Abstract
Elevated LDL/HDL ratio is an important risk factor for predicting atherosclerosis. Paraoxonase-1 protects LDLs from oxidative modifications and has a protective effect against atherosclerosis. Two common polymorphisms, Q192R and L55M, in PON1 gene can affect PON1 levels and function. The aim of this study was to evaluate the frequency PON1 polymorphisms in individuals with high and normal LDL/HDL ratios. To evaluate Q192R and L55M polymorphisms in Iranian case group (n = 70) with high LDL/HDL ratio, and control group (n = 80) with normal LDL/HDL ratio, we used PCR–RFLP method. Genotype frequencies for Q192R were 52.2 % for QQ, 46.3 % for QR, and 1.5 % for RR in case group (P = 0.003). However, Genotype frequencies in case group for L55M were 17.5 % LL, 75.3 % LM, and 7.2 % MM (P = 0.001). PON1 L55M polymorphism was associated with high LDL/HDL ratios in case group. So, the L55M polymorphisms can contribute in reducing the antioxidant function and decreasing level of HDL particles. In conclusion, PON1 L55M polymorphism can affect lipid metabolism and may be related to atherosclerosis in Iranian individuals.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Many risk factors are involved in atherosclerosis. Among these, dyslipidemias play an essential role in pathogenesis of atherosclerotic cardiovascular diseases (CVDs), and become a target for preventive and therapeutic approaches [1]. Elevated low density lipoproteins (LDL) and/or decreased high density lipoproteins (HDL) are two responsible factors in atherosclerosis due to different dyslipidemias [1–3]. LDL particles, which are heterogeneous in their composition, shape, size, and density, are the major carriers of cholesterol in human plasma. These particles are known as atherogenic lipoprotein particles and represent a strong cardiovascular risk factor [2–4]. HDL mediates reverse cholesterol transport and has several other anti-atherogenic effects [1, 2]. Oxidative modifications of LDL in arterial walls is the main pathogenic mechanism in initiation and acceleration of coronary artery disease (CAD) followed by atherosclerosis [5]. Human serum paraoxonase 1 (PON1), encoded by the polymorphic gene PON1 on chromosome 7q21.3, is produced mainly in the liver and is associated with HDL particles [6]. Indeed, one of the main protective effects of HDL is due to the enzyme activity of PON1, and probably prevents atherosclerosis by preventing LDL from oxidative modifications [7]. PON1 can hydrolyze some organophosphates such as paraoxon and sarin [8]. The PON1 gene has nearly 200 single nucleotide polymorphisms (SNPs). Of this Q192R and L55M polymorphisms located in the coding region are the most commonly studied SNPs [8]. The Q192R and L55M polymorphisms are the commonest polymorphic regions in this gene that are related to enzyme activity and serum levels of PON1 [8, 9]. There are several studies demonstrating an association between Q192R and L55M polymorphisms and susceptibility to CAD in cardiovascular patients and/or people who have been prone to atherosclerosis because having high LDL/HDL and or ApoB/ApoA-I ratios [10–13]. However some studies have failed to find such an association [10, 14] LDL levels and or calculated ratios such as LDL/HDL are useful predictors of risk of CVD in the primary and secondary prevention setting of these diseases [15]. However, some evidences suggest that the ratio of the corresponding apolipoproteins (ApoB/ApoA-I) might be superior or better predictors in some populations [4, 15, 16].
This highlights the need to evaluate the association between PON1 polymorphisms and LDL/HDL ratio, which is one of the important risk factors to develop coronary heart diseases, in general populations. Thus, we evaluated the association between LDL/HDL ratio by determination of lipid profile and two PON1 polymorphisms include L55M and Q192R, which may modulate serum lipids in Iranian population with no diagnosis of cardiovascular illness.
Materials and Methods
Study Population
A case–control study was performed in Tehran (at North) and Zanjan (at North-west), Iran during 1 year. Informed consent was obtained from each individuals included in the study. The study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a priori approval by the institution’s human research committee. Eligible participants were 150 unrelated individuals who were matched according to age and sex, aged 30–80 years, without coronary heart disease were admitted to enroll with present study. According to LDL/HDL normal and risk ranges, 70 patients with LDL/HDL ratio higher than 3.4, and 80 individuals with the ratios lower than 3.4 were purposed as case and control groups, respectively [17, 18]. Some information asked from participants by trained personnel. Structured questionnaire included age, sexuality, smoking habits, familial history of CVD, history of refer to cardiology clinic, history of hospitalization due to heart problems, history of bypass, history of angioplasty, hypertension and drug usage especially anti lipid and anti oxidant drugs. Individuals with liver, renal/and thyroid problems, diabetes mellitus and participants who had taken anti hypertensive, lipid lowering/and anti oxidant drugs during 1 month before sampling, excluded from study.
Sampling
Blood samples were collected after overnight fasting in EDTA coated tubes to analyze PON1 genotypes and without anticoagulant for serum lipid profile determinations. Collected samples were stored at −20 °C until analysis.
Determination of PON1 Q192R and L55M Polymorphisms
Genomic DNA was extracted using Genomic DNA Extraction Kit (Qiagen, Germany) according to the protocol of the manufacturers. After extraction, all DNA samples were measured by NanoDrop (ND-1000, Thermoscientific) and their purity was checked by A260/A28 ratio. PON1Q192R and L55M polymorphisms were determined by PCR, lyophilized premix PCR tubes (Bioneer, South Korea) and followed by restriction fragment length polymorphism analysis (RFLP). Primers [19, 20], annealing temperature and length of PCR products for each polymorphism explored in this study are given in Table 1. For quality control of PCR method, it was used an extra microtube containing all master mix components without DNA. This negative control was loaded on agarose electrophoresis post PCR.
According to our experiences previously [10], each restriction endonuclease mixture (total volume 24 μl) contained 15 μl amplified fragment, 5 μl distilled water, 2 μl appropriate buffer for each restriction endonuclease (Fermentas Life Science) and 4 unit of AlwI for Q192R and 4 unit Hin1II for L55M polymorphism (1 unit of restriction enzyme is the amount of enzyme required to digest 1 μg of lambda DNA in 1 h at 37 °C for Hin1II or at 55 °C for AlwI). Restriction products were separated by electrophoresis on a 4.5 % agarose gel in TBE buffer and stained with ethidium bromide.
Lipid Profile, LDL and HDL Determination
Serum total cholesterol (TC) and triglycerides (TG) were determined by enzymatic methods (Pars Azmoon, Iran/Hitachi 902). HDL-C and LDL-C were measured using turbidimetric method (Randox©, England/Hitachi 902).
Statistical Analyses
SPSS software (version 17) was used for statistical analysis. Comparisons of allele and genotype frequencies between case and control groups were analyzed using the Chi squared test. P values lower than 0.05 were considered statistically significant.
Results
General characteristic and lipid profile of participants are shown in Table 2. The mean of LDL/HDL ratio in case and control groups were 4.7 ± 1.0 and 2.5 ± 0.5, respectively. Participants mean ages in case and control groups were 57.5 ± 9.5 and 51.6 ± 10.2, respectively. The power of study was estimated around 80 %.
PON1 Q192R and L55M Genotypes Determination
According to Hardy–Weinberg Equilibrium (HWE) law, there are two allele for PON1 gene at 192 region include Q and R. In addition, there are two allele at 55 region include L and M.
Q192R Polymorphisms
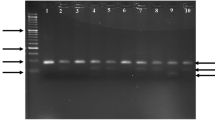
Undigested fragment (99 bp) was detected in homozygotes for Q192 allele (genotype QQ), digested fragments (66 and 33 bp) were detected in homozygotes for R192 allele (genotype RR) and both digested and undigested fragments (99, 66 and 33 bp) were detected in heterozygotes (genotype QR) (Fig. 1).
L55M Polymorphisms
Undigested fragment (172 bp) was detected in homozygotes for L55 allele (LL genotype), digested fragments (103 and 69 bp) were detected in homozygotes for M55 allele (genotype MM) and digested and undigested fragments (172, 103 and 69 bp) were detected in heterozygotes (genotype LM) (Fig. 2).
Association between PON1 Q192R and L55M Polymorphisms and LDL/HDL Ratios
Table 3 shows the PON1 genotype frequencies in case and control groups. Frequencies for Q192R one allele polymorphism (QR) in case and control groups were 46.3 and 32.2 %, respectively; but the frequencies of two allele polymorphism (RR) in case and control groups were 1.5 and 17.2 %, respectively (P = 0.003) (Fig. 3).
Frequencies for L55M one allele polymorphism (LM) in case and control groups were 75.3 and 46.8 %, respectively. Frequencies for L55M two allele polymorphism (MM) in case and control groups were also 7.2 and 8.6 %, respectively (P = 0.001) (Fig. 4).
In ANOVA analysis, the Q192R and L55M polymorphism signification between case and control groups were 0.112 and 0.007, separately. In investigation of polymorphism odd ratios in case group, the odd ratios (CI 95 %) of Q192R and L55M polymorphisms in one or two allele were 0.98 and 2.85, respectively.
Discussion
CVD is the most important cause of death in the world. Several studies have been reported about the sharply increasing of this disease over the next 10 years [21].
Atherosclerosis, as the one of the major causes of the CVD occurrence, is developed as the consequence different environmental and genetic factors; however lipoproteins have a especial role of its pathogenesis [21]. Changes in serum lipid profile include decrease of HDL-C and increased LDL-C, TG and TC can contribute to the pathogenesis of atherosclerosis [22]. Some studies suggest that the role of genetic factors on lipid profile is better than environmental factors for considering of atherosclerosis [23, 24].
In the different types of lipoproteins, LDL particles can damage coronary arteries, and HDL particles can prevent the damage. Hence, plasma levels of LDL and HDL are, positive and negative risk factors for CAD, respectively. In addition, the evaluation of plasma levels of apolipoprotein B and apolipoprotein A-I, which are the main surface proteins on LDL and HDL particles, is important [25].
The recent National Cholesterol Education Program Adult Treatment Panel III (ATP-III) guidelines recommend specific target levels of LDL cholesterol and HDL cholesterol for determining the risk of CVD and evaluating the effectiveness of lipid-lowering therapies. Some evidences show that the LDL-C/HDL-C ratio continues to be a valuable and standard tool to evaluate CVD risk in all populations [17].
Different studies have been shown that the oxidative change in LDL particles in the arterial walls is the main pathogenic mechanism in developing of atherosclerosis, and thus the CAD. In contrast, HDL particles have a protective effect by reverse cholesterol transport from tissues to liver [1]. Some studies reported that one of the important protective roles of HDL is due to the paraoxonase 1 (PON1) enzyme activity that bound to HDL surface. PON1 can prevent the LDL peroxidation and hydrolyze oxidized LDL particles and may have a role in preventing of atherosclerosis [23, 26, 27]. Studies have shown that PON1 knockout mice cannot hydrolyze the oxidized LDL and have increased risk of developing CAD [8, 28].
There are a wide variation in PON1 activity and genotypes in different ethnic groups and even within individuals in the same ethnic group. The PON1 activity is decreased in patients with familial hypercholesterolemia and diabetes mellitus, who are more prone to atherosclerosis. Hence, the lower PON1 activity may increase the oxidative modification in LDLs and the risk of CAD [8, 29, 30]. In addition to PON1 activity, some genetic polymorphisms can affect on the PON1 concentration and its activity. Two common polymorphisms in the coding region of PON1 gene have been reported: (1) replacement of leucine (allele L) by methionine (allele M) (Leu [L]/Met [M]) at 55 position, which is related with differences in PON1 activity, and (2) polymorphism at position 192 that recognized by replacement of glutamine (allele Q) by arginine (allele R) (Gln [Q]/Arg [R]), has been described as substrate-dependent and affects the catalytic efficiency of PON1 [8, 31].
There are several studies which have investigated the relationship between PON1 gene polymorphisms and the risk of atherosclerosis. Frequencies of PON1 alleles widely vary across human populations. Different studies have been shown that there is an association between Q192R and L55M polymorphisms and susceptibility to CAD and atherothrombosis due to oxidant stress [12, 13, 43]. However, other reports have failed to find such an association [14, 15, 32]. The concentrations of serum total cholesterol, HDL-C, and LDL-C and enzyme activity of PON1 may depend on the PON1-L55M and PON1-Q192R polymorphisms [31].
The association of PON1 Q192R polymorphism with the development of CVD is yet unresolved. Durringtone et al. in their epidemiological study suggested that PON1Q192R polymorphism, in particular the 192RR genotype, may represent a risk factor to develop CHD [33]. In addition, some other evidences suggest that the PON1 RR allele is associated with increased risk of CAD [12, 22]. Mackness et al. are observed an increased frequency of the PON1 QR and RR alleles in CAD patients [34]. Schmidt et al. reported that atherosclerotic lesions were more common in RR genotype [12]. Gupta et al. found that one or two polymorphism(s) in Q192R position were associated with increased risk of CAD after adjusting all conventional risk factors [9]. However, these observations have been challenged by Antikainen et al. who didn’t find any association in a Finnish population [32]. In addition, some other studies have failed to find this relationship [14, 19]. Ombres et al. also evaluated the Q192R genotype frequencies in angiographically defined CAD individuals; but there were no indication that this polymorphism is significantly associated with the risk of coronary atherosclerosis [14]. In addition to these reports, Schmidt et al. failed to detect a significant association between the Q192R polymorphism and atherosclerotic lesions in patients with carotid disease. In this study [13], we also didn’t observe the association between PON1 Q192R polymorphism and the risk of CVD, by defined with higher LDL/HDL ratios in case group. Therefore, there were no conclusions about a possible role of codon 192 PON1 polymorphism as a marker of CAD risk in the general population of coronary patients.
Nus et al. reported that individuals with Q192 had lower HDL levels than R192 carriers [31]. We are observed that individuals with RR genotype have higher LDL/HDL ratio (P = 0.003). Nus et al., in addition, found a positive correlation between arylesterase activity of PON1 and HDL-C levels, and negative correlation between arylesterase activity, oxidized LDL (ox-LDL) concentration and the ox-LDL/LDL ratio. This evidence supports the protective effect of PON1 on LDL oxidation [31]. Aviram et al. shown that the protective activity of PON1 against oxidation is impaired in 192R alloenzyme [35]. Therefore, low PON1 activity in patients with cardiovascular illness or other adverse oxidative stress condition may be related to their genotype [26].
Pérez-Herrera et al. reported that PON1 Q192R allelic frequencies were different from those reported in several Caucasian populations and our results [14, 23, 36]; but were similar to those in Chinese, Afro-American and other Mexican populations [37–39]. In addition, individuals with 192RR genotype had lower levels of HDL-C than 192QQ. Hence, this may suggest that PON1 polymorphisms could affect the serum lipid levels, particularly HDL. This report is in agreement with the study of Hegele et al. who found that 192QQ individuals in an isolated population from Canada had higher levels of HDL and lower levels of TC, LDL and TG than heterozygotes or homozygotes for the 192R allele [24]. However, we did not a significant change in 192QQ genotype between case and control groups. Pérez-Herrera et al. reported that 42 % of individuals had HDL values lower than 40 mg/dL, from which 36 % of individuals were 192RR homozygotes and 13 % were homozygote for the 192Q allele [23]. Considering that high values of HDL may act as a protective mechanism against CVDs. Our data suggest that there may be more 192RR individuals at risk for developing these pathologies. However, we didn’t find these similarity and 17 % of individuals in our control group with normal levels of LDL/HDL ratios, had 192RR genotype. Our study, likes Pérez-Herrera et al., was conducted in healthy individuals with no clinical diagnosis of CVDs.
The association between PON1 L55M polymorphism and lipid profile has been less studied. Deakin et al. shown that L55M polymorphism may be related to vascular disease and impaired glucose metabolism [40]. Some studies with inconsistent results have been shown the relationship between the L55M polymorphism and CAD [30]. Gupta et al. could not observe any significant effect of L55M polymorphisms with CAD and PON1 activity [9]. However, Schmidt et al. have been observed an independent association of CAD with L55M polymorphism in Austrian CAD patients [13]. In addition, they suggest that the 55LL genotype is a significant and independent predictor of carotid atherosclerosis in a middle-aged and elderly population [13]; but in this study, the L55M polymorphism, especially in one allele, was correlated with case group who had higher LDL/HDL ratios.
Pérez-Herrera et al. did not observe an association between PON1 L55M polymorphism and HDL, but significant higher levels of LDL in 55LM heterozygote subjects [23]. They reported that PON1 L55M allelic frequencies were similar to those reported in Afro-Americans and other Mexican populations [37, 39], but different from those in Caucasians [36]. Like this result, in presence study, the 55LM heterozygote individuals had higher LDL/HDL ratios.
Deakin et al. and Fanella et al. reported a trend of higher LDL and lower HDL levels according to PON1 L55M polymorphism, 55MM subjects had the less favorable profile [40, 41]. However, we didn’t observe this significant relationship between individuals with higher LDL/HDL ratios and 55MM genotype.
Grubisa et al. have shown that there is no significant difference in allele and genotype frequencies of PON1 polymorphisms between the atherosclerotic patients who had angiography and healthy controls [42].
Authors are aware that lipid profile is a multifactorial characteristic that cannot be explained by only one factor. Therefore, caution must be taken when comparing data from different populations.
In summary, as respects the increased LDL/HDL ratios, especially higher than 3.4, are important for developing of atherosclerosis, L55M polymorphism may be helpful for prediction of atherosclerosis. L55M polymorphisms in PON1 gene may decrease the antioxidant capability of HDL. This can increase the LDL/HDL ratio, even in individuals without higher amounts of LDL. In conclusion, it is important to explore the effect of PON1 L55M genetic polymorphisms on lipid metabolism and atherosclerosis in diverse populations with different ethnicities.
References
von Eckardstein A. HDL- a difficult friend. Drug Discov Today. 2008;5(3):315–24.
Pöss J, Custodis F, Werner C, Weingärtner O, Böhm M, Laufs U. Cardiovascular disease and dyslipidemia: beyond LDL. Curr Pharm Des. 2011;17(9):861–70.
Shah AS, Urbina EM, Khoury PR, Kimball TR, Dolan LM. Lipids and lipoprotein ratios: contribution to carotid intima media thickness in adolescents and young adults with type 2 diabetes mellitus. J Clin Lipidol. 2013;7(5):441–5.
Ren G, Rudenko G, Ludtke SJ, Deisenhofer J, Chiu W, Pownall HJ. Model of human low-density lipoprotein and bound receptor based on CryoEM. Proc Natl Acad Sci USA. 2010;107(3):1059–64.
von Eckardstein A, Assmann G. Prevention of coronary heart disease by raising high-density lipoprotein cholesterol? Curr Opin Lipidol. 2000;11(6):627–37.
Navab M, Ananthramaiah GM, Reddy ST, Van Lenten BJ, Ansell BJ, Fonarow GC, et al. The oxidation hypothesis of atherogenesis: the role of oxidized phospholipids and HDL. J Lipid Res. 2004;45(6):993–1007.
Martínez C, Molina JA, Alonso-Navarro H, Jiménez-Jiménez FJ, García-Martín E. Two common nonsynonymous paraoxonase 1 (PON1) gene polymorphisms and brain astrocytoma and meningioma. BMC Neurol. 2010;10:71.
Mackness MI, Durrington PN, Mackness B. The role of paraoxonase 1 activity in cardiovascular disease: potential for therapeutic intervention. Am J Cardiovasc Drugs. 2004;4(4):211–7.
Gupta N, Singh S, Maturu VN, Sharma YP, Gill KD. Paraoxonase 1 (PON1) polymorphisms, haplotypes and activity in predicting CAD risk in North-West Indian Punjabis. PLoS One. 2011;6(5):e17805.
Khoshi A, Mortazavi Y, Akbari A, Sokhanvar S, Kalantari S. Relationship between PON1L55M and Q192R gene polymorphisms and high ApoB/ApoA-I ratios. Indian J Clin Biochem. 2009;24(4):381–7.
Brophy VH, Jampsa RL, Clendenning JB, McKinstry LA, Jarvik GP, Furlong CE. Effects of 5′ regulatory-region polymorphisms on paraoxonase-gene (PON1) expression. Am J Hum Genet. 2001;68(6):1428–36.
Ruiz J, Blanché H, James RW, Garin MC, Vaisse C, Charpentier G, et al. Gln-Arg192 polymorphism of paraoxonase and coronary heart disease in type 2 diabetes. Lancet. 1995;346(8979):869–72.
Schmidt H, Schmidt R, Niederkorn K, Gradert A, Schumacher M, Watzinger N, et al. Paraoxonase PON1 Polymorphism Leu-Met54 is associated with carotid atherosclerosis: results of the Austrian stroke prevention study. Stroke. 1998;29(10):2043–8.
Ombres D, Pannitteri G, Montali A, Candeloro A, Seccareccia F, Campagna F, et al. The Gln-Arg192 polymorphism of human paraoxonase gene is not associated with coronary artery disease in Italian patients. Arterioscler Thromb Vasc Biol. 1998;18(10):1611–6.
Arca M, Ombres D, Montali A, Campagna F, Mangieri E, Tanzilli G, et al. PON1 L55M polymorphism is not a predictor of coronary atherosclerosis either alone or in combination with Q192R polymorphism in an Italian population. Eur J Clin Investig. 2002;32(1):9–15.
Andrikoula M, McDowell IF. The contribution of ApoB and ApoA1 measurements to cardiovascular risk assessment. Diabetes Obes Metab. 2008;10(4):271–8.
Fernandez ML, Webb D. The LDL to HDL cholesterol ratio as a valuable tool to evaluate coronary heart disease risk. J Am Coll Nutr. 2008;27(1):1–5.
Millán J, Pintó X, Muñoz A, Zúñiga M, Rubiés-Prat J, Pallardo LF, Masana L, et al. Lipoprotein ratios: physiological significance and clinical usefulness in cardiovascular prevention. Vasc Health Risk Manag. 2009;5:757–65.
Ko YL, Ko YS, Wang SM, Hsu LA, Chang CJ, Chu PH, et al. The Gln-Arg 191 polymorphism of the human paraoxonase gene is not associated with the risk of coronary artery disease among Chinese in Taiwan. Atherosclerosis. 1998;141(2):259–64.
Grdic M, Barisic K, Rumora L, Salamunic I, et al. Genetic frequencies of paraoxonase 1 gene polymorphisms in Croatian population. Croat Chem Acta. 2008;81(1):105–11.
Kastelein JJ, van der Steeg WA, Holme I, Gaffney M, Cater NB, Barter P, et al. Lipids, apolipoproteins, and their ratios in relation to cardiovascular events with statin treatment. Circulation. 2008;117(23):3002–9.
Serrato M, Marian AJ. A variant of human paraoxonase/arylesterase (HUMPONA) gene is a risk factor for coronary artery disease. J Clin Investig. 1995;96(6):3005–8.
Pérez-Herrera N, May-Pech C, Hernández-Ochoa I, Castro-Mañé J, Rojas-García E, Borja-Aburto VH, et al. PON1Q192R polymorphism is associated with lipid profile in Mexican men with Mayan ascendancy. Exp Mol Pathol. 2008;85(2):129–34.
Hegele RA, Brunt JH, Connelly PW. A polymorphism of the paraoxonase gene associated with variation in plasma lipoproteins in a genetic isolate. Arterioscler Thromb Vasc Biol. 1995;15(1):89–95.
Walldius G, Jungner I, Aastveit AH, Holme I, Furberg CD, Sniderman AD. The apoB/apoA-I ratio is better than the cholesterol ratios to estimate the balance between plasma proatherogenic and antiatherogenic lipoproteins and to predict coronary risk. Clin Chem Lab Med. 2004;42(12):1355–63.
Mackness B, Durrington PN, Mackness MI. The paraoxonase gene family and coronary heart disease. Curr Opin Lipidol. 2002;13(4):357–62.
Cao H, Girard-Globa A, Berthezene F, Moulin P. Paraoxonase protection of LDL against peroxidation is independent of its esterase activity towards paraoxon and is unaffected by the Q→R genetic polymorphism. J Lipid Res. 1999;40(1):133–9.
Shih DM, Xia YR, Wang XP, Miller E, Castellani LW, Subbanagounder G, et al. Combined serum paraoxonase knockout/apolipoprotein E knockout mice exhibit increased lipoprotein oxidation and atherosclerosis. J Biol Chem. 2000;275(23):17527–35.
Mackness MI, Harty D, Bhatnagar D, Winocour PH, Arrol S, Ishola M, et al. Serum paraoxonase activity in familial hypercholesterolaemia and insulin dependent diabetes mellitus. Atherosclerosis. 1991;86(2–3):193–9.
Mackness MI, Durrington PN, Mackness B. How high-density lipoprotein protects against the effects of lipid peroxidation. Curr Opin Lipidol. 2000;11(4):383–8.
Nus M, Frances F, Librelotto J, Canales A, Corella D, Sánchez-Montero JM, et al. Arylesterase activity and antioxidant status depend on PON1-Q192R and PON1-L55M polymorphisms in subjects with increased risk of cardiovascular disease consuming walnut-enriched meat. J Nutr. 2007;137(7):1783–5.
Antikainen M, Murtomaki S, Syvanne M, Pahlman R, Tahvanainen E, Jauhiainen M, et al. The Gln-Arg191 polymorphism of the human paraoxonase gene (HUMPONA) is not associated with the risk of coronary artery disease in Finns. J Clin Investig. 1996;98(4):883–5.
Durrington PN, Mackness B, Mackness MI. Paraoxonase and atherosclerosis. Arterioscler Thromb Vasc Biol. 2001;21(4):473–80.
Mackness B, Davies GK, Turkie W, Lee E, Roberts DH, Hill E, et al. Paraoxonase status in coronary heart disease: are activity and concentration more important than genotype? Arterioscler Thromb Vasc Biol. 2001;21(9):1451–7.
Aviram M, Billecke S, Sorenson R, Bisgaier C, Newton R, Rosenblat M, et al. Paraoxonase active site required for protection against LDL oxidation involves its free sulfhydryl group and is different from that required for its arylesterase/paraoxonase activities: selective action of human paraoxonase alloenzymes Q and R. Arterioscler Thromb Vasc Biol. 1998;18(10):1617–24.
Tomás M, Sentí M, Elosua R, Vila J, Sala J, Masià R, et al. Interaction between the Gln-Arg 192 variants of the paraoxonase gene and oleic acid intake as a determinant of high-density lipoprotein cholesterol and paraoxonase activity. Euro J Pharmacol. 2001;432(2–3):121–8.
Rojas-García AE, Solís-Heredia MJ, Piña-Guzmán B, Vega L, López-Carrillo L, Quintanilla-Vega B. Genetic polymorphisms and activity of PON1 in a Mexican population. Toxicol Appl Pharmacol. 2005;205(3):282–9.
Gamboa R, Zamora J, Rodríguez-Pérez JM, Fragoso JM, Cardoso G, Posadas-Romero C, et al. Distribution of paraoxonase PON1 gene polymorphisms in Mexican populations. Its role in the lipid profile. Exp Mol Pathol. 2006;80(1):85–90.
Chen J, Kumar MW, Chan M, Berkowitz G, Wetmur JG. Increased influence of genetic variation on PON1 activity in neonates. Environ Health Perspect. 2003;111(11):1403–9.
Deakin S, Leviev I, Nicaud V, Meynet MCB, Tiret L, James RW. Paraoxonase-1 L55M polymorphism is associated with an abnormal oral glucose tolerance test and differentiates high risk coronary disease families. J Clin Endocrinol Metab. 2002;87(3):1268–73.
Fanella S, Harris SB, Young TK, Hanley AJ, Zinman B, Connelly PW, et al. Association between PON1 L/M55 polymorphism and plasma lipoprotein in two Canadian aboriginal populations. Clin Chem Lab Med. 2000;38(5):413–20.
Grubisa I, Otasević P, Dimković N, Nedeljković I, Toljić B, Vucinić N. Genetic polymorphisms of paraoxonase 1 and susceptibility to atherogenesis. Srp Arh Celok Lek. 2013;141(9–10):629–33.
Cozzi L, Campolo J, Parolini M, De Maria R, Patrosso MC, Marocchi A, et al. Paraoxonase 1 L55M, Q192R and paraoxonase 2 S311C alleles in atherothrombosis. Mol Cell Biochem. 2013;374(1–2):233–8.
Acknowledgments
This study was financially supported by the Zanjan Metabolic Disorders Research Center. We also appreciate to Endocrinology and Metabolism Research Center of Shariati Hospital, Tehran.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Khoshi, A., Mortazavi, Y., Sokhanvar, S. et al. Determination of PON1 Gene Polymorphisms in Iranian Individuals with High LDL/HDL Ratios. Ind J Clin Biochem 30, 449–456 (2015). https://doi.org/10.1007/s12291-015-0479-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12291-015-0479-z