Abstract
Molecular targets in prostate cancer are continually being explored, for which there are currently few therapeutic options. Rapamycin (RPM) is an antifungal macrolide antibiotic isolated from Streptomyces hygroscopicus which can inhibit the G1 to S transition. URGCP (upregulator of cell proliferation) is a novel gene located on chromosome 7p13. We aimed to investigate the role of URGCP gene expression changes in PC3, DU145, and LNCAP cell lines with/out RPM. Average cell viability and cytotoxic effect of rapamycin were investigated at 24 h intervals for three days by using Trypan blue dye exclusion test and XTT assay. Cytotoxic effects of rapamycin in DU145, PC3 and LNCAP cells were detected in time and dose dependent manner with the IC50 doses within the range of 1–100 nM. As the results were evaluated, IC50 doses in the DU145, PC3, and LNCaP cells were detected as 10, 25, and 50 nM, respectively. The mean relative ratios of URGCP gene expression in DU145, LNCAP and PC3 cells were found as −1.48, 6.59 and −13.00, respectively, when compared to rapamycin-free cells. The False Discovery Rate adjusted p value in DU145, LNCAP and PC3 were 1.25 × 10−5, 2.20 × 10−8 and 6.20 × 10−9, respectively. When the URGCP gene expression level is compared between the dose and control group, we found that URGCP gene expression was significantly decreased in dose groups of DU145 and PC3 cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Recently, rapid developments have taken place in the genomics and proteomics research, which has allowed us to understand some of the cellular events regarding to carcinogenesis. This finding of the molecular basis of cancer enabled us to defining pathways and also it might be targeted selectively by novel agents or genes for preventing or treating cancer. As research into various urological cancers progresses, new molecular targets and genes are identified, and agents against these pathways are developed rapidly.
Prostate cancer is one of the leading causes of cancer death among men. When prostate cancer is diagnosed early, treatment options include prostatectomy along with androgen therapy. Although cancer is diagnosed early, it can often metastasize primarily to bone. Once prostate cancer metastasizes, the 5-year survival rate drops dramatically from approximately 100 to 32 % [2, 21]. Most patients who responded initially to resect of androgen through orchiectomy (orchidectomy), luteinizing hormone-releasing hormone agonists and non-steroidal anti-androgens will eventually develop progressive disease [4]. Once androgen-independent prostate cancer occurs, prognosis is poor, with a median survival time from 7 to 12 months [9, 10]. However chemotherapy has a role in the treatment of prostate cancer and various strategies have been followed up to improve the outcomes of prostate cancer, more effective combined treatment may be developed to accomplish this malignancy (Temsirolimus, Everolimus, Valproic Acid etc.) [3, 16].
Rapamycin (RPM) is a macrolide lactone which isolated from the soil bacteria Streptomyces hygroscopicus. The drug RPM is widely used as an immunosuppressant in organ transplants, and is also in clinical trials for various types of cancers [12]. Anticancer activity of RPM was identified for cancer therapy and many efforts are being developed to synthesize derivatives with lower toxicities. There have been significant studies into the effects of RPM on carcinogenesis. RPM can inhibit the G1 to S transition by stimulating the degradation of cyclin D1 [5]. It can also down regulate phospho-p70 S6 kinase, which is considered as an indicator of the activated Akt-mTOR pathway; the Akt-mTOR pathway is central to prostate cancer progression [11, 14]. Inhibition of the mTOR signaling pathway by RPM was also reported to be effective and showed the antitumor activity in the treatment of advanced prostate cancer [1]. RPM was reported to increase the cytotoxicity of cisplatin, and sensitize human pro-myelocytic leukemia cells and ovarian cancer cells to cisplatin treatment by inducing apoptosis; however, receptor tyrosine kinase inhibitors only had additive anti-proliferative effects in combination with RPM against prostate cancer [13]. RPM was rather quickly recognized to have potent and broad anti-tumor activity, entering the scene for cancer therapy. RPM and a number of its structural derivatives (temirolimus, everolimus, and AP-23573) are currently under robust and encouraging investigational use in Phase I and II Clinical Trials for a variety of cancers [3].
Upregulator of cell proliferation (URGCP), GeneBank NM_017920), a novel gene located on 7p13 and originally identified by Satiroglu-Tufan NL, encodes 922 amino acid in cytoplasm. Over-expression of URGCP in HepG2 cells promoted hepatocellular growth and survival in tissue culture and in soft agar, and accelerated tumor development in nude mice [19]. URGCP is strongly expressed in hepatocellular carcinoma, gastric cancer and osteosarcoma. Over-expression of URGCP stimulated cyclin D1 mRNA expression, and RNAi-mediated URGCP silencing also diminished cyclin D1 mRNA expression in HepG2 cells. These results suggest that cyclin D1 up-regulation contributes importantly to the mechanism of URGCP -mediated hepatocellular growth [17]. Hence, URGCP may be a putative oncogene that contributes importantly to multistep carcinogenesis and cell cycle regulation.
This preliminary study was undertaken to evaluate the effects of RPM treatment on URGCP gene expression on three prostate cancer cell lines and possible association of URGCP and RPM on prostate cancer has been analyzed for the first time.
Materials and methods
Tumor cell line
PC-3 (Androgen Independent Phenotype), DU145 (Androgen Independent Phenotype) and LNCAP (Androgen Dependent Phenotype) prostate cell lines were used as a model cell line in this study which was obtained from ATCC.
Cell culture and preparation of cytotoxicity experiments
PC-3, DU145 and LNCAP cell lines grown in DMEM: Ham’s F12 and RPMI 1640 Basal medium containing 2 mM l-glutamine supplemented with 10 % fetal bovine inactivated serum (FBS) and 1 % penicillin/streptomycin were maintained at a density of 6×105 cells/ml in a standard cell culture incubator at 37 °C, humidified 95 % air, and 5 % CO2 atmosphere. RPM diluted in RPMI 1640 and DMEM: Ham’s F12 mediums were used in treatments of 1–150 μM with all cell lines.
Cytotoxicity assay
Cytotoxic assays and determination of IC50 doses of RPM in prostate cells were performed by using trypan blue dye exclusion test and XTT assay as indicated in manufacturers’ instruction.
Viability was calculated as follows:
XTT assay
Cells were seeded in 96-well tissue culture plates and incubated for 24 h without reagent. After addition of reagents, cells were incubated for 24, 48 and 72 h and cell viability was assessed by using XTT-PMS mixture (XTT sodium salt; [2,3-bis (2-Methoxy-4-nitro-5-sulfophenyl) 2Htetrazolium- 5-carboxanilide inner salt], Phenazine Methosulfate (N-Methylphenazonium methyl sulfate salt)], as recommended by supplier. Formazan formation was quantified spectrophotometrically at 450 nm (reference wavelength 630 nm) using a microplate reader (Multiscan FC, Thermo Scientific). IC50 values were determined by nonlinear regression analysis [Y = bottom + (top – bottom)/(\(1 + 10^{({\text{Log IC}}_{50}\,-\, {\text{X}})\,{\text{x}}\,{\text{HillSlope}}}\)), GraphPad Prism software].
Isolation of total RNA and cDNA synthesis
Fifty microliters of total RNA was isolated from cell culture of prostate cells treated with RPM in IC50 doses for 24, 48 and 72 h and control cells by using High Pure RNA Isolation Kit (Roche, Germany). Reverse transcription procedure was performed for cDNA synthesis by using Transcriptor First Strand cDNA Synthesis Kit (Roche, Germany) according to the manufacturers’ instructions.
Relative quantification of URGCP
Real-time quantitative RT-PCR analyses of gene URGCP was performed with Lightcycler instrument and software. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH “housekeeping” gene) was chosen as a standard to control the variability in amplification. PCR was performed by using TaqMan Master Kit (Roche Diagnostics) according to the instructions of the manufacturer. Studied genes target probe was labeled at the 5′ end with the reporter dye molecule 6-carboxyfluorescein (FAM). The GAPDH target probe was labeled with 6- carboxyfluorescein. Both probes were labeled with the quencher flour 6-carboxytetramethylrhodamine (TAMRA) at the 3′ end. To quantify genes mRNA from cell culture, we constructed a calibration curve (Error: 0.100 Efficiency: 2.0) using copy number (108, 107, 106, 105, 104, 103, 102 and 10) of GAPDH. Relative ratio of gene expressions was calculated using the formula:
Results
The expression profile of URGCP gene is evaluated by treating 1 to 100 nM with doses of RPM at the 24th, 48th and 72nd h to DU145, PC3 and LNCaP cells. Expression analysis is performed by using the Real Time online PCR method and expression rates detect in time and dose dependent manner.
In this study, as the results are evaluated, Trypan blue staining showed that dose and time dependent decreased viability. DU145, PC3 and LNCaP cell lines were 50 % reduction in viability with 10, 25 and 50 nM RPM respectively at 72nd h (Fig. 1). IC50 doses of RPM in the DU145, PC3 and LNCaP cell lines are detected in the 72nd h as 11.08, 50.80 and 1.24 nM, respectively by XTT assay (Fig. 2). The ΔΔCT method is used to evaluate differences in the relative expression levels [15]. The mean relative ratios of URGCP gene expression in DU145, PC3 and LNCAP which were treated with concentrations of RPM that reduced cell viability to 50 %, are found −1.48, −13.00 and 6.59 fold change, respectively. The False Discovery Rate adjusts p value in DU145, LNCAP and PC3 are 1.25 × 10−5, 2.20 × 10−8 and 6.20 × 10−9, respectively.
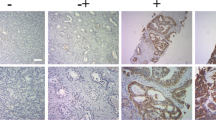
When the URGCP gene expression level compared to prostate cancer cells’ dose group and control group, were significantly decreased in dose group DU145 and PC3 cells but URGCP gene expression was significantly increased in dose group of LNCaP cells (Fig. 3).
Discussion
The view for patients with hormone resistant prostate cancer is poor. Current chemotherapeutic reagents can only offer a few months survival. Preclinical studies show RPM to be most effective when combined with other inhibitory compounds. Actually, given the level of cross-talk between mitogenic cell-signaling pathways, it could be unrealistic to expect a single specific inhibitory molecule to have significant efficacy. Thus, combinatorial therapy using RPM may prove to be the most efficient approach for further clinical investigation.
The molecular mechanism of RPM is complex and it is not known the mechanism about the antitumor effects. The mTOR pathway is approved as a main regulator in various malignant tumors and investigators suppose that RPM can suppress the expression of mTOR or phosphorylation of its downstream effectors [7]. Some studies indicated that RPM could down-regulate the expression of hypoxia-inducible factor-1α (HIF-1α) and vascular endothelial cell growth factor (VEGF) and also RPM induces apoptosis with activation of caspase 3, up-regulation of Bcl-xl, and down-regulation of Bcl-2 [22, 23]. But there are some alterations in the expression of many genes and chemotherapeutic agent resistances are detected both androgen-dependent to androgen-independent prostate cancer [6].
In the past decade, a very large number of proto-oncogenes and tumor suppressor genes have been found. Recently, URGCP, a novel gene up regulated by HBxAg in human hepatocellular carcinoma, has been identified (Gen Bank accession no. NM_017920). Previous data suggested that over-expression of URGCP in HepG2 cells promoted hepatocellular growth and survival in tissue culture and nude mice. Hence, URGCP may be an oncogene operating in hepatocarcinogenesis [20].
The mechanism of URGCP biologic activity in normal and malignant cells is not yet fully understood. Song et al. [18] showed that URGCP was up regulated in human gastric cancer tissues and also in gastric cancer cell lines and over-expression of URGCP could promote cell proliferation. Huang et al. [8] described in their data URGCP was highly expressed in 40 of 46 (86.96 %) osteosarcoma specimens with cytoplasmic staining, and also increased in the specimens with recurrence (p < 0.05) and metastasis (p < 0.05). They thought that URGCP may play important roles in the development of osteosarcoma, and might be a useful molecular marker for predicting the prognosis of osteosarcoma.
In this study, we investigated the effect of RPM on human prostate cells viabilities and effects of RPM on a novel gene URGCP expression in prostate cells. It was found that RPM diminishes URGCP gene expression in DU145 and PC3 cells, but opposite to this repression, we were found increase URGCP gene expression in dose group of LNCaP cells.
This preliminary study is the first research to show the correlation between RPM and URGCP gene expression in prostate cancer cell lines. These results demonstrated that rapamycin affects the URGCP gene expression level without androgen hormone related cancer cell lines.
References
Amato RJ, Jac J, Mohammad T, Saxena S (2008) Pilot study of rapamycin in patients with hormone-refractory prostate cancer. Clin Genitourin Cancer 6:97–102
Chin JL, Reiter RE (2004) Molecular markers and prostate cancer prognosis. Clin Prostate Cancer 3:157–164
Garcia JA, Danielpour D (2008) Mammalian target of rapamycin inhibition as a therapeutic strategy in the management of urologic malignancies. Mol Cancer Ther 7(6):1347–1354
Goktas S, Crawford ED (1999) Optimal hormonal therapy for advanced prostatic carcinoma. Semin Oncol 26:162–173
Hashemolhosseini S, Nagamine Y, Morley SJ, Desrivières S, Mercep L, Ferrari S (1998) Rapamycin inhibition of the G1 to S transition is mediated by effects on cyclin D1 mRNA and protein stability. J Biol Chem 273:14424–14429
Holzbeierlein J, Lal P, LaTulippe E, Smith A, Satagopan J, Zhang L et al (2004) Gene expression analysis of human prostate carcinoma during hormonal therapy identifies androgen responsive genes and mechanisms of therapy resistance. Am J Pathol 164:217–227
Hou G, Xue L, Lu Z, Fan T, Tian F, Xue Y (2007) An activated mTOR/p70S6K signaling pathway in esophageal squamous cell carcinoma cell lines and inhibition of the pathway by rapamycin and siRNA against mTOR. Cancer Lett 253:236–248
Huang J, Zhu B, Lu L, Lian Z, Wang Y, Yang X, Satiroglu-Tufan NL, Liu J, Luo Z (2009) The expression of novel gene URG4 in osteosarcoma: correlation with patients’ prognosis. Pathology 41:49–54
Hussain M, Wolf M, Marshall E, Crawford ED, Isenberger M (1994) Effects of continued androgen-deprivation therapy and other prognostic factors on response and survival in phase II chemotherapy trials for hormone-refractory prostate cancer: a southwest oncology group report. J Clin Oncol 12:1868–1875
Kantoff PW, Halabi S, Conaway M (1999) Hydrocortisone with or without mitoxantrone in men with hormone-refractory prostate cancer: results of the cancer and leukemia group B 9182 study. J Clin Oncol 17:2506–2513
Kremer CL, Klein RR, Mendelson J, Browne W, Samadzedeh LK, Vanpatten K et al (2006) Expression of mTOR signaling pathway markers in prostate cancer progression. Prostate 66:1203–1212
Lee CH, Inoki K, Guan KL (2007) mTOR pathway as a target in tissue hypertrophy. Annu Rev Pharmacol Toxicol 47:443–467
Masiello D, Mohi MG, McKnight NC, Smith B, Neel BG, Balk SP et al (2007) Combining an mTOR antagonist and receptor tyrosine kinase inhibitors for the treatment of prostate cancer. Cancer Biol Ther 6:195–201
Nozawa H, Watanabe T, Nagawa H (2007) Phosphorylation of ribosomal p70 S6 kinase and rapamycin sensitivity in human colorectal cancer. Cancer Lett 251:105–113
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29(9):e45
Sandler HM, Narayan S, Smith DC (2003) Combined modality treatment for prostate cancer: role of chemotherapy. Semin Oncol 30:95–100
Satiroglu-Tufan NL, Dodurga Y, Gok D, Cetinkaya A, Feitelson MA (2010) RNA interference-mediated URG4 gene silencing diminishes cyclin D1 mRNA expression in HepG2 cells. Genet Mol Res 9(3):1557–1567
Song J, Xie H, Liany Z, Yang G, Du R, Du Y, Zou X, Jin HF, Gao J, Liu J, Fan D (2006) Enhanced cell survival of gastric cancer cells by a novel gene URG4. Neoplasia 8:995–1002
Tufan NL, Lian Z, Liu J, Pan J, Arbuthnot P, Kew M, Clayton MM, Zhu M, Feitelson MA (2002) Hepatitis Bx antigen stimulates expression of a novel cellular gene, URG4, that promotes hepatocellular growth and survival. Neoplasia 4:355–368
Tufan NL, Lian Z, Liu J, Pan J, Arbuthnot P, Kew M, Clayton MM, Zhu M, Feitelson MA (2002) Hepatitis Bx antigen stimulates expression of a novel cellular gene, URG4, that promotes hepatocellular growth and survival. Neoplasia 4:355–368
Velcheti V, Karnik S, Bardot SF, Prakash O (2008) Pathogenesis of prostate cancer: lessons from basic research. Ochsner J 8(4):213–218
Wang W, Jia WD, Xu GL, Wang ZH, Li JS, Ma JL et al (2009) Antitumoral activity of rapamycin mediated through inhibition of HIF-1 alpha and VEGF in hepatocellular carcinoma. Dig Dis Sci 54:2128–2136
Zhang JF, Liu JJ, Lu MQ, Cai CJ, Yang Y, Li H et al (2007) Rapamycin inhibits cell growth by induction of apoptosis on hepatocellular carcinoma cells in vitro. Transpl Immunol 17:162–168
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dodurga, Y., Avcı, Ç.B., Susluer, S.Y. et al. The expression of URGCP gene in prostate cancer cell lines: correlation with rapamycin. Mol Biol Rep 39, 10173–10177 (2012). https://doi.org/10.1007/s11033-012-1891-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-012-1891-6