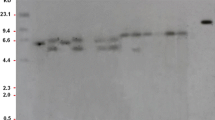Abstract
Glyphosate quashes the synthesis of 5-enolpyruvylshikimate-3- phosphate synthase (EPSPS) enzyme which intercedes the functioning of shikimate pathway for the production of aromatic amino acids. Herbicide resistant crops are developed using glyphosate insensitive EPSPS gene isolated from Agrobacterium sp. strain CP4, which give farmers a sustainable weed control option. Intentions behind this study were to design and characterize the synthetic herbicide resistant CP4-EPSPS gene in a model plant system and check the effectiveness of transformed tobacco against application of glyphosate. Putative transgenic plants were obtained from independent transformation events, and stable plant transformation, transgene expression and integration were demonstrated respectively by PCR, qRT-PCR and Southern hybridization. Gene transcript level and gene copy number (1–4) varied among the tested transgenic tobacco lines. Herbicide assays showed that transgenic plants were resistant to glyphosate after 12 days of spraying with glyphosate, and EPSPS activity remained at sufficient level to withstand the spray at 1000 ppm of the chemical. T1 plants analyzed through immunoblot strips and PCR showed that the gene was being translated into protein and transmitted to the next generation successfully. This codon optimized synthetic CP4-EPSPS gene is functionally equivalent to the gene for glyphosate resistance available in the commercial crops and hence we recommend this gene for transformation into commercial crops.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Weeds cause severe crop yield losses ranging from 30 to 45% worldwide. Integrated weed control management programs using classical and chemical methods have been successfully used for decades. However, many weed species could quite easily escape these control measures (Gianessi 2013), unless the issue was resolved by the advent of broad-spectrum chemical herbicide and subsequent development of herbicide resistant crops harnessing the shikimate pathway. Shikimate pathway is critical for biosynthesis of aromatic amino acids such as tyrosine, tryptophan and phenylealanine in plants, fungi, and bacteria. The enzyme 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS, EC2.5.1.19) catalyzes the transfer of enolpyruvyl moiety of phosphoenolpyruvate (PEP) to the 5-hydroxyl group of shikimate-3-phosphate (S3P) for the formation of 5-enolpyruvylshikimate-3-phosphate (EPSP) in the penultimate step of the shikimate pathway. EPSPS acts as a target of N-phosphonomethyl-glycine (glyphosate). Glyphosate resembles phosphoenolpyruvate (PEP) in transition state, and forms a dead end complex with plant chloroplast bound EPSPS enzyme, resulting in the complete inhibition of shikimate pathway. As a result biosynthesis of aromatic amino acids ceases and results in subsequent wilting of the plant and ultimate death (Sammons and Gaines 2014).
Genetic engineering enabled scientists to employ DNA manipulation techniques to transfer genes conferring herbicide resistance (HR), and successfully produce herbicide resistant crops. One of such HR genes is CP4-EPSPS that is isolated from Agrobacterium CP4 strain. The enzyme EPSP synthase is endogenously produced by all plants, but its conformation is quite different from that of the CP4-EPSPS product. This difference in conformation of CP4-EPSPS enzyme decreases its affinity for glyphosate, and helps the transgenic plants to survive (Dill 2005). The gene encoding CP4-EPSPS protein must express uniformly and continuously for the maintenance of sufficient level of EPSPS enzyme in the plant after herbicide application, and high transgene expression can be attributed to the adjustments in the GC contents, codon usage bias, and secondary structures (Liu et al. 2015). CP4-EPSPS genes can exhibit a thousand time enhanced gene expression in the eukaryotic system, and such codon optimized genes have already paved the way for agricultural revolution in some countries of the world.
The present study describes the development and characterization of a synthetic CP4-EPSPS gene for possible applications in crop plants. The gene was codon optimized according to cotton preferred codons as the trait was basically intended to be incorporated into cotton, and got synthesized commercially. We decided to characterize this newly synthesized gene in a model plant to check the expression and translational efficiencies of the transformed construct before proceeding for transformation of a construct into cotton and possibly other commercially important crops as well.
Materials and methods
Development of synthetic CP4-EPSPS gene and its cloning in pGreen0029 plant expression vector
Nucleotide sequence of CP4-EPSPS gene was modified and codon optimized according to preferred codon usage in cotton using Geneious software (http://www.geneious.com) in a way that amino acid sequence of the EPSPS protein remained unaltered. The CP4-EPSPS gene (1584 bp) was designed using codon usage frequency based on highly expressed cotton nuclear encoded genes. The CP4-EPSPS gene encodes EPSPS protein of 55.48 kDa containing 527 amino acids. As bacterial CP4-EPSPS gene is expressed in the cytoplasm and lacks chloroplast transit peptides (CTP), therefore, CTP signal (296 bp) sequence was added to CP4-EPSPS gene sequence for transportation of protein to the chloroplast. Furthermore, cauliflower mosaic virus (CaMV) promoter sequence (561 bp) and the E9 terminator sequence (201 bp) from yeast cofactors were incorporated respectively at 5′ and 3′ ends of the optimized CP4-EPSPS gene. The expression cassette including CaMV promoter, CP4-EPSPS gene, CTP signal and E9 terminator, respectively was synthesized and cloned into pUC57 vector having the EcoRI and HindIII restrictions sites at the 5′ and 3′ ends of expression cassette by DNA2 (Newark, California, USA). This expression cassette was removed from pUC57 plasmid using EcoR1 and HindIII restriction enzymes and sub-cloned into pGreen0029 expression vector at the same sites. The EPSPS expression cassette was confirmed by restriction analysis and the resultant plasmid (7278 bp) was named as pGEPC (Supplementary Fig. 1).
Plant transformation and molecular analysis
The plant expression vector was transformed into Agrobacterium tumefaciens strain LBA4404 using electroporation methods following the standard protocol (Main et al. 1995). Transformation of Nicotiana tabacum L. cv. Samsun was carried out using protocol as described by Horsch et al. (1984). Culture was diluted with LB broth in 1:1 ratio after overnight incubation at 150–250 rpm and 28 °C, and the transformed cells were allowed to grow to cell density of 1.0 at A600. Leaf discs were cut from fully expanded leaves of 4–5 weeks old in vitro grown plants, and used for Agrobacterium mediated transformation. Following infection with A. tumefaciens, leaf discs were transferred to co-cultivation medium [modified MS (Murashige and Skoog 1962) medium 4.2 g/L, sucrose 30 g/L, 6-benzylaminopurine 2.0 mg/L, α- naphthaleneacetic acid 0.1 mg/L, B5 vitamins 0.2 mg/L, phytagar 3.0 g/L] and incubated in dark for 2–3 days at 28 °C. Leaf discs were washed with cefotaxime 250 mg/L and cultured on co-cultivation medium containing kanamycin 50 mg/L and cefotaxime 250 mg/L) for differentiation. When the shoots were 2–3 cm, they were transferred to rooting medium (MS salts 4.2 g/L, sucrose 30 g/L, phytagar 4.2 g/L, cefotaxime 250 mg/L and kanamycin 100 mg/L) in jars. Regenerated plants formed the roots within 1–2 weeks of transfer and were shifted to the pots containing sterilized peat moss. Plants were acclimatized in standard growth room condition with artificial lighting providing a photo system of vertically active irradiance of 170 µM photons m−2 s−1 on a 16 h photoperiod.
All of the transgenic lines raised from independent transformation events were screened by PCR analysis using gene specific EPSPS primers PF5-5′CCTCACCTCCTGCGAGACGGA3′, and PR5-5′CGCCGCTACCGGATGCAGATT 3′. Southern hybridization analysis was carried out using AlkPhos Direct Labeling and Detection Kit (GE Health Care, Germany) following manufacturer’s instructions. About 20 µg genomic DNA extracted from transgenic and wild type tobacco was digested with EcoRI, and was immobilized on Hybond-N+ membrane. Amplified fragment of 569 bp using primers from EPSPS CaMV promoter region (CaMV-F TGAGGATACAACTTCAGAGA, EPSPS6-R TCCATTTCCAACGCCGTCAA T) was used for preparing the probe. Since there is only one EcoRI site in the plasmid used for transformation, so digestion of genomic DNA from the transgenic plants is expected to produce transgenes bearing bands of different sizes which will indicate the integration pattern and may also indicate the number/copies of transgenes integrated in the plant genome.
Herbicide resistance assay
From the PCR confirmed transformants, 22 lines were selected for herbicide screening. Commercial herbicide Roundup Ready glyphosate (Monsanto, USA) 1.0% (v/v) was applied over the leaf surface of individual plants with a canvass brush under controlled conditions of light, temperature and humidity in the containment. Data on vegetative injury was recorded 12 days after the herbicide application by visual observation. Glyphosate resistance levels were assessed according to the method described by Ye et al. (2001).
Expression analysis of CP4-EPSPS gene using qRT-PCR
Total RNA was extracted from newly emerged leaves of 30 days old transgenic and wild type tobacco plants using trizol method (Invitrogen, USA) followed by DNase1 (Invitrogen, USA) treatment. Total RNA (1 µg) was reverse transcribed to cDNA using commercial cDNA synthesis kit (Advanced Biosystems, USA). Quantitative real- time PCR (qRT-PCR) was performed using cDNA as template. Gene specific primers EPSPS1 (F-TGGGTTTGGTTGGTGTTT, R-AAGTTA TGGGAGTGGGAG) and housekeeping gene GAPDH primers (F-CACGGCCACTGGAAGCA, R-TCCTCAGGG TTCCTGATGCC) were designed to amplify 180 bp CP4-EPSPS and 150 bp GAPDH fragments. The reaction mixture consisted of 0.4 μl of each primer (EPSPS-F1 and EPSPS-R1; 4 pmol), 2 μl cDNA, 12.5 μl platinum SYBR Green Supermix (Thermo-Fisher Scientific, USA) with final volume reaction of 25 μl. PCR reactions were performed in Real-Time PCR Detection System (Advanced Biosystems, USA) in 96- well plates. The samples were run in triplicate. PCR machine was programmed at 94 °C for 5 min for 1 cycle followed by 40 cycles consisting of 20 s at 94 °C, 20 s at 60 °C and 20 s at 72 °C, followed by final extension of 10 min at 72 °C.
EPSPS activity assay
EPSP synthase activity depends upon the amount of inorganic phosphate release, which can be determined by recording the change in optical density. EPSPS activity was analyzed by determining inorganic phosphate release using malachite green assay according to method described by Lanzetta et al. (1979).
Inheritance of CP4-EPSPS gene in T1 generation and CP4-EPSPS protein analysis
For progeny analysis, T1 seeds were harvested from glyphosate resistant T0 plants and incubated at 25 °C for germination in trays containing peat moss. After two weeks of germination, T1 plants were analyzed for presence of CP4-EPSPS gene by PCR. Some of the PCR positive plants from each of five independent transgenic lines were subjected to immunoblot strip (EnviroLogix, USA) assays.
Data analyses
Expression of mRNA among different HR-transgenic lines was compared using three-way Analyses of Variance (ANOVA). Afterwards, the means were compared using Least Significant Difference (LSD) test. Similarly, the CP4-EPSPS enzyme activity in different transgenic lines and the control was compared by ANOVA. The means of different lines were compared using LSD test. Inheritance of transgene in T1 generation was assessed by comparison of observed and expected ratios. Goodness of fit to the Mendelian ratio of 3:1 was tested using Chi square analysis. Correlations and regression analyses were also performed to assess the influence of different parameters on each other. All statistical analyses were performed using computer software MS Excel Version 13 (Microsoft, USA) and SPSS Version 16 (IBM, USA).
Results
Molecular analyses of transgenic plants
The plasmid pGEPC (Supplementary Fig. 1) was transformed into N. tabacum plants using Agrobacterium mediated approach. A total of 42 putative transgenic plants obtained from independent transformation events, were confirmed by PCR using gene specific primers. CP4-EPSPS fragment was amplified according to the expected band size of 565 bp (Supplementary Fig. 2), which suggested that EPSPS gene had been successfully transferred in the host genome. Southern hybridization was performed to see the integration pattern of CP4-EPSPS gene in the genome of transgenic tobacco lines. Southern hybridization analysis showed that transgenic lines GTT-7, GTT-8, GTT-9 and GTT-10 had single inserts, while transgenic lines GTT-1 GTT-2, GTT-3, GTT-4, GTT-5 and GTT-6 contained multiple inserts. There was no hybridizing band detected in case of WT plant (Fig. 1).
Southern hybridization analysis of T0 transgenic tobacco plants transformed with CP4-EPSPS gene. Genomic DNA (20 µg) from wild type and transgenic plants was digested with EcoRI and resolved for 14 h on 0.8% agarose gel electrophoresis. The resolved DNA was shifted on to the nylon membrane. CaMV-EPSPS 569 bp labelled fragment was probed with the blot. Lane 1 shows the linearized plasmid used as positive control and lane 2 shows the untransformed wild type plant while 10 independent transgenic plants are shown in lanes 3, 4, 5, 6, 7, 8, 9, 10, 11 and 12 representing lines GTT-1, GTT-2, GTT-3, GTT-15 and GTT-16, GTT-6, GTT-7, GTT-8, GTT-9 and GTT-10 respectively. Lanes 9, 10, 11 and 12 showed single copy integration, while lane 3 showed double copy integration. Lanes 4, 5, 6, 7 and 8 showed multiple copy integration (3–4)
Herbicide resistance assay
All the PCR positive putative tobacco transgenic plants were shifted to the soil. Twenty-two independent PCR positive T0 lines were selected for glyphosate resistance assay. Transgenic tobacco plants along with non-transformed wild-type control were treated with 1.0% (v/v) herbicide Roundup ready (41.0%, w/v isopropylamine glyphosate salt as an active ingredient). Under the field conditions, 0.2% glyphosate is sufficient to kill most of the weed plants (Cakmak et al. 2009). Wild type untransformed plants showed severe chlorosis, and subsequently lead to stunting and wilting within 5 days following treatment, and died 7–12 days after treatment as shown in Fig. 2. Response to glyphosate spray was variable among transgenic plants. However, majority of the transgenic lines (GTT-6, GTT-7, GTT-8, GTT-9, GTT-10, GTT-13, GTT-15, GTT-16, GTT-20) showed no change in agronomic characteristics and produced the seeds normally, giving the notion that CP4-EPSPS expression in tobacco plants have conferred glyphosate resistance and that this trait is conserved strictly in their genome. While agronomic characteristics of some of the transgenic lines (GTT-2, GTT-19, GTT-21) changed after glyphosate treatment with appearance of slight chlorosis of newly emerged leaves and delay in flowering time. Few transgenic lines (GTT-18, GTT-22) showed severe stunting and wilted permanently (Table 1).
Herbicide assay of T0 transgenic tobacco plants. Tobacco plants were transferred to the pots. 1% (v/v) commercial Roundup ready (Monsanto) was applied on the PCR positive transgenic lines. Photographs were taken 12 days post herbicide application. Glyphosate treated plants were scored for resistance using a scale ranging of 1–4; 4 (highly resistant; normal plant vigor, fertile, typical maturity and seed setting), through to 3 (small chlorotic symptoms, fertile, typical maturity and seed setting), 2 (a part of leaf wilted after one week of glyphosate application, fertile, typical maturity and seed setting), 1 (two or more leaves wilted after one week of glyphosate application, stunted growth, delayed maturity and abnormal seed setting, and 0 (permanently wilted, dead plants). A and B represents highly resistant plants (scale 4), C and D represents resistant plants with little stress (scale 3), E shows the plants with part of leaf wilted (scale 2), F represent susceptible plants with 2 or more leaves wilted (scale1), G represents permanently wilted plants (scale 0), while H shows the wild type control which wilted after 10 days of herbicide treatment
CP4-EPSPS expression analysis using qRT-PCR
CP4-EPSPS gene expression analysis in various transgenic tobacco lines was carried out using qRT-PCR. All of the transgenic lines showed accumulation of transcript, however, there was some variation in gene expression among different transgenic lines. ANOVA results show the expression of mRNA varied significantly (F = 393.569, p < 0.05) between different transgenic lines at 95% confidence level. The LSD comparison of means (95% confidence) showed that the highest expression was recorded in line GTT-9, followed by line GTT-6, while the lowest expression was recorded in line GTT-7 as shown in Fig. 3.
CP4-EPSPS expression analysis of T0 transgenic tobacco plants using qRT-PCR. GAPDH gene was used as internal control and ΔCt values were calculated using difference in the Ct mean of the target gene and reference gene. Bars are the mean standard deviation calculated from three technical and three biological replicates. X axis shows different transgenic lines, and Y axis shows normalized expression of transgene
EPSPS enzyme assay
ANOVA was performed to assess the difference in the enzymatic activity among different transgenic lines. The EPSPS activity varied significantly among different lines (F = 25.226, p < 0.05, confidence 95%). Moreover, the comparison of means using LSD showed that the highest activity was in line GTT-9 (2.25 µmol min−1 mg−1) followed by GTT-6 (1.71 µmol min−1 mg−1) while the lowest activity was found in the wild type plant (0.2 µmol min−1 mg−1) as shown in Fig. 4.
EPSPS activity (µmol min−1 mg−1 + SE) of five transgenic tobacco T0 lines and a wild type control line. Bars are the mean standard deviation calculated from five technical and three biological replicates. Different letters indicate a significant statistical difference between means at 95% confidence with the LSD test (N = 15)
Correlation between enzyme activity and mRNA transcripts of transgenic plants
When the values of enzymatic activity of different transgenic lines were plotted against the expression level, positive correlation was found between these variables. EPSPS activity and mRNA transcript levels were positively correlated to each other at 0.9927 as shown in Fig. 5.
Mendelian inheritance pattern and CP4-EPSPS protein analysis
For the analysis of T1 generation, five lines (GTT-6, GTT-7, GTT-8, GTT-9 and GTT-10) were selected. Mendelian inheritance pattern in T1 progeny was analyzed using PCR (Fig. 6) and immunoblots (Supplementary Fig. 3). The observed and expected ratios of the PCR positive and negative plants were tested by Chi square test. The results (Supplementary Table 2) showed that the progeny of lines GTT-7, GTT-8, GTT-9 and GTT-10 containing one copy of transgene were within the ratio of 3:1 (X 2 = 0.516, X 2 = 0.550,X 2 = 0.09 and X 2 = 0.550, respectively at 95% confidence level). While, the progeny of line GTT-6 (X 2 = 0.001, confidence 95%) differed significantly from Mendelian ratio. This line segregated at the ratio of 15:1, probably due to multiple T-DNA insertions. The PCR positive plants from each of five independent transgenic lines subjected to immunoblot strip (EnviroLogix, USA) assays gave the protein bands suggesting that gene translated in the T1 inherited plants.
T1 Progeny analysis of representative transgenic tobacco line by PCR verification. PCR amplification of 565 bp product using CP4-EPSPS sequence specific primers; Lane M, 1 kbp DNA ladder; lane 1 negative control (water); lane 2 negative (non-transformed plant DNA) control; lane 3 empty well; lane 4 positive control (pGEPC); lanes 5-23, PCR amplified products from genomic DNA of representative transgenic tobacco line
Discussion
The CP4-EPSPS gene expression has been found to confer glyphosate resistance in various crop plants and facilitate more effective weed management via post emergence herbicide application. Unfortunately, number of transgenic crop cultivars raised with the herbicide resistance traits are very limited, and thus a vast space exists for extending this technology to diverse cultivars of various crops particularly for under-developed and developing countries. Keeping this in mind, we designed CP4-EPSPS gene so that local crop cultivars with herbicide resistance trait can be developed. Optimization of codon usage is mandatory to imitate the highly expressed endogenous genes of the host plant for achieving higher levels of heterologous expression of the transgene. While designing the synthetic CP4-EPSPS gene, use of rare codons was avoided at the expense of GC contents and codon adaptation index. Modification in the coding sequence, such as elimination of polyadenylation signals and potential RNA processing sequences is also an important pre-requisite. Codon optimization and precise location of signal sequence in the gene design are critical for increasing stability and accumulation of protein in receptor cells.
This codon optimized synthetic CP4-EPSPS gene was used to develop glyphosate resistant transgenic tobacco plants. PCR analysis confirmed the integration of coding sequence of transgene in genome of putative transgenic tobacco plants. Southern analysis revealed that transgene copy number varied from 1 to 4 in different transgenic plants and these results were in accordance with the findings of Wang et al. (2014). Detection of multiple copy number can be mainly attributed to the integration of transgene at more than one insertion sites. Usually preferences are given to transgene events with single copy number and Agrobacterium mediated transformation is considered a method of choice for gene transformation because it offers low copy insertions, simple and precise integration patterns, and higher transgene expression. However, correlation between copy number and gene expression has also been declined in literature (Tizaoui and Kchouk 2012). The qRT-PCR analysis confirmed that CP4-EPSPS gene was expressing at elevated levels in some of the tested transgenic lines, while low in others. The findings of this study were in compliance with the result of Bhullar et al. (2009) who reported variable expression levels among plants transformed with the same expression cassette.
There are number of factors responsible for variable transgene expression, including transgene copy number, rearrangement or recombination prior to transgene integration, position effect and DNA methylation (Garg et al. 2015). The most probable reason for lower gene expression in some of the lines might be due to partial transgene inactivation. The site of transgene integration in the plant genome is also critical for mRNA expression, for example, integration of gene in highly repetitive DNA region may results in the full inhibition of gene expression. Gene methylation or occurrence of any other homologous gene in host genome may lead to complete or partial silencing of the transgene as well. One percent glyphosate spray (v/v) used for herbicide assay in this study was equivalent to the concentration used by Chhapekar et al. (2015) and remarkably high in comparison to 0.1 to 0.5% (v/v) concentration used by Te et al. (2011). HR tobacco plants expressing CP4-EPSPS gene conferred variable resistance against glyphosate at routine field dosage. The level of resistance was directly proportional to the level of expression of the mRNA, observed in qRT-PCR. Majority of PCR positive plants conferred high level of resistance while some showed low or no resistance at all. The lines with lower gene expression and HR showed characteristic chlorosis in all plant tissues and some of these lines recovered from glyphosate stress within few weeks. Lower EPSPS gene expression has been previously reported by Dun et al. (2014) in transgenic crops including soybean, maize, potato, mustard, sugar beet, and tomato which displayed such chlorotic symptoms. The enzyme activity exhibited direct correlation with the amount of mRNA detected in the qRT-PCR. Thus, the transgenic tobacco lines showing relatively higher transgene expression offered higher enzymatic activity after one week of herbicide application. These results are in accordance with findings of Cao et al. (2012) who reported higher shikimate levels after 5 days of glyphosate treatment. After initial surge, the shikimate level begins returning to normal, probably due to maintenance of over-expressed EPSPS protein (Liu et al. 2015). In four out of five tested T1 lines, transgene was inherited according to Mendelian rules giving segregation ration of 3:1, however, the fifth line exhibited a segregation ration of 15:1, and these results were similar to the finding of Guo et al. (2003) who reported 3:1 segregation ratio for lines with 1–2 copy number and 15:1 segregation ratios for lines with 3–4 transgene inserts in transgenic tobacco plants. This abnormal segregation pattern was due to multiple copy numbers of the transgene in T0 parent, as segregation ratios are determined by the number of functional transgene inserts integrated into T0 plants. The greater the copy number of independently segregating transgene in the genome, the greater the probability of transgene gametes and thus the ratio of transgenic plants increase (Tizaoui and Kchouk 2012). The presence of multiple copy number was confirmed by the Southern analysis. Immunoblot strip assay showed that T1 transgenic progenies were expressing the CP4-EPSPS protein at sufficient level.
The study presented the development of new synthetic herbicide resistant CP4-EPSPS gene, its integration, expression and translation, and resistance assay against the target glyphosate herbicide in transgenic tobacco lines, and hence we recommend this trait to be harnessed in developing herbicide resistant commercial cultivars.
References
Bhullar S, Chakravarthy S, Pental D, Burma PK (2009) Analysis of promoter activity in transgenic plants by normalizing expression with a reference gene: anomalies due to the influence of the test promoter on the reference promoter. J Biosci 34(6):953–962
Cakmak I, Yazici A, Tutus Y, Ozturk L (2009) Glyphosate reduced seed and leaf concentrations of calcium, manganese, magnesium, and iron in non-glyphosate resistant soybean. Eur J Agron 31:114–119
Cao G, Liu Y, Zhang S, Yang X, Chen R, Zhang Y, Lin M (2012) A novel 5-enolpyruvylshikimate-3-phosphate synthase shows high glyphosate tolerance in Escherichia coli and tobacco plants. PLoS ONE 7:e38718
Chhapekar S, Raghavendrarao S, Pavan G, Ramakrishna C, Singh VK, Phanindra MLV, Kumar PA (2015) Transgenic rice expressing a codon-modified synthetic CP4-EPSPS confers tolerance to broad-spectrum herbicide, glyphosate. Plant Cell Rep 34:721–731
Dill GM (2005) Glyphosate-resistant crops: history, status and future. Pest Manag Sci 61:219–224
Dun B, Wang X, Lu W, Chen M, Zhang W, Ping S, Lin M (2014) Development of highly glyphosate-tolerant tobacco by coexpression of glyphosate acetyltransferase gat and EPSPS G2-aroA genes. Crop J 2:164–169
Garg R, Chevala VN, Shankar R, Jain M (2015) Divergent DNA methylation patterns associated with gene expression in rice cultivars with contrasting drought and salinity stress response. Sci Rep 5:14922
Gianessi LP (2013) The increasing importance of herbicides in worldwide crop production. Pest Manag Sci 69:1099–1105
Guo X, Han H, Zhang J, Wang H (2003) The viral resistance of transgenic tobacco plants containing untranslatable PVYN coat protein gene. Shi Yan Sheng Wu Xue Bao 36:176–184
Horsch RB, Fraley RT, Rogers SG, Sanders PR, Lloyd A, Hoffman H (1984) Inheritance of functional genes in plants. Science 223:496–498
Lanzetta PA, Alvarez LJ, Reinach PS, Candia OA (1979) An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem 100:95–97
Liu Y, Cao G, Chen R, Zhang S, RenY LuW, Wang G (2015) Transgenic tobacco simultaneously overexpressing glyphosate N-acetyltransferase and 5-enolpyruvylshikimate-3-phosphate synthase are more resistant to glyphosate than those containing one gene. Transgenic Res 24:753–763
Main GD, Reynolds S, Gartland JS (1995) Electroporation protocols for Agrobacterium. Methods Mol Biol 44:405–412
Sammons RD, Gaines TA (2014) Glyphosate resistance: state of knowledge. Pest Manag Sci 70:1367–1377
Te Z, Lin CY, Shen ZC (2011) Development of transgenic glyphosate-resistant rice with G6 gene encoding 5-enolpyruvylshikimate-3-phosphate synthase. Agr Sci China 10:1307–1312
Tizaoui K, Kchouk ME (2012) Genetic approaches for studying transgene inheritance and genetic recombination in three successive generations of transformed tobacco. Genet Mol Biol 35:640–649
Wang J, Zuo K, Lu W, Zhu Y, Ye C, Lin M, Tang K (2014) Over-expression of the Gr5 aroA gene from glyphosate-contaminated soil confers high tolerance to glyphosate in tobacco. Mol Breeding 33:197–208
Ye GN, Hajdukiewicz PT, Broyles D, Rodriguez D, Xu CW, Nehra N, Staub JM (2001) Plastid-expressed 5-enolpyruvylshikimate-3-phosphate synthase genes provide high level glyphosate tolerance in tobacco. Plant J 25:261–270
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Imran, M., Asad, S., Barboza, A.L. et al. Genetically transformed tobacco plants expressing synthetic EPSPS gene confer tolerance against glyphosate herbicide. Physiol Mol Biol Plants 23, 453–460 (2017). https://doi.org/10.1007/s12298-017-0424-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-017-0424-0