Abstract
Since the 2000s, the Y439 lineage of H9N2 avian influenza virus (AIV) has been the predominant strain circulating in poultry in Korea; however, in 2020, the Y280 lineage emerged and spread rapidly nationwide, causing large economic losses. To prevent further spread and circulation of such viruses, rapid detection and diagnosis through active surveillance programs are crucial. Here, we developed a novel H9 rRT-PCR assay that can detect a broad range of H9Nx viruses in situations in which multiple lineages of H9 AIVs are co-circulating. We then evaluated its efficacy using a large number of clinical samples. The assay, named the Uni Kor-H9 assay, showed high sensitivity for Y280 lineage viruses, as well as for the Y439 lineage originating in Korean poultry and wild birds. In addition, the assay showed no cross-reactivity with other subtypes of AIV or other avian pathogens. Furthermore, the Uni Kor-H9 assay was more sensitive, and had higher detection rates, than reference H9 rRT-PCR methods when tested against a panel of domestically isolated H9 AIVs. In conclusion, the novel Uni Kor-H9 assay enables more rapid and efficient diagnosis than the “traditional” method of virus isolation followed by subtyping RT-PCR. Application of the new H9 rRT-PCR assay to AI active surveillance programs will help to control and manage Korean H9 AIVs more efficiently.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The first case of poultry infected with low pathogenicity avian influenza virus (LPAIV) H9N2 was officially reported on a turkey farm in Wisconsin, USA, in 1966 (Homme & Easterday, 1970). Since the mid-1990s, H9N2 avian influenza virus (AIV) has been identified worldwide, and the disease has not been eradicated in many countries in Asia and the Middle East (Alexander, 2007; Davidson et al., 2014; Guo et al., 2000). H9N2 is a major subtype of AIV that has adverse effects on the global poultry industry (Alexander, 2007; Guo et al., 2000; Peacock et al., 2019). Although H9N2 AIV is classified as low pathogenicity, it causes mortality or egg drop depending on the breeding environment or co-infection with other pathogens, leading to severe economic losses (Guo et al., 2000; Lai et al., 2021; Lee et al., 2007; Mo et al., 2003). In general, H9N2 viruses are divided into North American and Eurasian types, and the Eurasian type is further classified into G1, Y280, and Y439 lineages based on the sequence of the hemagglutinin (HA) gene (Dong et al., 2011; Guo et al., 2000).
In Korea, the H9N2 AIV belonging to the Y439 lineage was first reported in poultry in 1996. Subsequently, it spread nationwide and became endemic in the 2000s; these viruses are now classified as the Korean lineage (Lee & Song, 2013; Lee et al., 2007; Mo et al., 2003, 2016). Vaccination against Y439 lineage H9N2 AIV was introduced in 2007, the main aim being to reduce economic losses to the layer and broiler breeder chicken industries. However, despite the H9N2 vaccination program, the virus continues to circulate, mainly within small-scale native Korean chicken farms and live bird markets (LBMs) (Choi et al., 2008; Lee et al., 2010; Youk et al., 2020). In June 2020 (and for the first time in Korea), the Y280 lineage of H9N2 AIV was isolated from chickens at an LBM (Youk et al., 2020). Since then, this lineage of H9N2 AIV has spread quickly nationwide, and is now the predominant strain among domestic poultry (Lai et al., 2021).
The AI national surveillance program in Korea relies on conventional real-time reverse transcription PCR (rRT-PCR) targeting the M, H5, or H7 genes, which enables rapid detection and diagnosis of high pathogenicity AIV (HPAIV). Although South Korea uses a subtyping RT-PCR assay and full-sequencing of the HA segment to identify the H9N2 virus, many other countries also employ various H9-specific rRT-PCR assays to identify the detailed distribution of various H9N2 lineages, including viruses of wild bird origin (El Khantour et al., 2020; Monne et al., 2008). Thus, occurrence of multiple H9N2 AIV lineages in South Korea means that we need to develop and evaluate the sensitivity and specificity of conventional H9 AIV-specific diagnostic assays that can detect multiple lineages of H9 viruses. To address this unmet need, we developed and evaluated a novel H9 AIV rRT-PCR assay (called the Uni Kor-H9 assay) based on highly specific universal primer/probe sets. This assay enables rapid, accurate, and efficient detection of the Y280 and Y439 lineages of Korean H9N2 AIVs.
Materials and Methods
LBM Surveillance and Virus Isolation
The nationwide LBM surveillance system in Korea is based on laboratory testing of clinical samples from live poultry on sale; these samples are collected from every registered poultry store two to four times a year depending on location. Briefly, clinical specimens are collected by the Livestock Health Control Association (LHCA) and sent to the Animal and Plant Quarantine Agency (APQA) or to the provincial veterinary service laboratories (VSLs), where they are tested for AIVs. Oropharyngeal (OP) swabs and cloacal (CL) swabs (or feces) are collected from the Korean native chickens, broiler chickens, and domestic ducks, which are the main species sold at LBMs. Detection of H9 AIV was confirmed by a subtyping RT-PCR assay after isolating the virus from embryonated eggs; positive samples (cycle threshold (Ct) ≤ 40) were screened using the AIV M gene detection kit (Median Diagnostics, VR8100).
Primer and Probe Design
To differentiate the specific lineages of H9N2 viruses, several primers and probes were designed based on lineage-specific sequence information. Briefly, 67 complete HA gene sequences of H9 AIVs isolated in Korea were downloaded from the Korea Animal Health Integrated System (KAHIS) (https://home.kahis.go.kr) and the Global Initiative on Sharing All Influenza Database (GISAID) (https://gisaid.org): these sequences comprised 29 representative H9 AIVs belonging to the Y280 lineage, isolated since June 2020; 19 representative viruses selected from the Y439 lineage, isolated from 1996 to 2018; and 19 representative H9Nx AIVs, isolated from wild birds during 2020–2021. All sequences were edited and analyzed using the CLC Main Workbench program (version 6.9.1).
RNA Extraction and rRT-PCR Assay
Viral RNA was extracted from each reference strain and clinical isolate using the Maxwell RSC SimplyRNA Tissue Kit and the Maxwell RSC 48 instrument (Promega, AS1340). All H9 rRT-PCR analyses were performed using the CFX96 Touch Real-time PCR Detection System (C1000 Touch, Bio-Rad) and the QuantiNova Probe RT-PCR Kit (Qiagen, 208354). The volume of the reaction mixture for the H9 rRT-PCR assay was 20 µl, comprising 2× QuantiNova Probe RT-PCR Master Mix (10 µl), QN Probe RT-Mix (0.2 µl), 20× primer-probe mix (0.8 µM each forward and reverse primer and 0.2 µM TaqMan Probe; 1 µl of each), template RNA (5 µl; ≤ 400 ng/reaction), and 3.8 µl of RNase-Free Water. The H9 rRT-PCR conditions were as follows: 45 °C for 10 min, followed by 95 °C for 5 min, and then 40 cycles of 5 sec at 95 °C and 30 sec at 60 °C. The amplification data were collected and analyzed using Bio-Rad CFX Maestro 1.1 software (Version: 4.1.2433.1219).
Analytical Specificity
The specificity of the novel H9 rRT-PCR assay was tested using each reference H1–H15 subtype virus. In addition, other major avian viruses such as Newcastle disease virus (NDV), Infectious bursal disease virus (IBDV), Infectious Bronchitis virus (IBV), Fowl Pox virus (FPV), and Chicken infectious anemia virus (CIAV), as well as major bacterial pathogens, were tested. Furthermore, 18 AIV-negative specimens were used as controls. The complete list of samples tested for analytical specificity is provided in the Supplementary data 3.
Analytical Sensitivity
The sensitivity of the novel H9 rRT-PCR assay was evaluated using viruses representative of the Y280 and Y439 lineages, which were isolated from poultry and wild birds. The full-size HA genes of each representative virus were amplified using the primers and method proposed by Hoffmann et al. (2001). The amplicon was then confirmed and purified using the QIAquick Gel Extraction Kit (Qiagen). The concentration of the final purified product was measured using a NanoDrop One Spectrophotometer (ThermoFisher Scientific). Finally, the copy number of the full-size HA gene was quantified using the method described by CEL-seq (http://cels.uri.edu/gsc/cndna.html). These copy numbers were serially diluted ten-fold to obtain a “standard” sample for each concentration. To evaluate the standard rRT-PCR assay, each sample was tested three times.
Comparison with Other H9 rRT-PCR Assays
The sensitivity of the novel H9 rRT-PCR assay for the H9 subtype was compared with that of other diagnostic methods reported by IZSVe (Instituto Zooprofilattico Sperimentale delle Venezie, Italy) and NIID (National Institute of Infectious Diseases, Japan) (Monne et al., 2008; Saito et al., 2019). The rRT-PCR assay conditions and the primer/probe sets used have been reported previously. Fifty-six viruses isolated from 2016 to 2021 were used, all belonging to the Y439 and Y280 lineages isolated from poultry in LBMs or from wild birds.
Agreement Between the New H9 rRT-PCR Assay and Virus Isolation Method
Clinical samples obtained from LBMs were used to evaluate the correlation between the sensitivity of the virus isolation method and the rRT-PCR assay. Briefly, 301 clinical samples, including OP and CL swabs and feces, collected from Korean native chickens in 2020–21 were used, and the diagnostic results were compared. Of these, 142 samples (118 OP swabs and 24 feces samples) identified as AIV M gene-positive were selected and tested using the two methods: virus isolation after egg inoculation, and the new H9 rRT-PCR assay. The virus isolation results were confirmed by a hemagglutination test, and the HA subtype of the isolates was determined using the LiliF AIV Multi-tube RT-PCR Kit (iNtRON Biotechnology, IPC11009).
Analytical Reproducibility
To evaluate the performance of the novel H9 rRT-PCR assay in a field setting, 29 clinical samples (including 13 AIV genome-positive swabs) were shared and tested independently at the APQA and two VSLs (Gyeonggi and Gyeongsangbuk-do). The results were expressed as a correlation coefficient (CC), as described previously (Mirzaei et al., 2018).
Statistical Analysis
Data were analyzed using Prism version 5.0 software (GraphPad Software). Comparison of Ct values between groups was made by one-way analysis of variance (ANOVA). The threshold of Ct value was analyzed using the log-rank test. P < 0.05 was considered statistically significant.
Results
Circulation of Multiple Lineages of H9 AIVs in Korea
The Y439 lineage of Korean H9 AIV was detected in poultry in South Korea from 1996 to 2018. Although the Y439 lineage of H9N2 AIV has not been detected in domesticated poultry since 2018, the total number of H9N2 detections in Korea has increased due to emergence of the Y280 lineage of H9 AIV in LBMs in 2020. Since then, the Y280 lineage has spread gradually to domestic poultry farms, including layer chickens and Korean native chickens, and become the dominant H9 AIV strain nationwide (Fig. 1). In addition, the Y439 lineage of wild bird-origin (Eurasian lineage) H9 AIVs (Han et al., 2019; Suttie et al., 2019) has been detected continuously during annual wild bird surveillance; this virus is another potential source of infection of poultry (Supplementary data 1).
Design and Evaluation of Universal Primer and Probe Set for H9 AIV
To design the universal primer and probe specific for H9N2 AIVs, we first constructed phylogenetic trees based on the complete HA gene sequences of H9Nx AIVs isolated from Korea and other countries (these sequences were obtained from the KAHIS and GISAID). As shown in Fig. S1, the H9 subtype AIVs were classified genetically into three groups: Y280, Korean Y439, and wild bird-origin Eurasian Y439 H9 lineages. Based on sequence analysis, we designed a new diagnostic assay (the Uni Kor-H9 assay) that can detect Y280, Korean Y439, and wild bird-origin Eurasian Y439 H9 lineages simultaneously (Table 1).
To evaluate the specificity of the new Uni Kor-H9 assay, we tested different subtypes of AIV (n = 14), other avian viruses (n = 5), and bacteria found in poultry (n = 18) (see Supplementary data 3). As expected, all H9N2 viruses generated a positive signal for the M and H9 genes, regardless of lineage. However, all AIV subtypes other than H9 showed a positive signal only for the M gene (Ct value 15.38 ± 1.4). Furthermore, the new assay did not show any cross-reactivity with other major avian respiratory bacterial or viruses (i.e., NDV, IB, IBV, and CIAV; Table 2).
The analytical sensitivity of the Uni Kor-H9 assay was determined by measuring the limit of detection using H9N2 viruses that are representative of each lineage (i.e., A/chicken/Korea/LBM261/2020 [Y280], A/chicken/Korea/LBM294/2018 [Y439], A/chicken/Korea/01310/2001 [Vaccine strain of Y439], and A/wild bird/Korea/H276/2021 [Eurasian Y439]). Quantitative rRT-PCR analysis demonstrated that the R2 values from standard curve analyses ranged from 0.993 to 0.998 (Table 3). The virus belonging to the Y280 lineage, A/chicken/Korea/LBM261/2020, was detected at a copy number as low as 7.3, while the A/chicken/Korea/LBM294/2018, A/chicken/Korea/01310/2001 and A/wild bird/Korea/H276/2021 were detected at copy numbers of 478.5, 88.04 and 19.93, respectively (Table 3).
Comparison with Other H9 rRT-PCR Assays
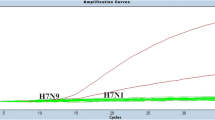
To evaluate the efficacy of the Uni Kor-H9 assay, we tested a panel of viruses using different rRT-PCR methods. A comparison of the relative sensitivity of these H9 rRT-PCR assays revealed that the Uni Kor-H9 assay had the highest sensitivity for all three lineages: Y280, Korean Y439, and wild bird-origin Eurasian Y439 viruses (Fig. 2 and Supplementary data 2). The Uni Kor-H9 assay detected all 56 viruses, whereas the NIID and IZSVe methods detected only 47 and 41 viruses, respectively. In addition, the deviation of the detection values per virus group for these says was greater than that of the Uni Kor-H9 assay (based on a positive Ct value of ≤ 40). The sensitivity of the NIID method (Saito et al., 2019) for the Y280 lineage of H9 AIV was comparable with that of the Uni Kor-H9 assay, whereas the sensitivity of the NIID for both Y439 lineages in poultry and wild birds was quite low. The IZSVe method (Monne et al., 2008) showed relatively low sensitivity for all three lineages.
Comparison of the relative sensitivity of the H9 rRT-PCR assay with that of two different rRT-PCR methods. Fifty-six H9N2 viruses were tested, including 26 belonging to the Y280 lineage, 15 belonging to the Y439 lineage isolated from poultry, and 15 belonging to the Y439 lineage isolated from wild birds. Blue bars indicate the Ct value for the Uni Kor-H9 assay, orange bars indicate the Ct value for the H9 rRT-PCR assay developed by NIID (Saito et al., 2019), and gray bars indicate the Ct value for the H9 rRT-PCR assay developed by IZSVe (Monne et al., 2008)
The reproducibility of the Uni Kor-H9 assay was further tested at the APQA and two VSLs (Gyeonggi and Gyeongsangbuk-do). Briefly, 29 known samples (13 positive and 16 negative) were tested in the Uni Kor-H9 assay, and all three laboratories obtained the same results with respect to positivity or negativity. The standard deviation of the Ct value for positive samples among laboratories ranged from 0.78 to 2.21 (Table 4). The CC for pairwise comparison of positive samples was 0.87–0.93 (Table 5).
Agreement Between the H9 rRT-PCR Assay and the Virus Isolation Method
Finally, 142 clinical specimens (118 OP swabs and 24 feces specimens, all of which tested positive using the M rRT-PCR detection kit) were tested using the Uni Kor-H9 assay to evaluate its efficacy. Of these, 136 tested positive in the H9 rRT-PCR, whereas only 76 specimens (66 OP swabs and 10 feces) tested positive in the virus isolation assay (Supplementary data 4). When compared with the results obtained using the conventional AIV M gene detection kit, the sensitivity of the Uni Kor-H9 assay was 95.8% (based on 100% positivity of 142 samples), which is much higher than that of the virus isolation method (53.5%).
Discussion
Since 2008, a large-scale AI active surveillance program has been implemented in South Korea for early detection of HPAI viruses (Mo et al., 2016; Sagong et al., 2023). To this end, wild birds (feces, captured wild birds, and carcasses), domestic poultry (chickens, domestic ducks, and other poultry), LBMs, and poultry traders have been targeted for periodic testing by rRT-PCR of the viral M, H5, and H7 genes. This national AI active surveillance program has detected high pathogenicity H5Nx AIVs in wild birds at the early stages of their migration, before outbreaks occur in poultry (Baek et al., 2021; Sagong et al., 2022). The program also detected the Y280 lineage of H9N2 AIV in LBMs (Choi et al., 2005; Heo et al., 2021; Liu et al., 2003). The Y280 lineage of H9N2 AIV began circulating in LBMs in China and Southeast Asia, from where it spread to neighboring regions and countries (Chen et al., 2016, 2017; Lin et al., 2017; Nomura et al., 2012; Novianti et al., 2019; Suttie et al., 2019). The genetic diversity of H9N2 AIV increases during circulation in LBMs, which may increase the chances of viral transmission to other species, including humans (Chen et al., 2015; Lee et al., 2010; Li et al., 2014). Therefore, continuous and comprehensive monitoring of poultry farms and LBMs, as well as genetic characterization of H9 AIVs, is essential for early detection of AIVs and for raising the alarm regarding public health concerns. To achieve this, a more accurate, simple, and sensitive diagnostic assay is required.
The standard method for H9 subtyping for AI surveillance in Korea involves a combination of virus isolation and RT-PCR; however, this is both time-consuming and labor-intensive. The rRT-PCR assay used to detect the M, H5, and H7 genes has been used for AI surveillance; however, the H9 rRT-PCR assay was not introduced until recently (Kim et al., 2013).
Here, we developed a new H9 rRT-PCR assay that was more sensitive, specific, and reproducible than current detection methods, including the virus isolation method. The new assay detected both Y439 and Y280 lineage AIVs in clinical specimens obtained from poultry and wild birds. There were no false positives or cross-reactions with other pathogens, including other AIV subtypes, avian viruses, and avian bacteria (Table 2). Furthermore, the diagnostic sensitivity of the Uni Kor-H9 assay kit (i.e., a relative sensitivity of 95.8% for clinical samples collected for LBM surveillance) was comparable with that of a commercial AIV M gene detection assay (Table 6). In addition, when using selected samples with detectable M genes, we found that detection rate of the Uni Kor-H9 assay was higher than that of the conventional diagnostic method (i.e., virus isolation) (Mirzaei et al., 2018). However, the Uni Kor-H9 assay also yielded false negative results for clinical samples containing very low concentrations (100 to 101 EID50/ml) of virus (data not shown).
Although the analytical sensitivity of the Uni Kor-H9 assay for Y439 lineage viruses appeared to be lower than that for the Y280 lineage, the assay demonstrated a broad detection range of H9 lineage, and showed particularly high sensitivity for the Y280 lineage, which has recently become dominant in Korea (Fig. 1 and Table 3). In contrast to representative viruses from other lineages that did not exhibit any mismatches in the primer/probe sequences, the two representative H9 AIVs belonging to the Y439 lineage showed a single mismatch at the reverse primer binding site. This mismatch may have contributed to the lower limit of detection for the Y439 lineage (copy number, 88.04–478.5).
To verify the efficacy of the Uni Kor-H9 assay for field application, it was tested in two field surveillance studies conducted during the 2022 season, which showed it to be a useful method of AI surveillance. Furthermore, the Uni Kor-H9 assay was more efficient than previously reported diagnostic methods used to identify H9N2 viruses, including recent H9 AIVs detected in poultry or wild birds in Korea (Fig. 2). Although our new H9 rRT-PCR assay may need further evaluation in large-scale studies, and with a single set of primers and probes, to determine whether it can identify all variations or mutations occurring during continuous virus circulation (especially of H9 AIVs) (Slomka et al., 2013), the results clearly demonstrate that the assay is more suitable than other methods for monitoring H9 AIVs circulating recently in Korea.
To summarize, we developed a highly efficient rRT-PCR assay that detects a broad range of H9 AIVs, including Y280 and various Y439 lineages, and evaluated its efficacy by comparing it with currently used methods. Furthermore, we used the assay to test a large number of clinical specimens from AI surveillance programs, and found that it was as sensitive and specific as commercial AIV M diagnostic kits, and more sensitive and specific than the virus isolation method. These results suggest that this new assay can be used to detect and identify Korean H9 AIV quickly and efficiently, making it an effective replacement for existing diagnostic methods. Although it may need continuous development and updates, we believe that this assay is suitable for large-scale surveillance of a broad range of H9 AIVs present on domestic poultry farms and in wild bird habitats.
Data Availability
Change history
27 June 2024
An Erratum to this paper has been published: https://doi.org/10.1007/s12275-024-00149-6
References
Alexander, D. J. (2007). An overview of the epidemiology of avian influenza. Vaccine, 25, 5637–5644.
Baek, Y. G., Lee, Y. N., Lee, D. H., Shin, J. I., Lee, J. H., Chung, D. H., Lee, E. K., Heo, G. B., Sagong, M., Kye, S. J., et al. (2021). Multiple reassortants of H5N8 clade 2.3.4.4b highly pathogenic avian influenza viruses detected in South Korea during the winter of 2020–2021. Viruses, 13, 490.
Chen, J., Ma, J., White, S. K., Cao, Z., Zhen, Y., He, S., Zhu, W., Ke, C., Zhang, Y., Su, S., et al. (2015). Live poultry market workers are susceptible to both avian and swine influenza viruses, Guangdong Province, China. Veterinary Microbiology, 181(3–4), 230–235.
Chen, L. J., Lin, X. D., Guo, W. P., Tian, J. H., Wang, W., Ying, X. H., Wang, M. R., Yu, B., Yang, Z. Q., Shi, M., et al. (2016). Diversity and evolution of avian influenza viruses in live poultry markets, free-range poultry and wild wetland birds in China. The Journal of General Virology, 97, 844–854.
Chen, L. J., Lin, X. D., Tian, J. H., Liao, Y., Ying, X. H., Shao, J. W., Yu, B., Guo, J. J., Wang, M. R., Peng, Y., et al. (2017). Diversity, evolution and population dynamics of avian influenza viruses circulating in the live poultry markets in China. Virology, 505, 33–41.
Choi, J. G., Lee, Y. J., Kim, Y. J., Lee, E. K., Jeong, O. M., Sung, H. W., Kim, J. H., & Kwon, J. H. (2008). An inactivated vaccine to control the current H9N2 low pathogenic avian influenza in Korea. Journal of Veterinary Science, 9, 67–74.
Choi, Y. K., Seo, S. H., Kim, J. A., Webby, R. J., & Webster, R. G. (2005). Avian influenza viruses in Korean live poultry markets and their pathogenic potential. Virology, 332, 529–537.
Davidson, I., Fusaro, A., Heidari, A., Monne, I., & Cattoli, G. (2014). Molecular evolution of H9N2 avian influenza viruses in Israel. Virus Genes, 48, 457–463.
Dong, G., Luo, J., Zhang, H., Wang, C., Duan, M., Deliberto, T. J., Nolte, D. L., Ji, G., & He, H. (2011). Phylogenetic diversity and genotypical complexity of H9N2 influenza A viruses revealed by genomic sequence analysis. PLoS ONE, 6, e17212.
El Khantour, A., Soulaymani, A., Salek, M., Maltouf, A. F., Darkaoui, S., El Mellouli, F., Ducatez, M. F., & Fellahi, S. (2020). Molecular characterization of the hemagglutinin gene of H9N2 avian influenza viruses isolated from broiler flocks in Morocco from 2016 to 2018. Veterinary Archives, 90, 477–484.
Guo, Y. J., Krauss, S., Senne, D. A., Mo, I. P., Lo, K. S., Xiong, X. P., Norwood, M., Shortridge, K. F., Webster, R. G., & Guan, Y. (2000). Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology, 267, 279–288.
Han, L., He, W., Yan, H., Li, X., Wang, C., Shi, Q., Zhou, T., & Dong, G. (2019). The evolution and molecular characteristics of H9N2 avian influenza viruses in Jiangxi of China. Journal of Medical Virology, 91, 711–716.
Heo, G. B., Kye, S. J., Sagong, M., Lee, E. K., Lee, K. N., Lee, Y. N., Choi, K. S., Lee, M. H., & Lee, Y. J. (2021). Genetic characterization of H9N2 avian influenza virus previously unrecognized in Korea. Journal of Veterinary Science, 22, e21.
Hoffmann, E., Stech, J., Guan, Y., Webster, R. G., & Perez, D. R. (2001). Universal primer set for the full-length amplification of all influenza A viruses. Archives of Virology, 146, 2275–2289.
Homme, P. J., & Easterday, B. C. (1970). Avian influenza virus infections. I. Characteristics of influenza A-Turkey-Wisconsin-1966 virus. Avian Diseases, 14, 66–74.
Kim, H. R., Oem, J. K., Bae, Y. C., Kang, M. S., Lee, H. S., & Kwon, Y. K. (2013). Application of real-time reverse transcription polymerase chain reaction to the detection the matrix, H5 and H7 genes of avian influenza viruses in field samples from South Korea. Virology Journal, 10, 85.
Lai, V. D., Kim, J. W., Choi, Y. Y., Kim, J. J., So, H. H., & Mo, J. (2021). First report of field cases of Y280-like LPAI H9N2 strains in South Korean poultry farms: Pathological findings and genetic characterization. Avian Pathology, 50, 327–338.
Lee, D. H., & Song, C. S. (2013). H9N2 avian influenza virus in Korea: Evolution and vaccination. Clinical and Experimental Vaccine Research, 2, 26–33.
Lee, H. J., Kwon, J. S., Lee, D. H., Lee, Y. N., Youn, H. N., Lee, Y. J., Kim, M. C., Jeong, O. M., Kang, H. M., Kwon, J. H., et al. (2010). Continuing evolution and interspecies transmission of influenza viruses in live bird markets in Korea. Avian Diseases, 54(1 Suppl), 738–748.
Lee, Y. J., Shin, J. Y., Song, M. S., Lee, Y. M., Choi, J. G., Lee, E. K., Jeong, O. M., Sung, H. W., Kim, J. H., Kwon, Y. K., et al. (2007). Continuing evolution of H9 influenza viruses in Korean poultry. Virology, 359, 313–323.
Li, X., Shi, J., Guo, J., Deng, G., Zhang, Q., Wang, J., He, X., Wang, K., Chen, J., Li, Y., et al. (2014). Genetics, receptor binding property, and transmissibility in mammals of naturally isolated H9N2 avian influenza viruses. PLoS Pathogens, 10, e1004508.
Lin, T. N., Nonthabenjawan, N., Chaiyawong, S., Bunpapong, N., Boonyapisitsopa, S., Janetanakit, T., Mon, P. P., Mon, H. H., Oo, K. N., Oo, S. M., et al. (2017). Influenza A(H9N2) virus, Myanmar, 2014–2015. Emerging Infectious Diseases, 23, 1041–1043.
Liu, M., He, S., Walker, D., Zhou, N., Perez, D. R., Mo, B., Li, F., Huang, X., Webster, R. G., & Webby, R. J. (2003). The influenza virus gene pool in a poultry market in South central China. Virology, 305, 267–275.
Mirzaei, S. G., Shoushtari, A., & Nouri, A. (2018). Development and evaluation of Real-Time RT-PCR test for quantitative and qualitative recognition of current H9N2 subtype avian influenza viruses in Iran. Archives of Razi Institute, 73, 177–182.
Mo, I. P., Bae, Y. J., Lee, S. B., Mo, J. S., Oh, K. H., Shin, J. H., Kang, H. M., & Lee, Y. J. (2016). Review of avian influenza outbreaks in South Korea from 1996 to 2014. Avian Diseases, 60(1 Suppl), 172–177.
Mo, I. P., Song, C. S., Kim, K. S., & Rhee, J. C. (2003). An occurrence of non-highly pathogenic avian influenza in Korea. Avian Diseases, 47, 379–383.
Monne, I., Ormelli, S., Salviato, A., De Battisti, C., Bettini, F., Salomoni, A., Drago, A., Zecchin, B., Capua, I., & Cattoli, G. (2008). Development and validation of a one-step real-time PCR assay for simultaneous detection of subtype H5, H7, and H9 avian influenza viruses. Journal of Clinical Microbiology, 46, 1769–1773.
Nomura, N., Sakoda, Y., Endo, M., Yoshida, H., Yamamoto, N., Okamatsu, M., Sakurai, K., Hoang, N. V., Nguyen, L. V., Chu, H. D., et al. (2012). Characterization of avian influenza viruses isolated from domestic ducks in Vietnam in 2009 and 2010. Archives of Virology, 157, 247–257.
Novianti, A. N., Rahardjo, K., Prasetya, R. R., Nastri, A. M., Dewantari, J. R., Rahardjo, A. P., Estoepangestie, A. T. S., Shimizu, Y. K., Poetranto, E. D., Soegiarto, G., et al. (2019). Whole-genome sequence of an avian influenza A/H9N2 virus isolated from an apparently healthy chicken at a live-poultry market in Indonesia. Microbiology Resource Announcements, 8, e01671-18.
Peacock, T. H. P., James, J., Sealy, J. E., & Iqbal, M. (2019). A global perspective on H9N2 avian influenza virus. Viruses, 11, 620.
Sagong, M., Lee, K. N., Lee, E. K., Kang, H., Choi, Y. K., & Lee, Y. J. (2023). Current situation and control strategies of H9N2 avian influenza in South Korea. Journal of Veterinary Science, 24, e5.
Sagong, M., Lee, Y. N., Song, S., Cha, R. M., Lee, E. K., Kang, Y. M., Cho, H. K., Kang, H. M., Lee, Y. J., & Lee, K. N. (2022). Emergence of clade 2.3.4.4b novel reassortant H5N1 high pathogenicity avian influenza virus in South Korea during late 2021. Transboundary and Emerging Diseases, 69, e3255–e3260.
Saito, S., Takayama, I., Nakauchi, M., Nagata, S., Oba, K., Odagiri, T., & Kageyama, T. (2019). Development and evaluation of a new real-time RT-PCR assay for detecting the latest H9N2 influenza viruses capable of causing human infection. Microbiology and Immunology, 63, 21–31.
Slomka, M. J., Hanna, A., Mahmood, S., Govil, J., Krill, D., Manvell, R. J., Shell, W., Arnold, M. E., Banks, J., & Brown, I. H. (2013). Phylogenetic and molecular characteristics of Eurasian H9 avian influenza viruses and their detection by two different H9-specific RealTime reverse transcriptase polymerase chain reaction tests. Veterinary Microbiology, 162, 530–542.
Suttie, A., Tok, S., Yann, S., Keo, P., Horm, S. V., Roe, M., Kaye, M., Sorn, S., Holl, D., Tum, S., et al. (2019). The evolution and genetic diversity of avian influenza A(H9N2) viruses in Cambodia, 2015–2016. PLoS ONE, 14, e0225428.
Youk, S. S., Lee, D. H., Jeong, J. H., Pantin-Jackwood, M. J., Song, C. S., & Swayne, D. E. (2020). Live bird markets as evolutionary epicentres of H9N2 low pathogenicity avian influenza viruses in Korea. Emerging Microbes & Infections, 9, 616–627.
Acknowledgements
We thank Jeong-Eui Lee, Byeong-Suk Jeon, and Chae-Rin Lee for technical assistance. We also thank our colleagues worldwide for their laboratory contributions, which were made available through GISAID. This research was supported by a grant from the Animal and Plant Quarantine Agency (N-1543085-2017-36-0107) of the Republic of Korea.
Funding
Animal and Plant Quarantine Agency, N-1543085-2107-36-0107.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare no conflicts of interest.
Additional information
The original online version of this article was revised: Acknowledgment and Funding information has been updated.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sagong, M., Kang, YM., Kim, N.Y. et al. Development of a Novel Korean H9-Specific rRT-PCR Assay and Its Application for Avian Influenza Virus Surveillance in Korea. J Microbiol. 61, 929–936 (2023). https://doi.org/10.1007/s12275-023-00088-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-023-00088-8