Abstract
Warfarin is the most commonly used drug for chronic prevention of thromboembolic events, it also ranks high among drugs that cause serious adverse events. The variability in dose requirements has been attributed to inter-individual differences in medical, personal, and genetic factor. Cytochrome P-450 2C9 is the principle enzyme that terminates the anticoagulant effect of warfarin by catalyzing the conversion of the pharmacologically more potent S-enantiomer to its inactive metabolites. Warfarin exerts its effect by inhibition of vitamin K epoxide reductase. This protein is encoded by vitamin K epoxide reductase complex subunit 1 gene (VKORC1). The current study aimed to investigate the pharmacogenetic effect of CYP2C9 and VKORC1 gene polymorphisms on the patients response to warfarin. One hundred cases starting warfarin treatment and 20 healthy controls were enrolled. The mean daily dose of warfarin was calculated from patient’s medical records. For each patient, less than 10 % variability in warfarin dose and a target international normalized ratio (INR) within the therapeutic target range were required for at least 3 months for one of the following indications (deep vein thrombosis, pulmonary embolism, cerebrovascular stroke and myocardial infarction) prior to inclusion in the study. Tetraprimer amplification refractory mutation system PCR was performed to determine CYP2C9*2, CYP2C9*3, and the VKORC1 1639 G > A genetic polymorphisms. Plasma warfarin determination was performed using rapid fluorometric assay. Plasma warfarin concentration ranged from 2.19 to 10.98 μg/ml with a median 3.52 μg/ml. Supratherpeutic INR was observed in 11 % of the cases. Thromboembolic complications occurred in 7 % of the cases and 8 % of cases experienced major bleeding. High maintenance dose (>7 mg/day) was associated with the combined non VKORC1*2 and homozygous wild type CYP2C9 (CYP2C9*1*1) alleles, while low maintenance dose was associated with the Variant (AG + AA)/Wild (*1/*1). (p value <0.001). CYP2C9 variant was a risk factor for supratherapeutic INR in the multivariate logistic regression model. Thromboembolic complication and incidence of supratherapeutic INR were observed in patients carrying combined VKORC1 Variant (AG + AA) and CYP2C9 Variant (*2/*3). Data from our study suggest that together with clinical factors, VKORC1 and CYP2C9 polymorphisms are important contributors to warfarin dosing and may help predict adverse effects in Egyptian patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Warfarin is the most widely prescribed oral anticoagulant. It’s a Vitamin K antagonist, which have been the main agents used for prolonged anticoagulation therapy to prevent and treat thromboembolic disease [1, 2]. Warfarin is a vitamin K antagonist that produce their anticoagulant effect by interfering with the cyclic interconversion of vitamin K and its 2,3 epoxide (vitamin K epoxide) [3]. Vitamin K is a cofactor for the posttranslational carboxylation of glutamate residues to gamma carboxyglutamates on the N-terminal regions of vitamin K-dependent proteins [4, 5].
When prescribing warfarin, notable challenges face the clinicians to determine the optimal and safest dosage, which needs to be individualized for each patient. One of the most important and cornerstone actions to improve the management of warfarin therapy, is the introduction of the International Normalized Ratio (INR) in the early 1990s, where the effectiveness and safety of warfarin is critically dependent on maintaining the prothrombin time, expressed as the international normalized ratio (INR), within the therapeutic range [3]. Which allowed a more consistent and universal means to measure the intensity of anticoagulation.
The mysteries behind the complexity of warfarin dose–response pushed the investigators worldwide to search for clues to unravel. There are multiple influencing factors that affect the individual response to warfarin therapy including age, body size, vitamin K intake, comorbidities and co-medications and this creates more challenges in grouping approaches to anticoagulation management [6, 7]. The ability to measure or predetermine patient-specific variables can create an opportunity to initiate or modify dosing that minimizes over anticoagulation and bleeding, as well as under anticoagulation and thrombosis.
One of the most important and influential recent advancement is the recognition of genotype differences that influence patients’ response to warfarin. Single nucleotide polymorphisms (SNPs) have been identified that clearly influence warfarin metabolism and sensitivity [8, 9]. This includes SNP variants of the gene encoding cytochrome P-450 isoenzyme 2C9 (CYP2C9)-cytochrome P450, family 2, subfamily C, polypeptide 9, which is an isoenzyme that is primarily involved in the metabolism of the more potent S-isomer of warfarin [10, 11]. Moreover, polymorphism of Vitamin K epoxide reductase gene VKORC1 is thought to alter sensitivity to warfarin and is being investigated for its association with patients’ warfarin-response [12–15]. For this reason it is now suggested that initiation of warfarin guided by pharmacogenetics would improve clinical outcome.
Patients and Methods
Patients
This study was carried out on 100 cases starting warfarin treatment. The patients were selected from the Hematology department, Medical Research Institute Hospital, Alexandria University. The median age was 49 years. Males represented 56 % of the cases while 44 % were females. The body mass index of the patients ranged from 19.81 to 42.44.
Informed consent was obtained from each subject or patient prior to participating in this work. The mean daily dose of warfarin (mg) was calculated from patient’s medical records. For each patient, <10 % variability in warfarin dose and a target international normalized ratio (INR) within the therapeutic target range [2, 3] and for patients having valve replacement and recurrent pulmonary embolism target was [3, 4], were required for at least 3 months for one of the following indications (deep vein thrombosis, pulmonary embolism, cerebrovascular stroke and Myocardial infarction) prior to inclusion in the study. Patients were stratified into three groups based on their maintenance dose (>2.5 mg is the low dose group, ≤2.5 to 7 mg is the standard dose group and >7 is the high dose group). The Exclusion criteria were comorbid conditions (liver and/or kidney diseases), or patients requiring chronic therapy on drugs that can influence hepatic cytochrome enzymes.
Methods
The patients were subjected to complete clinical evaluation including detailed genetic history taking and Complete clinical examination. Also, all the included cases were subjected to laboratory studies involving complete blood count, INR measurement, liver and kidney function tests. Molecular genetic test was performed on EDTA anticoagulated venous blood samples. DNA was isolated and purified from peripheral blood using QIAmp minikit (Qiagen, Hilden, Germany).
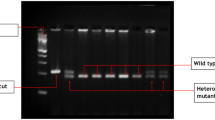
Three single-nucleotide polymorphisms (SNPs) affecting warfarin sensitivity, which are CYP2C9 (cytochrome P450, family 2, subfamily C, polypeptide 9) *2, CYP2C9 *3, and the VKORC1 (vitamin K epoxide reductase complex 1)-1639G > A genetic polymorphism, were assayed. The genotypes were determined with the tetraprimer amplification refractory mutation system (T-ARMS) using specific outer and inner primers in 3 PCR tubes for each patient. Primer sequences, reaction components and cyclic conditions were carried out as previously reported by Poe et al. [16]. The assay was developed with Primus 25 thermal cycler (Peqlab, Germany, CAT No: 95-4002). PCR products were visualized by submarine agarose gel electrophoresis to determine the corresponding genotype (Fig. 1).
Agarose gel electrophoresis for 3 PCR products for single patient. Lane 1 bands (235 and 133 bp) corresponding to VKORC1 AA genotype. Lane 2 bands (154 + 206 + 307 bp) corresponds to CYP2C9 *1*2. Lane 3 bands (110 + 286 + 340 bp) corresponds to CYP2C9 *1*3. Lane M represents 100 bp ladder base pair marker
Plasma warfarin concentration was assayed on EDTA anticoagulated samples from all patients. The plasma was separated by centrifugation of blood samples at 3000 rpm for 10 min and stored at −20 °C until analysis was done. Warfarin was analyzed using rapid fluoremetric assay as described by Milton corn and Robert Berberish [17] with slight modification involving the use of 1 ml of 1 N HCl instead of 0.05 ml of 0.1 N HCl as described by the paper to decrease the fluorescence.
Results
The demographic data of patients, the indications for warfarin therapy and frequencies of concomitant diseases were described in Table 1. Homozygotes of the VKORC1 1639 G > A polymorphism (AA) were the least frequent (21 %), whereas the AG and GG genotypes represented 47 and 32 % of patients. The frequencies of A and G alleles were 0.68 and 0.79, respectively. On the other hand, the CYP2C9 wild type (*1/*1) homozygotes were the most frequent (56 %), while heterozygotes (*1/*2, 1*/3*) and homozygotes (*2/*2, 2*/3*, 3*/3*) were 33 and 11 % of patients, respectively (Table 1).
Plasma warfarin concentration ranged from 2.19 to 10.98 µg/ml with a median 3.52 µg/ml. This warfarin level was significantly higher in patients on high daily maintenance dose compared to standard and low dose groups (p value <0.001).
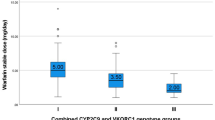
Patients with AA genotype required lower warfarin maintenance doses compared to other VKORC1 genotypes (p < 0.001). No one of the studied patients with the GG genotype received low warfarin maintenance dose and on the contrary, no single carrier of the A allele received high warfarin maintenance dose. The CYP2C9 genotypes did not affect the warfarin maintenance dose (Table 2) and (Fig. 2). Patients with the wild (GG) VKORC1 genotype and patients with the wild (*1/*1) CYP2C9 genotype reached the target INR upon warfarinization in a shorter duration when compared to other genotypes (p value <0.001 for both) (Table 3).
The risk of suffering bleeding complications was significantly lower among patients with homozygous wild CYP2C9 genotype (*1/*1) compared to other studied variants (p < 0.02). On the other hand, this risk was not affected by VKORC1 genotypes. The risk of rethrombosis was significantly lower among patients with wild genotypes for both VKORC1 and CYP2C9 (p = 0.093 and 0.042 respectively). None of the studied patients with the VKORC1 wild genotype experienced thromboembolic complications. Patients with CYP2C9 variant were at higher risk of experiencing supratherapeutic INR compared to those with the wild type (p < 0.001) (95 % CI 1.982–132.051) (Table 4).
On studying the combined effect of the variants of both genes, patients with the VKORC1 variants (AG + GG) and CYP2C9 wild genotype required a significantly lower maintenance dose compared to other combination of genotypes (p < 0.001) (Table 2).
Patients with combined variant CYP2C9 and VKORC1 genotypes experienced higher risk of supratherapeutic INR (p = 0.011) (95 % CI 2.344–180.548). However, these combined genotypes did not increase the risk of bleeding while it significantly increased the risk of thromboembolic complications (p = 0.004) (95 % CI 13.389–229.37) (Table 4).
Discussion
Warfarin has been approved as the most commonly used oral anticoagulant for the treatment and prevention of thromboembolic events since 1950s [14]. Even with available alternatives (thrombin and factor Xa inhibitors) presently [18–21], several issues related to cost, indication, antidote, and physicians experience still favor an extensive and ongoing use of warfarin. However, several conditions largely limited the application of warfarin including the narrow therapeutic window, the inconvenient monitoring, and especially the risk for hemorrhagic and thromboembolic complications.
Individual responses to anticoagulation using warfarin are now believed a reflection of a number of clinical and genetic factors. The contributory effect of pharmacogenetics is increasingly investigated, however, up till now genetic testing and personalized medicine haven’t been adequately adopted into clinical practice. This study has examined the impact of CYP2C9 and VKORC1 polymorphisms on both laboratory and clinical outcome variables in Egyptian population. Among the studied 100 patients receiving warfarin treatment, CYP2C9 *1/*1 and VKORC1 (AG) were the most prevalent genotypes. A study performed by Eldin et al. [22] reported similar findings among Egyptians.
In concordance with multiple previously published studies (Bazan et al. [16], Lund et al. [11], Gage et al. [23], Cho et al. [24], Namazi et al. [25], Kimura et al. [26], Wells et al. [27], Moreau et al. [28], this study showed an association between the presence of the variant A allele of the VKORC1 gene and the lower warfarin maintenance daily dose. This is explained by the production of less VKORC1 in individuals with the A allele compared to those lacking it. Hence, lower warfarin doses are needed to inhibit VKORC1 and to produce an anticoagulant effect in carriers of the A allele.
This study did not find any association between the warfarin maintenance dose and the CYP2C9 variants. This comes in agreement with the reports of Bazan et al. [6] and Lund et al. [11]. However, Zohir et al. [29] reported border line significance enforced by the work of Scordo et al. [30] and Herman et al. [31]. This contradiction can be attributed to differences in the characteristics of the studied patients in these reports as regards number of cases, median age and exclusion criteria. However, in the present work, the lack of such an association might provide a justification for further evaluation of warfarin hepatic metabolism among Egyptians.
In the present study, higher daily warfarin maintenance dose was associated with higher plasma warfarin concentrations (p = < 0.001), this result is in agreement with the study of Lombardi et al. [32]. With respect to the impact of VKORC1 or CYP2C9 variants on plasma warfarin concentration, this study showed that carriers of the CYP2C9 variant either in homozygous or heterozygous state in each dose group had significantly higher plasma warfarin concentration compared to those with wild genotype. These results are in agreement with a study of Herman et al. [31] who found significant increase in the plasma warfarin concentration with those carrying CYP2C9 wild genotype and concluded that CYP2C9*2 and CYP2C9*3 alleles were associated with a reduction in S-warfarin clearance.
The present study revealed that the presence of either variant CYP2C9 or variant VKORC1 genotypes was significantly associated with higher risk of experiencing supratherapeutic INR. This is in accordance with the reports of Krajciova et al. [33], Schwarz et al. [34], Higashi et al. [35] and Ferder et al. [36]. However, Bazan et al. [6] reported lack of such an association in their cohort. This may be attributed to relative smaller sample size of Bazan et al. [6] study cohort.
Our study suggests that CYP2C9*2 and CYP2C9*3 increase the risk of bleeding complication, and this risk increased in the homozygous state. This may be linked to increased risk of supratherapeutic INR in these carriers. These findings were in agreement with multiple previously published studies (Higashi et al. [35], Bazan et al. [6], Aithal et al. [37], Ogg et al. [38], Limdi et al. [39] and Ferder et al. [36]. On the other hand, Roth et al. [40] reported that CYP2C9 *2/*3 variants did not confer increased major bleeding risk. Moreover, the study population of Roth et al. [40] was of the same ethnic origin like those of Ferder et al. [36] and Higashi et al. [35]. A finding that percludes the role of ethinity in explanation of this contradiction. However, the possibility of the presence of other genetic polymorphic variants affecting the response to warfarin could be a candidate explanation waiting for further evaluation. As regards VKORC1 variant, it didn’t significantly increase the risk of bleeding which is in concordance with studies of Roth et al. [40] and Bazan et al. [6]. This might be explained by the probable clinician response towards the supratherapeutic INR by lowering the warfarin dose before bleeding occurs. This explanation might be extended to justify our finding regarding the risk of recurrent thromboembolic events while on warfarin therapy. We found, in this study, a significant increase in the risk of thromboembolic complication in carriers of both CYP2C9 and VKORC1 variants compared to wild genotypes. These findings are in concordance with Bazan et al. [6] and Lund et al. [11]. This highlights the need for genotype based individualized approach for dealing with these patients with supratherapeutic INR receiving warfarin therapy.
In agreement with Bazan et al. [6], Schwarz et al. [34], Limdi et al. [39] and Takeuchi et al. [41], our study revealed that the presence of either VKORC1 or CYP2C9 variants was associated with a significantly more time to achieve stable dosing and reach therapeutic INR when compared to the wild type. On the other hand, Lund et al. [11] found out that patients with the VKORC1 AA genotype reached an INR of 2 more quickly when compared independently to the VKORC1 GA and GG genotypes and they concluded that warfarin-sensitizing SNPs reduced the time taken to reach therapeutic INR. Same explanation could be through what we have reported above concerning the studied genotypes.
The present work demonstrated the effect of the presence of combined genetic variants on the risk of developing adverse events associated with warfarin therapy. All patients with variant CYP2C9 and variant VKORC1 genotypes experienced supratherapeutic INR. However, these combined genotypes didn’t significantly increase the bleeding risk. This may be explained through the clinician’s decision of warfarin dose reduction for these patients before the occurrence of bleeding episode.
In line with this explanation, our study revealed that the risk of rethrombosis was high in these patients. This highlights the need for genotype-based individualized dosing to ensure reaching target INR on a stabilized warfarin dose in short time. Up to our best knowledge, no previous studies investigated the influence of combined genetic variants on various clinical outcomes of warfarin therapy.
The present work explored the value of VKORC1 and CYP2C9 genotyping as an important determinant of individualized warfarin dosing and as a predictor of warfarin-related adverse effects in Egyptian patients. These genotypes were revealed to affect warfarin dose, time to reach target INR and the risk of complications.
Change history
19 March 2018
The aim of this erratum is to acknowledge that the original version of this article was inadvertently published with incorrect author name and affiliation for a co-author.
References
Wang L, McLeod HL, Weinshilboum RM (2011) Genomics and drug response. N Engl J Med 364(12):1144–1153
Lee AYY, Peterson EA (2013) Treatment of cancer-associated thrombosis. Blood 122(14):2310–2317
De Caterina R, Husted S, Wallentin L, Andreotti F, Arnesen H, Bachmann F et al (2013) Vitamin K antagonists in heart disease: current status and perspectives (Section III). Thromb Haemost 110(6):1087–1107
Azuma K, Ouchi Y, Inoue S (2014) Vitamin K: novel molecular mechanisms of action and its roles in osteoporosis. Geriatr Gerontol Int 14(1):1–7
Falcone TD, Kim SS, Cortazzo MH (2011) Vitamin K: fracture prevention and beyond. PM&R 3(6):S82–S87
Bazan N, Sabry N, Rizk A, Mokhtar S, Badary O (2014) Factors affecting warfarin dose requirements and quality of anticoagulation in adult Egyptian patients: role of gene polymorphism. Ir J Med Sci 183(2):161–172
Jorgensen AL, FitzGerald RJ, Oyee J, Pirmohamed M, Williamson PR (2012) Influence of CYP2C9 and VKORC1 on patient response to warfarin: a systematic review and meta-analysis. PLoS ONE 7(8):e44064
Eckman MH, Rosand J, Greenberg SM, Gage BF (2009) Cost-effectiveness of using pharmacogenetic information in warfarin dosing for patients with nonvalvular atrial fibrillation. Ann Intern Med 150(2):73–83
Yin T, Miyata T (2007) Warfarin dose and the pharmacogenomics of <i> CYP2C9 </i> and <i> VKORC1 </i>—rationale and perspectives. Thromb Res 120(1):1–10
Wei M, Ye F, Xie D, Zhu Y, Zhu J, Tao Y et al (2012) A new algorithm to predict warfarin dose from polymorphisms of CYP4F2, CYP2C9 and VKORC1 and clinical variables: derivation in Han Chinese patients with non valvular atrial fibrillation. Thromb Haemost 107(6):1083
Lund K, Gaffney D, Spooner R, Etherington AM, Tansey P, Tait RC (2012) Polymorphisms in VKORC1 have more impact than CYP2C9 polymorphisms on early warfarin international normalized ratio control and bleeding rates. Br J Haematol 158(2):256–261
D’Andrea G, D’Ambrosio RL, Di Perna P, Chetta M, Santacroce R, Brancaccio V et al (2005) A polymorphism in the VKORC1 gene is associated with an interindividual variability in the dose-anticoagulant effect of warfarin. Blood 105(2):645–649
Wang D, Chen H, Momary KM, Cavallari LH, Johnson JA, Sadée W (2008) Regulatory polymorphism in vitamin K epoxide reductase complex subunit 1 (VKORC1) affects gene expression and warfarin dose requirement. Blood 112(4):1013–1021
Johnson J, Gong L, Whirl-Carrillo M, Gage B, Scott S, Stein C et al (2011) Clinical pharmacogenetics implementation consortium guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clin Pharmacol Ther 90(4):625–629
Miyagata Y, Nakai K, Sugiyama Y (2011) Clinical significance of combined CYP2C9 and VKORC1 genotypes in Japanese patients requiring warfarin. Int Heart J 52(1):44–49
Poe BL, Haverstick DM, Landers JP (2012) Warfarin genotyping in a single PCR reaction for microchip electrophoresis. Clin Chem 58(4):725–731
Corn M, Berberich R (1967) Rapid fluorometric assay for plasma warfarin. Clin Chem 13(2):126–131
Connolly SJ, Ezekowitz MD, Yusuf S, Eikelboom J, Oldgren J, Parekh A et al (2009) Dabigatran versus warfarin in patients with atrial fibrillation. N Engl J Med 361(12):1139–1151
Patel MR, Mahaffey KW, Garg J, Pan G, Singer DE, Hacke W et al (2011) Rivaroxaban versus warfarin in nonvalvular atrial fibrillation. N Engl J Med 365(10):883–891
Granger CB, Alexander JH, McMurray JJ, Lopes RD, Hylek EM, Hanna M et al (2011) Apixaban versus warfarin in patients with atrial fibrillation. N Engl J Med 365(11):981–992
Yang J, Chen Y, Li X, Wei X, Chen X, Zhang L et al (2013) Influence of CYP2C9 and VKORC1 genotypes on the risk of hemorrhagic complications in warfarin-treated patients: a systematic review and meta-analysis. Int J Cardiol 168(4):4234–4243
El Din M, Amin D, Ragab S, Ashour E, Mohamed M, Mohamed A (2012) Frequency of VKORC1 (C1173T) and CYP2C9 genetic polymorphisms in Egyptians and their influence on warfarin maintenance dose: proposal for a new dosing regimen. Int J Lab Hematol 34(5):517–524
Gage BF, Lesko LJ (2008) Pharmacogenetics of warfarin: regulatory, scientific, and clinical issues. J Thromb Thrombolysis 25(1):45–51
Cho H-J, Sohn K-H, Park H-M, Lee K-H, Choi B, Kim S et al (2007) Factors affecting the interindividual variability of warfarin dose requirement in adult Korean patients. Pharmacogenomics 8(4):329–337
Namazi S, Azarpira N, Hendijani F, Khorshid MB, Vessal G, Mehdipour AR (2010) The impact of genetic polymorphisms and patient characteristics on warfarin dose requirements: a cross-sectional study in Iran. Clin Ther 32(6):1050–1060
Kimura R, Miyashita K, Kokubo Y, Akaiwa Y, Otsubo R, Nagatsuka K et al (2007) Genotypes of vitamin K epoxide reductase, γ-glutamyl carboxylase, and cytochrome P450 2C9 as determinants of daily warfarin dose in Japanese patients. Thromb Res 120(2):181–186
Wells P, Majeed H, Kassem S, Langlois N, Gin B, Clermont J et al (2010) A regression model to predict warfarin dose from clinical variables and polymorphisms in CYP2C9, CYP4F2, and VKORC1: derivation in a sample with predominantly a history of venous thromboembolism. Thromb Res 125(6):e259–e264
Moreau C, Bajolle F, Siguret V, Lasne D, Golmard J-L, Elie C et al (2012) Vitamin K antagonists in children with heart disease: height and VKORC1 genotype are the main determinants of the warfarin dose requirement. Blood 119:861–867
Zohir N, Afifi R, Ahmed A, Aly Z, Elsobekey M, Kareem H et al (2013) Role of CYP2C9, VKORC1 and calumenin genotypes in monitoring warfarin therapy: an Egyptian study. Maced J Med Sci 6(4):414–420
Scordo MG, Pengo V, Spina E, Dahl ML, Gusella M, Padrini R (2002) Influence of CYP2C9 and CYP2C19 genetic polymorphisms on warfarin maintenance dose and metabolic clearance[ast]. Clin Pharmacol Ther 72(6):702–710
Herman D, Locatelli I, Grabnar I, Peternel P, Stegnar M, Mrhar A et al (2005) Influence of CYP2C9 polymorphisms, demographic factors and concomitant drug therapy on warfarin metabolism and maintenance dose. Pharmacogenomics J 5(3):193–202
Lombardi R, Chantarangkul V, Cattaneo M, Tripodi A (2003) Measurement of warfarin in plasma by high performance liquid chromatography (HPLC) and its correlation with the international normalized ratio. Thromb Res 111(4):281–284
Krajciova L, Deziova L, Petrovic R, Luha J, Turcani P, Chandoga J (2013) Frequencies of polymorphisms in CYP2C9 and VKORC1 genes influencing warfarin metabolism in Slovak population: implication for clinical practice. Bratisl Lek Listy 115(9):563–568
Schwarz UI, Ritchie MD, Bradford Y, Li C, Dudek SM, Frye-Anderson A et al (2008) Genetic determinants of response to warfarin during initial anticoagulation. N Engl J Med 358(10):999–1008
Higashi MK, Veenstra DL, Kondo LM, Wittkowsky AK, Srinouanprachanh SL, Farin FM et al (2002) Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA 287(13):1690–1698
Ferder NS, Eby CS, Deych E, Harris JK, Ridker PM, Milligan PE et al (2010) Ability of VKORC1 and CYP2C9 to predict therapeutic warfarin dose during the initial weeks of therapy. J Thromb Haemost 8(1):95–100
Aithal GP, Day CP, Kesteven PJL, Daly AK (1999) Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet 353(9154):717–719
Ogg MS, Brennan P, Meade T, Humphries SE (1999) CYP2C9* 3 allelic variant and bleeding complications. Lancet 354(9184):1124
Limdi N, McGwin G, Goldstein J, Beasley T, Arnett D, Adler B et al (2008) Influence of CYP2C9 and VKORC1 1173C/T genotype on the risk of hemorrhagic complications in African-American and European-American patients on warfarin. Clin Pharmacol Ther 83(2):312–321
Roth JA, Boudreau D, Fujii MM, Farin FM, Rettie AE, Thummel KE et al (2014) Genetic risk factors for major bleeding in patients treated with warfarin in a community setting. Clin Pharmacol Ther 95(6):636–643
Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N et al (2009) A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet 5(3):e1000433
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declare that they have no conflict of interests.
Ethical Standards
All procedures performed were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Rights and permissions
About this article
Cite this article
Bedewy, A.M.L., Showeta, S., Mostafa, M.H. et al. The Influence of CYP2C9 and VKORC1 Gene Polymorphisms on the Response to Warfarin in Egyptians. Indian J Hematol Blood Transfus 34, 328–336 (2018). https://doi.org/10.1007/s12288-016-0725-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12288-016-0725-4