Abstract
Background
Upregulated gene 4 (URG4) is a recently described oncogene that upregulates cell proliferation. Its overexpression has been identified in many malignancies, and it is thought to be related to tumour progression, angiogenesis, metastasis and the recurrence of many cancers. This is the first study to show its expression in breast cancer patients and its association with clinicopathological characteristics in these patients.
Methods
Fifty invasive ductal breast carcinoma cases and 25 control cases were included in the study. Tumourous tissues and control tissues were assessed molecularly for quantification of mRNA expression of URG4 and immunohistochemically for protein expression of URG4.
Results
The mean ages of the patients and controls were 54.3 ± 11.3 and 38.9 ± 9.7 years, respectively. The expression levels of URG4 mRNA in tumour tissues were higher compared to control breast tissues (p = 0.023). An immunohistochemical assessment suggested that URG4 is strongly expressed in normal breast tissues and lower-grade (grades I and II) ductal carcinomas of the breast, but it is weakly expressed in high-grade (grade III) ductal breast carcinomas. Additionally, the immunohistochemical and molecular expression results of URG4 were relevant to most prognostic parameters (tumour size, oestrogen and progesterone receptor status, HER2 status and Ki67 proliferative index) for breast cancer. However, unlike the immunohistochemical studies, the molecular studies revealed that there was no significant difference in URG4 expression for different grades of tumour tissues.
Conclusion
The literature data suggest that URG4 overexpression is associated with poor prognosis in many types of cancer. Conversely, our results in breast cancer specimens indicate that URG4 overexpression in breast ductal carcinomas is significantly associated with good prognostic parameters. Nevertheless, these preliminary findings should be confirmed by further studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Upregulated gene 4 (URG4), which is also known as upregulator of cell proliferation (URGCP), is an oncogene that is located on chromosome 7p13 with an mRNA of 3.607 kb, which contributes to the progression of several tumours, including hepatocellular cancer [1], ovarian cancer [2], gastric cancer [3], bladder cancer [4], glioblastoma [5], non-small cell lung cancer [6], medullary thyroid cancer [7], neuroblastoma [8], prostate cancer [9] and leukaemia [10]. Previous studies have shown that URG4 activates Akt signalling pathways, inhibits the cell-cycle inhibitors p27 and p21 and facilitates angiogenesis by NF-κB activation [11]. Overexpression of URG4 not only results in tumour progression but also in metastasis and recurrence during the disease course [3]. To the best of our knowledge, the presence of URG4 in breast cancer has not been studied before. The aims of this study are to determine UGR4 expression immunohistochemically, including an evaluation of its associations with the clinical and pathological characteristics of breast cancer patients, and to compare the molecular gene expressions, via real-time polymerase chain reaction (PCR) analyses, of breast cancer versus normal breast tissues.
Materials and methods
This study included 50 females who had undergone radical mastectomy and axilla dissection procedures for breast cancer and had been diagnosed with invasive ductal carcinoma via pathological assessments between 2013 and 2017 at Balıkesir University Hospital and 25 females who had undergone mammoplasty. None of the patients in the study had undergone either radiotherapy, chemotherapy or hormonal therapy prior to the surgery.
The histopathological parameters were evaluated in paraffin blocks of patients that were stained with haematoxylin–eosin. The tumours were graded based on the Nottingham modification of the Bloom–Richardson grading system.
The relationships between URG4 expression and the prognostic parameters [age, tumour grade, tumour size, presence of lymph node involvement, oestrogen (ER) and progesterone (PR) receptor status, HER2 status, Ki67 proliferative index, breast cancer subtype (Luminal A, Luminal B, triple-negative/basal-like and HER2-over) and Nottingham Prognostic Index (NPI)] were evaluated.
This study included two methodological stages:
-
Molecular demonstration of the gene expression
-
Immunohistochemical demonstration of the protein expression of URG4 in paraffin blocks
Gene expression
The RNAs were isolated from the paraffin blocks of 25 breast cancer patients and 25 healthy controls (tumour-free, normal breast tissues from 25 mammoplasty patients). The total RNA was extracted using RNeasy FFPE (QIAGEN), per the manufacturer’s protocol. Complementary DNA isolation from RNA (Gene All, Hyperscript first-strand synthesis kit, Cat no.: 601-005, Lot no.: FS015B04002) was performed in two steps. Applied Biosystem Step One Plus equipment was used for the Real-Time PCR (GeneAllSybr Green Master Mix, Cat no.: 801-520, Lot no.: QP116G25001) analyses. Samples were analysed three times, and reactions were established using both Actin beta (ACTB) and their own genes. Gene expression levels were quantified using 7500 Fast Real-Time Sequence detection system Software (Applied Biosystems, Foster City, CA, USA). Gene expression was defined based on the threshold cycle (Ct), and ACTB was used as a reference gene that acts as an internal reference to normalise the RNA expression, which was calculated as 2−ΔΔCT.
The sequences of the primers were as follows:
ACTB forward: 5′ CCTGACTGACTACCTCATGAAGATCCTC 3′.
Reverse: 5′ CGTAGCACAGCTTCTCCTTAATGTCAC 3′ (103 bp).
URG-4 forward: 5′ GATACGCCAGTGAACCCCTTAGAC 3′.
Reverse: 5′ CAGCAGAAATGTATGGTAGTGGTTCTC 3′ (156 bp).
Immunohistochemistry
For the immunohistochemistry assessments, the paraffin blocks from 50 patients that included both the tumour and surrounding normal tissues in the same section were selected. In addition, 25 healthy controls (tumour-free normal breast tissues from 25 mammoplasty patients) were included in the immunohistochemistry analyses. Paraffin-embedded specimens were cut into 4-µm-thick sections and baked at 60 °C. They were deparaffinised in xylene and hydrated in alcohol. The samples were heated in a microwave oven with EDTA buffer for antigenic retrieval. Three percent hydrogen peroxidase was applied to eliminate any endogenous peroxidase activity. After administration of Phosphate Buffer Saline (PBS), a protein block (TA-125-PBQ Thermo Scientific) was applied to eliminate non-specific binding. Then, Primary Antibody (diluted 1:50; URG4 Abcam: ab103323) was dripped onto the sections and incubated for 60 min at room temperature. Following a wash with PBS, Amplifier (TL-125-QPH Thermo Scientific) was administered for 20 min at room temperature. After another PBS wash, HRP Polymer (TL-125-QPH Thermo Scientific) was administered for 30 min at room temperature. Following a third PBS wash, DAB Chromogen (TL-125-HD Thermo Scientific) was administered for 3 min. The sections were then washed with distilled water and stained with Mayer’s haematoxylin. URG4 was mainly expressed in the cytoplasm of breast epithelial cells and partly in the nuclei.
Because all the tumour tissue had been stained, the staining was only evaluated per its intensity in the cytoplasm, which was done by two independent pathologists. Evaluators were blinded to the clinical characteristics and outcomes of the patients. The intensity was graded from 1 to 3 as weak staining (light yellow), moderate staining (yellow–brown) and strong staining (brown), respectively.
Statistical analyses
The Number Cruncher Statistical System 2007 (Kaysville, Utah, USA) programme was used for statistical analyses. Descriptive statistical methods (frequency, rate, minimum and maximum) as well as quantitative data were used to evaluate the data. The Pearson Chi square test, Fisher–Freeman–Halton’s test and Fisher’s exact test were used to compare qualitative data. The Spearman’s correlation analysis was used to evaluate inter-variable relationships. In the molecular studies, for the statistical evaluation of the one-way ANOVA, the Student’s T test was used. Data are presented as mean percent ± SD. A p level of < 0.05 was accepted as statistically significant.
Results
In total, 75 patients (50 breast cancer patients and 25 control cases with tumour-free, normal breast tissues) were included in the analyses. URG4 immunohistochemistry staining was evaluated in all samples, and gene expression was evaluated in 25 selected breast cancer tissues as well as all control tissues. The mean ages of the patients and controls were 54.3 ± 11.3 years and 38.9 ± 9.7 years, respectively.
Immunohistochemistry assessments
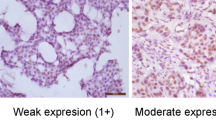
The URG4 staining index of all control tissues and non-tumourous breast tissue around tumours were scored on the highest score 3. For the breast cancer samples, the URG4 staining index comparisons between clinicopathological variables are presented in Table 1. In grade III tumour cells, the expression of URG4 was lower, whereas it was higher in grades I and II tumour cells. The results found a negative relationship between URG4 expression and the degree of tumour differentiation (p = 0.001). Figure 1 presents the strong expression of URG4 in normal breast tissue and lower-grade (grades I and II) ductal carcinoma, while it shows a weak expression in high-grade (grade III) ductal carcinoma.
a Strong URG4 expression in the normal breast tissue (URG4, × 400). b Strong URG4 expression in the normal breast epithelium at the upper left corner and in the grade I ductal breast carcinoma epithelium (URG4, × 400). c Strong URG4 expression in the grade II ductal breast carcinoma epithelium (URG4, × 400). d Weak URG4 expression in the grade III ductal breast carcinoma epithelium (URG4, × 400)
URG4 expression was also related to tumour size ≤ 2 cm (p = 0.019), ER positivity (p = 0.002), PR positivity (p = 0.003) and HER2 negativity (p = 0.001). Higher URG4 expression was found in luminal A tumours. There was no relationship between URG4 expression and age (p = 0.208), lymph node metastases (p = 0.368) and the Ki67 proliferative index (p = 0.294). The Ki67 proliferative index was also statistically evaluated by the Spearman’s correlation test by dividing it into two categories: > 15% and ≤ 15%. As a result, a negative correlation was observed with respect to the variables. Accordingly, expression of URG4 was found to be negatively correlated with the Ki67 proliferative index (r: − 0.321; p = 0.023) and the Nottingham prognostic index (r: − 0.506; p = 0.001). The correlations between the URG4 staining index, the Ki67 proliferative index and the Nottingham prognostic index are presented in Table 2.
Gene expression assessment
The quantitative distribution of the URG4 expression among the prognostic parameters is presented in Fig. 2. Quantitative gene expression analyses by real-time PCR studies revealed that the expression levels of URG4 mRNA in tumour tissues were higher compared to control breast tissues (p = 0.023), but there was no significant difference in URG4 expression for different grades of tumour tissues (grades I and II: p = 0.313; grade III: p = 0.607) when compared to the control.
The URG4 mRNA level was measured by qRT-PCR and normalised to ACTB levels. a Expression levels of URG4 mRNA in tumour tissues were higher compared to control breast tissues (n = 25, p = 0.023). b The expression levels of the URG4 mRNA were higher in smaller size (tumour size ≤ 2 cm) (p = 0.03) tumours. c The expression levels of URG4 mRNA were higher in negative lymph node metastasis (p = 0.016) (lymph node none *p = 0.016, n = 11; lymph node present p = 0.53, n = 14). d There was no significant difference in URG4 expression for different grades of tumour tissues (grades I and II: p = 0.313; grade III: p = 0.607). e The expression levels of the URG4 mRNA Ki67 proliferative index were not statistically significant (Ki67 > 15%: p = 0.243, n = 6; Ki67 ≤ 15%: p = 0.122, n = 19). f The expression levels of the URG4 mRNA were higher in ER(+) (p = 0.006) tumours (ER positive *p = 0.006, n = 19; ER negative p = 0.113, n = 6). g The expression levels of the URG4 mRNA were higher in PR(+) (p = 0.04) (progesterone positive *p = 0.04, n = 15; progesterone negative p = 0.406, n = 10). h In HER2 negative patients, URG4 expression increased statistically significantly (HER2 negative *p = 0.028, n = 17; HER2 positive p = 0.55, n = 8)
The expression levels of URG4 mRNA were higher in smaller size (tumour size ≤ 2 cm) (p = 0.03) negative lymph node metastasis (p = 0.016), ER (+) (p = 0.006), PR (+) (p = 0.04) and HER2(−) (p = 0.028) tumours. Moreover, the expression of URG4 mRNA was higher in the Ki67 proliferative index ≤ 15% subgroup, whereas it was lower in the Ki67 proliferative index > 15% subgroup; however, there was no statistical significance (Ki67 < 15%: p = 0.243; Ki67 ≥ 15%: p = 0.122). Similarly, the expression of URG4 mRNA was higher in older patients, but there was no statistical significance (age > 50: p = 0.079; age ≤ 50: p = 0.153).
Discussion
The role of the URG4 gene in the progression of several cancers, while discussed in previous studies, has not been investigated in breast cancer to date. Notably, our immunohistochemical and molecular expression results for URG4 were relevant to most prognostic parameters (tumour size, ER and PR receptor status, HER2 status and the Ki67 proliferative index) for breast cancer. Our immunohistochemical assessment revealed that URG4 was strongly expressed in normal breast tissues and lower-grade (grades I and II) ductal carcinomas of the breast, but it was weakly expressed in high-grade (grade III) ductal breast carcinomas.
Our results corresponded to the study investigating the relationship between tumour differentiation and URG4 expression in gastric tumours. This study stated that poorly differentiated gastric tumours have a lower immunohistochemical expression than well-differentiated tumours [3]. However, many other cancers show opposite trends to our results. For example, Yu et al. [11] reported that increased URG4 expression was significantly correlated with advanced clinical stage, larger tumour size and increased lymph node involvement, and they associated it with poor survival in their sample of patients with nasopharyngeal carcinoma. Another study by Hong et al. [12] on glioma patients found that URGCP was gradually increased in low-grade disease and became markedly increased in high-grade disease, which was also reflected in poor survival for patients with high URGCP expression. This association was consistently reported in other cancer types, including bladder cancer [4], non-small cell lung cancer [6] and cervical cancer [13].
The primary effect of upregulation of URG4 gene is generally considered to be the activation of both Akt signalling and the NF-κB pathway, which result in cellular proliferation and angiogenesis, respectively. In breast cancer, Akt signalling is regarded as one of major pathways following growth factor receptor activation. Previous studies suggested that Akt phosphorylation can be considered a surrogate marker for HER2 and EGFR activation [14, 15]. The Akt activation in breast cancer yields mTOR activation, cellular proliferation, decreased cellular death, increased cellular motility and in vitro resistance to tamoxifen and doxorubicin [16,17,18,19,20]. Thus, our results seem contradictory to the correlation between URG4 and Akt activation, observed in aforementioned previous studies. Ahmad et al. [21] reported that oestrogen and IGF-1 stimulate Akt phosphorylation and its kinase activity in breast cancer cells. One may suppose that oestrogens and IGF-1 are readily available in women without breast cancer, and these stimulants may also increase Akt phosphorylation in normal breast epithelial cells. If Akt phosphorylation is directly associated with URG4 gene signals, this evidence regarding the presence of Akt in normal tissues may partly explain our findings of the high frequency of URG4 positivity in tumour-free mammoplasty specimens. However, this raises a question about the poor expression of URG4 in high-grade ductal carcinoma of the breast, which was supposed to be an opposite finding. The answer to this question may be associated with the effects of hormonal interactions on cellular expression of URG4. Because the URG4 staining index was much higher in hormone-receptor positive cases, the overexpression of URG4 may be associated with the basal hormonal status of the women studied. In addition, other mechanisms involving URG4 and its roles in breast cancer can be existed and should be examined in the future.
In our study, we also found that there may be a discrepancy between the immunohistochemistry results and the molecular results. For example, unlike the molecular results, the immunohistochemical results did not demonstrate the association of URG4 with lymph node metastasis. In addition, the molecular study revealed that there was no significant difference in URG4 expression for different grades of tumour tissues, which did not agree with the immunohistochemical results. These results may be due to a small number of samples examined. In addition, these differences between the gene and protein expression patterns of URG4 might suggest its distinct characteristics in breast cancer.
Despite scientific evidence that URG4 overexpression is associated with poor prognoses in many types of cancer, our results with breast cancer specimens indicate that URG4 overexpression in mammary ductal carcinomas is significantly associated with good prognostic parameters. Therefore, URG4 can be used as a marker for the evaluation of the prognosis of breast cancer. Nevertheless, these preliminary findings should be confirmed with a larger sample size and more detailed studies.
References
Tufan NL, Lian Z, Liu J, Pan J, Arbuthnot P, Kew M, et al. Hepatitis Bx antigen stimulates expression of a novel cellular gene, URG4, that promotes hepatocellular growth and survival. Neoplasia. 2002;4:355–68.
Li W, Zhou N. URG4 upregulation is associated with tumor growth and poor survival in epithelial ovarian cancer. Arch Gynecol Obstet. 2012;286:209–15.
Song J, Xie H, Lian Z, Yang G, Du R, Zou X, et al. Enhanced cell survival of gastric cancer cells by a novel gene URG4. Neoplasia. 2006;8:995–1002.
Wu M, Chen J, Wang Y, Hu J, Liu C, Feng C, et al. URGCP/URG4 promotes apoptotic resistance in bladder cancer cells by activating NF-κB signalling. Oncotarget. 2015;6:30887–901.
Chen LC, Zhang HY, Qin ZY, Wang Y, Mao Y, Yao Y, et al. Serological identification of URGCP as a potential biomarker for glioma. CNS Neurosci Ther. 2014;20:301–7.
Cai J, Li R, Xu X, Zhang L, Wu S, Yang T, et al. URGCP promotes non-small cell lung cancer invasiveness by activating the NF-κB-MMP-9 pathway. Oncotarget. 2015;6:36489–504.
Dodurga Y, Eroğlu C, Seçme M, Elmas L, Avcı ÇB, Şatıroğlu-Tufan NL, et al. Anti-proliferative and anti-invasive effects of ferulic acid in TT medullary thyroid cancer cells interacting with URG4/URGCP. Tumour Biol. 2016;37:1933–40.
Dodurga Y, Gundogdu G, Koc T, Yonguç GN, Kucukatay V, Satıroğlu-Tufan NL, et al. Expression of URG4/URGCP, Cyclin D1, Bcl-2, and Bax genes in retinoic acid treated SH-SY5Y human neuroblastoma cells. Contemp Oncol (Pozn). 2013;17:346–9.
Dodurga Y, Avcı CB, Susluer SY, Satıroğlu Tufan NL, Gündüz C. The expression of URGCP gene in prostate cancer cell lines: correlation with rapamycin. Mol Biol Rep. 2012;39:10173–7.
Dodurga Y, Oymak Y, Gündüz C, Satıroğlu-Tufan NL, Vergin C, Çetingül N, et al. Leukemogenesis as a new approach to investigate the correlation between up regulated gene 4/upregulator of cell proliferation (URG4/URGCP) and signal transduction genes in leukemia. Mol Biol Rep. 2013;40:3043–8.
Yu G, Meng Q, Zhang T, Zeng C, He B, Zhang S. URG4 expression is a novel prognostic factor for the progression of nasopharyngeal carcinoma and overall survival of patient. Onco Targets Ther. 2016;9:3059–65.
Hong L, Qing O, Ji Z, Chengqu Z, Ying C, Hao C, et al. Downregulation of miR-16 via URGCP pathway contributes to glioma growth. Sci Rep. 2017;7:13470.
Zhang L, Huang H, Zhang L, Hou T, Wu S, Huang Q, et al. URG4 overexpression is correlated with cervical cancer progression and poor prognosis in patients with early-stage cervical cancer. BMC Cancer. 2014;14:885.
Morgensztern D, McLeod HL. PI3K/Akt/mTOR pathway as a target for cancer therapy. Anticancer Drugs. 2005;16:797–803.
Nahta R, Yu D, Hung MC, Hortobagyi GN, Esteva FJ. Mechanisms of disease: understanding resistance to HER2-targeted therapy in human breast cancer. Nat Clin Pract Oncol. 2006;3:269–80.
Likhite VS, Stossi F, Kim K, Katzenellenbogen BS, Katzenellenbogen JA. Kinase-specific phosphorylation of the estrogen receptor changes receptor interactions with ligand, deoxyribonucleic acid, and coregulators associated with alterations in estrogen and tamoxifen activity. Mol Endocrinol. 2006;20:3120–32.
Lee ER, Kim JY, Kang YJ, Ahn JY, Kim JH, Kim BW, et al. Interplay between PI3K/Akt and MAPK signaling pathways in DNA-damaging drug-induced apoptosis. Biochim Biophys Acta. 2006;1763:958–68.
Liang K, Lu Y, Li X, Zeng X, Glazer RI, Mills GB, et al. Differential roles of phosphoinositide-dependent protein kinase-1 and akt1 expression and phosphorylation in breast cancer cell resistance to Paclitaxel, Doxorubicin, and gemcitabine. Mol Pharmacol. 2006;70:1045–52.
Altomare DA, Testa JR. Perturbations of the AKT signaling pathway in human cancer. Oncogene. 2005;24:7455–64.
Knuefermann C, Lu Y, Liu B, Jin W, Liang K, Wu L, et al. HER2/PI-3K/Akt activation leads to a multidrug resistance in human breast adenocarcinoma cells. Oncogene. 2003;22:3205–12.
Ahmad S, Singh N, Glazer RI. Role of AKT1 in 17beta-estradiol- and insulin-like growth factor I (IGF-I)-dependent proliferation and prevention of apoptosis in MCF-7 breast carcinoma cells. Biochem Pharmacol. 1999;58:425–30.
Acknowledgements
This study was funded by Balıkesir University Coordinator of Scientific Research Projects. The authors would like to appreciate the help from Prof. Dr. Feray Koçkar and Dr. Esra Tokay, for their guidance on the processes in this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
About this article
Cite this article
Aslan, F., Avcıkurt, A.S. URG4 expression in invasive breast carcinoma and its relation to clinicopathological characteristics. Breast Cancer 26, 485–491 (2019). https://doi.org/10.1007/s12282-019-00947-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12282-019-00947-6