sulfite to nontoxic sulfate by improving SO activity in plants during SO2 stress (Li et al. 2016). Our findings revealed that SlHyPRP1 and MST1 might be functioning in mitochondria in imparting sulfite metabolism during abiotic (SO2) stress.
Snakin-2 is an antimicrobial cysteine rich protein known to localize in plasma membrane (Nahirñak et al. 2012). Tomato Snakin-2 whose expression elevates in response to fungal infection, mechanical wounding and external application of methyl jasmonate indicating its involvement in the JA-dependent defense response (Herbel et al. 2017). No interaction between DEA1 and Snakin-2 from our in vivo study indicates that, even though DEA1 is a cell membrane localized protein involved in programmed cell death (PCD) during biotic stress (Weyman et al. 2006a), it might have no role in JA-dependent defense responses. Though both proteins localized in plasma membrane, DEA1 and Snakin-2 do not interact with each other due to the functional dissimilarities.
The novel finding from our in vivo study is the strong interaction between SlHyPRP1 and DEA1 in cytoplasm and plasma membrane. As we observed from our subcellular localization study, both proteins localized to cytoplasm and plasma membrane, both belong to 8CM superfamily proteins associated with abiotic and biotic stress responses (Gujjar et al. 2018). Along with multiple abiotic stress responses, SlHyPRP1 is also sensitive to pathogen attack by degrading cell wall bound pathogens during biotic stress (Takahashi 2010; Li et al. 2016; Yang et al. 2018). DEA1 is involved in cold stress tolerance (Weyman et al. 2006a) and regulation of plant biotic stress responses (Klink et al. 2011). As a plasma membrane localized protein, DEA1 function in PCD regulation by inhibiting PCD-activating proteasome complex (Weyman et al. 2006b) similar to the function of other lipid transfer proteins such as TED-4. Our in vivo protein–protein interaction data showing a positive correlation between SlHyPRP1 and DEA1 proteins suggests that they both co-expressed and play a role in abiotic stress tolerance. Taken together, our findings indicate that SlHyPRP1 and DEA1 together functionally strongly connected in direct or indirect association with WRKY3 transcription factor and MST1 in regulating multiple abiotic and biotic stress responses in the tomato plant.
Conclusions
SlHyPRP1 and DEA1 express tissue specifically in different tomato (S. lycopersicum) cultivars and their expression levels are relatively higher in a stress susceptible cultivar than a resistant cultivar. The sub-cellular localization of SlHyPRP1 and DEA1, as well as their protein–protein interactions at cytoplasm and plasma membrane, indicate their sites of activity are cytoplasm and plasma membrane for stress response function. Strong positive interaction of SlHyPRP1 with stress responsive WRKY3 and MST1 proteins suggests that SlHyPRP1 is a key protein of the 8CM family and it might function together with DEA1, WRKY3 and MST1 in regulating multiple stress responses in the tomato plant.
Further genetic analysis through double knock-out studies of SlHyPRP1 and DEA1 or triple knock-out of HyPRP1, DEA1 and WRKY3/ MST1 would reveal their genetic mechanism and functional complementarities in imparting multi-stress responses in plants. A scheme of developing multi-stress tolerance through multi-gene editing of HyPRPs is reported in our recent review article (Saikia et al. 2020). In future, these findings would be helpful in precision dual or multiple gene editing for developing sustainable multi-stress tolerance in economically important crop plants under rapidly changing global climate.
Abbreviations
- PRP:
-
Proline rich proteins
- 8CM:
-
8-Cysteine motif
- Bi-FC:
-
Bi-molecular fluorescent complementation
- DEA1:
-
Differential expression of arachidonic acid
- CRISPR-Cas9:
-
Clustered regularly inter spaced short palindromic repeats/CRISPR-associated protein 9
- ORF:
-
Open reading frame
- MST1:
-
Mercaptopyruvate sulphur-trasnferase-like protein
References
Aamir M, Singh VK, Meena M et al (2017) Structural and functional insights into WRKY3 and WRKY4 transcription factors to unravel the WRKY – DNA ( W-Box ) complex interaction in tomato ( Solanum lycopersicum L.) A Computational Approach. Front Plant Sci 8:1–24. https://doi.org/10.3389/fpls.2017.00819
Baruah I, Baruah G, Sahu J, Singha DL (2020) Transient sub-cellular localization and in vivo protein-protein interaction study of multiple abiotic stress-responsive AteIF4A-III and AtALY4 proteins in arabidopsis thaliana, Plant Mol Biol Rep.
Brooks C, Nekrasov V, Lipppman ZB, Van Eck J (2014) Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol 166:1292–1297. https://doi.org/10.1104/pp.114.247577
Brychkova G, Grishkevich V, Fluhr R, Sagi M (2013) An essential role for tomato sul fi te oxidase and enzymes of the sul fi te network in maintaining leaf. Plant Physiol 161:148–164. https://doi.org/10.1104/pp.112.208660
Chikkaputtaiah C, Debbarma J, Baruah I et al (2017) Molecular genetics and functional genomics of abiotic stress-responsive genes in oilseed rape (Brassica napus L.): a review of recent advances and future. Plant Biotechnol Rep 11:365–384. https://doi.org/10.1007/s11816-017-0458-3
Cohen SP, Leach JE (2019) Abiotic and biotic stresses induce a core transcriptome response in rice. Sci Rep 9:1–11. https://doi.org/10.1038/s41598-019-42731-8
Das S, Bansal M (2019) Variation of gene expression in plants is influenced by gene architecture and structural properties of promoters. PLoS ONE 14:1–31. https://doi.org/10.1371/journal.pone.0212678
Debbarma J, Sarki YN, Saikia B et al (2019) Ethylene response factor (ERF) family proteins in abiotic stresses and CRISPR–Cas9 genome editing of ERFs for multiple abiotic stress tolerance in crop plants: a review. Mol Biotechnol 61:153–172. https://doi.org/10.1007/s12033-018-0144-x
Dvořáková Lenka CF and F (2007) Analysis of the hybrid proline-rich protein families from seven plant species suggests rapid diversification of their sequences and expression patterns. BMC Genomics https://doi.org/https://doi.org/10.1186/1471-2164-8-412
Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5:199–206. https://doi.org/10.1016/S1360-1385(00)01600-9
FAO (2019) Agriculture and climate change,FAO 2019. Rome
Gehl C, Waadt R, Kudla J et al (2009) New GATEWAY vectors for high throughput analyses of protein-protein interactions by bimolecular fluorescence complementation. Mol Plant 2:1051–1058. https://doi.org/10.1093/mp/ssp040
Gengqing H, Gong SW, Xu PL et al (2011) GhHyPRP4, a cotton gene encoding putative hybrid proline-rich protein, is preferentially expressed in leaves and involved in plant response to cold stress. Acta Biochim Biophys Sin 43:519–527. https://doi.org/10.1093/abbs/gmr040.Advance
Gerszberg A, Hnatuszko-Konka K, Kowalczyk T, Kononowicz AK (2015) Tomato (Solanum lycopersicum L.) in the service of biotechnology. Plant Cell Tissue Organ Cult 120:881–902. https://doi.org/10.1007/s11240-014-0664-4
Sarvajeet Singh Gill, Ritu Gill RT& NT (2014) Genetic engineering of crops: a ray of hope for enhanced food security. Plant Signal Behav ISSN1559–2324 2324:7–10. https://doi.org/https://doi.org/10.4161/psb.28545
Goodwin W, Pallas JA, Jenkins IG (1996) Transcripts of a gene encoding a putative cell wall-plasma membrane linker protein are specifically cold-induced in Brassica napus. Plant Mol Biol 31:771–781
Gothandam KM, Flower RÁ (2010) OsPRP3, a flower specific proline-rich protein of rice, determines extracellular matrix structure of floral organs and its overexpression confers cold-tolerance. Plant Mol Biol 72:125–135. https://doi.org/10.1007/s11103-009-9557-z
Gujjar RS, Karkute S, Rai A (2018) Proline-rich proteins may regulate free cellular proline levels during drought stress in tomato Proline-rich proteins may regulate free cellular proline levels during drought. Res Commun 114:. https://doi.org/https://doi.org/10.18520/cs/v114/i04/909-915
He CY, Zhang JS, Chen SY (2002) A soybean gene encoding a proline-rich protein is regulated by salicylic acid, an endogenous circadian rhythm and by various stresses. Theor Appl Genet 104:1125–1131. https://doi.org/10.1007/s00122-001-0853-5
Herbel V, Sieber-Frank J, Wink M (2017) The antimicrobial peptide snakin-2 is upregulated in the defense response of tomatoes (Solanum lycopersicum) as part of the jasmonate-dependent signaling pathway. J Plant Physiol 208:1–6. https://doi.org/10.1016/j.jplph.2016.10.006
Hichri I, Muhovski Y, Žižková E, et al (2017) The Solanum lycopersicum WRKY3 Transcription Factor SlWRKY3 Is Involved in Salt Stress Tolerance in Tomato. 8:1–18. https://doi.org/https://doi.org/10.3389/fpls.2017.01343
Höfler S, Lorenz C, Busch T et al (2016) Dealing with the sulfur part of cysteine: four enzymatic steps degrade l-cysteine to pyruvate and thiosulfate in arabidopsis mitochondria. Physiol Plant 157:352–366. https://doi.org/10.1111/ppl.12454
Huang G, Gong S, Xu W et al (2011) GhHyPRP4, a cotton gene encoding putative hybrid proline-rich protein, is preferentially expressed in leaves and involved in plant response to cold stress. Acta Biochim Biophys Sin (Shanghai) 43:519–527. https://doi.org/10.1093/abbs/gmr040
Jones D, Garcia BJ, Furches A et al (2019) Plant host-associated mechanisms for microbial selection. Front Plant Sci 10:862
José-Estanyol M, Gomis-Rüth FX, Puigdomènech P (2004) The eight-cysteine motif, a versatile structure in plant proteins. Plant Physiol Biochem 42:355–365. https://doi.org/10.1016/j.plaphy.2004.03.009
Jung HW, Tschaplinski TJ, Wang L, Glazebrook J, Greenberg JT (2012) Priming in systemic plant immunity. Science. https://doi.org/10.1126/science.1170025
Kapoor R, Kumar G, Arya P et al (2019) Genome-wide analysis and expression profiling of rice hybrid proline-rich proteins in response to biotic and abiotic stresses, and hormone treatment. Plants. https://doi.org/10.3390/plants8090343
Kerppola TK (2010) Nano-and_Micro_Eelectromechanical_Systems-S_E_Lyshevski.pdf. 465–487. https://doi.org/https://doi.org/10.1146/annurev.biophys.37.032807.125842.BIMOLECULAR
Klink VP, Hosseini P, Matsye PD et al (2011) Differences in gene expression amplitude overlie a conserved transcriptomic program occurring between the rapid and potent localized resistant reaction at the syncytium of the glycine max genotype Peking (PI 548402) as compared to the prolonged and potent. Plant Mol Biol 75:141–165. https://doi.org/10.1007/s11103-010-9715-3
Li J, Ouyang B, Wang T et al (2016) HyPRP1 gene suppressed by multiple stresses plays a negative role in abiotic stress tolerance in tomato. Front Plant Sci 7:1–14. https://doi.org/10.3389/fpls.2016.00967
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Lohani N, Jain D, Singh MB et al (2020) Engineering multiple abiotic stress tolerance in Canola. Brassica napus Front Plant Sci 11:3
Lu Y, Xie L, Chen J (2012) A novel procedure for absolute real-time quantification of gene expression patterns. Plant Methods 8:1–11. https://doi.org/10.1186/1746-4811-8-9
Marchler-Bauer A, Anderson JB, DeWeese-Scott C et al (2003) CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res 31:383–387. https://doi.org/10.1093/nar/gkg087
Marwein R, Debbarma J, Sarki YN, et al (2019) Genetic Engineering / Genome Editing Approaches to Modulate Signaling Processes in Abiotic Stress Tolerance. Elsevier Inc.
Shikata M, and Ezura H (2016) Micro-Tom tomato as an alternative plant model system: mutant collection and effi cient transformation. In: Methods in molecular biology (Clifton, N.J.). pp vii–x
Mellacheruvu S, Tamirisa S, Vudem DR, Khareedu VR (2016) Pigeonpea hybrid-proline-rich protein (CcHyPRP) confers biotic and abiotic stress tolerance in transgenic rice. Front Plant Sci 6:1–13. https://doi.org/10.3389/fpls.2015.01167
Nahirñak V, Almasia NI, Fernandez PV et al (2012) Potato Snakin-1 gene silencing affects cell division, primary metabolism, and cell wall composition. Plant Physiol 158:252–263. https://doi.org/10.1104/pp.111.186544
Nakamura T, Yamaguchi Y, Sano H (2000) Plant mercaptopyruvate sulfurtransferases molecular cloning, subcellular localization and enzymatic activities. Eur J Biochem 267:5621–5630. https://doi.org/10.1046/j.1432-1327.2000.01633.x
Neto LB, De ORR, Wiebke-strohm B et al (2013) Identification of the soybean HyPRP family and specific gene response to Asian soybean rust disease. Genet Mol Biol 224:214–224
Pandey P, Irulappan V, Bagavathiannan MV, Senthil-Kumar M (2017) Impact of combined abiotic and biotic stresses on plant growth and avenues for crop improvement by exploiting physio-morphological traits. Front Plant Sci 8:1–15. https://doi.org/10.3389/fpls.2017.00537
Parmar N, Singh KH, Sharma D et al (2017) Genetic engineering strategies for biotic and abiotic stress tolerance and quality enhancement in horticultural crops: a comprehensive review. 3 Biotech 7(4):239
Rahim MA, Jung HJ, Afrin KS et al (2018) Comparative transcriptome analysis provides insights into dwarfism in cherry tomato (Solanum lycopersicum var. Cerasiforme). PLoS ONE 13:1–21. https://doi.org/10.1371/journal.pone.0208770
Ray S, Lahiri S, Halder M et al (2015) An efficient method of isolation and transformation of protoplasts from tomato leaf mesophyll tissue using the binary vector pcambia 1302. Int Adv Res J Sci Eng Technol 2:146–150. https://doi.org/https://doi.org/10.17148/IARJSET.2015.2631
Saikia B, Singh S, Debbarma J et al (2020) Multigene CRISPR/Cas9 genome editing of hybrid proline rich proteins (HyPRPs) for sustainable multi-stress tolerance in crops: the review of a promising approach. Physiol Mol Biol Plants 26:857–869. https://doi.org/10.1007/s12298-020-00782-6
Savvides A, Ali S, Tester M, Fotopoulos V (2016) Chemical priming of plants against multiple abiotic stresses: mission possible? Trends Plant Sci 21:329–340. https://doi.org/10.1016/j.tplants.2015.11.003
Schwarz D, Thompson AJ, Kläring HP (2014) Guidelines to use tomato in experiments with a controlled environment. Front Plant Sci 5:1–16. https://doi.org/10.3389/fpls.2014.00625
Szklarczyk D, Gable AL, Lyon D et al (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613. https://doi.org/10.1093/nar/gky1131
Takahashi H (2010) Regulation of sulfate transport and assimilation in plants, 1st edn. Elsevier Inc.
Ueki S, Lacroix B, Krichevsky A et al (2009) Functional transient genetic transformation of Arabidopsis leaves by biolistic bombardment. Nat Protoc 4:71–77. https://doi.org/10.1038/nprot.2008.217
Ueki S, Magori S, Lacroix B, VC, (2013) Transient gene expression in epidermal cells of plant leaves by biolistic DNA delivery. Methods Mol Biol 940:17–26. https://doi.org/10.1007/978-1-62703-110-3
Upreti R, Thomas P (2015) Root-associated bacterial endophytes from Ralstonia solanacearum resistant and susceptible tomato cultivars and their pathogen antagonistic effects. Front Microbiol. https://doi.org/10.3389/fmicb.2015.00255
Weyman PD, Pan Z, Feng Q et al (2006a) A circadian rhythm-regulated tomato gene is induced by arachidonic acid and phythophthora infestans infection. Plant Physiol 140:235–248. https://doi.org/10.1104/pp.105.068874
Weyman PD, Pan Z, Feng Q et al (2006b) DEA1, a circadian- and cold-regulated tomato gene, protects yeast cells from freezing death. Plant Mol Biol 62:547–559. https://doi.org/10.1007/s11103-006-9039-5
Winicov I, Bastola DR (1999) Transgenic overexpression of the transcription factor Alfin1 enhances expression of the endogenous MsPRP2 gene in alfalfa and improves salinity tolerance of the plants. Plant Physiol 120:473–480. https://doi.org/10.1104/pp.120.2.473
Yang J, Zhang Y, Wang X et al (2018) HyPRP1 performs a role in negatively regulating cotton resistance to V. dahliae via the thickening of cell walls and ROS accumulation. BMC plant 1–18:339
Yeom SI, Seo E, Oh SK et al (2012) A common plant cell-wall protein HyPRP1 has dual roles as a positive regulator of cell death and a negative regulator of basal defense against pathogens. Plant J 69:755–768. https://doi.org/10.1111/j.1365-313X.2011.04828.x
Yu C, Cheng C, Su W, et al (2014) CELLO2GO: a web server for protein subcellular localization prediction with functional gene ontology annotation. 9:. https://doi.org/10.1371/journal.pone.0099368
Acknowledgements
The authors would acknowledge Director, CSIR-NEIST Jorhat for the facility and lab space. Authors also acknowledge Dr. M. Malligeppagol, ICAR-IIHR Bangalore for the seeds of tomato cultivars. The work was funded by Science and Engineering Research Board (SERB), Govt. of India as Early Career Research Award (ECR/2016/001288) and Ramanujan Fellowship (SB/S2/RJN-078/2014) to CC.
Author information
Authors and Affiliations
Contributions
CC has designed the concept, corrected the manuscript and involved in overall co-ordination of the project. BS performed all the experiments and drafted the manuscript, prepared the figures and tables and analysed the data. JD helped in qRT-PCR experiments, data analysis and corrected the manuscript. JM revised and improved the figures, reviewed and critically evaluated the manuscript. DLS, NV, HD, and KPA have provided critical comments, helped with analysis and corrected the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethics approval
This article does not contain any studies with human participants performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Saikia, B., Debbarma, J., Maharana, J. et al. SlHyPRP1 and DEA1, the multiple stress responsive eight-cysteine motif family genes of tomato (Solanum lycopersicum L.) are expressed tissue specifically, localize and interact at cytoplasm and plasma membrane in vivo. Physiol Mol Biol Plants 26, 2553–2568 (2020). https://doi.org/10.1007/s12298-020-00913-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-020-00913-z