Abstract
The Gaussian-based 3D-QSAR studies for 58 selective COX-2 (cyclooxygenase-2) inhibitors belonging to benzopyran chemical class were performed. Partial least squares analysis produced statistically significant model with (R 2training = 0.866) and predictability (Q 2training = 0.66, Q 2test = 0.846). The 3D-QSAR model includes steric, electrostatic, hydrophobic, and hydrogen bond acceptor field indicators, whereas the potential field contributions indicate that the steric and hydrophobic features of the molecules play an important role in governing their biological activity. A molecular docking simulation and protein–ligand interaction pattern analysis reveal the importance of Tyr-361 and Ser-516 of the COX-2 active site for X-ray crystal structures and this class of molecules. Thus the combined approach of ligand-based and structure-based models provided an improved understanding in the interaction between benzopyran chemical class and COX-2 inhibition, which will guide the future identification of more potent anti-inflammatory drugs.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Non-steroidal anti-inflammatory drugs (NSAIDs) are the most widely used drugs worldwide for the treatment of pain and edema associated with arthritis and inflammation. Through their anti-inflammatory, anti-pyretic and analgesic activities, they represent the choice treatment in various inflammatory diseases such as arthritis, rheumatism as well as relieving the pains of everyday life. In ancient times, the first NSAID with therapeutic benefits was aspirin, which has been used in pure form for more than 100 years as a NSAID (Fiorucci et al. 2001). NSAIDs are competitive inhibitors of cyclooxygenase (COX), the enzyme which mediates the bioconversion of arachidonic acid to inflammatory prostaglandins (PGs). COX was first identified as the therapeutic target of NSAIDs when it was revealed that these anti-inflammatory substances block the biosynthesis of prostaglandins (PGs) that contribute to a variety of physiological and pathophysiological functions (Vane 1971). There are two COX isoenzymes, referred to as COX-1 and COX-2, which catalyze identical reactions (Kujubu et al. 1991; O’Banion et al. 1992; Xie et al. 1991). COX-1 is thought to produce prostaglandins (PGs) important for homeostasis and certain physiological functions and is expressed constitutively in most tissues and cells (O’Neill and Ford-Hutchinson 1993) including the gastrointestinal (GI) tract, kidney, lung, brain, and platelets, where PGs produced by this enzyme play key roles in the maintenance of various physiological functions (Otto and Smith 1995). A second, inducible form of COX was hypothesized to exist on the basis of the finding of a glucocorticoid-regulated increase in COX activity observed in vitro and in vivo in response to inflammatory stimuli (Fu et al. 1990; Masferrer et al. 1992). COX-2 is expressed in small amounts in most tissues (except in the brain, where it is expressed constitutively), but is induced to high expression in inflamed tissue by inflammatory mediators including tumor necrosis factor (TNF) and interleukin-1 (IL-1) in cells such as macrophages and synoviocytes (Crofford 1997; Seibert et al. 1994). COX-2 expression is inhibited by glucocorticoids, and the catalytic activities of both COX-1 and COX-2 are about equally inhibited by all NSAIDs; thus, NSAIDs are mixed or non-selective inhibitors of COX-1 and COX-2 (Masferrer et al. 1992). Many traditional NSAIDs are effective in treating the pain and edema associated with inflammation but their GI side effects limit their widespread use. Selective inhibition of COX-2 usually avoids the GI side effects, while retaining its efficacy. The first approved selective COX-2 inhibitor (Celecoxib) opened the area for the design and optimization of several analogs with selective inhibition. In recent years, a number of selective COX-2 inhibitors have been designed, synthesized and evaluated for their activity. In 2010, Wang et al. reported the novel benzopyran class of selective COX-2 inhibitors displayed good analgesic, anti-inflammatory and oral bioavailability relative to diaryl heterocyclic series of COX-2 inhibitors (Celecoxib) (Wang et al. 2010b).
Modern approaches for finding new leads or optimizing molecules for therapeutic targets emphasize the use of computer-aided ligand/structure-based design to reduce the number of compounds that need to be synthesized and tested. Predicting the binding mode of an inhibitor to the binding pocket of its target and rationalizing the features governing its activity through the use of 3D-QSAR approaches like CoMFA and CoMSIA has been successfully used in many applications (Cramer et al. 1988). In the present study, the pharmacological profiles of the benzopyran class of selective COX-2 inhibitors were explored using Gaussian-based 3D QSAR. The binding modes and interaction pattern profiles of various inhibitors were characterized by the application of molecular docking simulations and protein–ligand interaction fingerprint (PLIF) analysis (Deng et al. 2004; Kelly and Mancera 2004). Furthermore, the validity and reliability of the 3D-QSAR model and molecular docking were further assessed by statistical parameters and receiver operating characteristics (ROC) curve.
Materials and methods
Dataset collection
The structure and biological activity of a set of 58 novel benzopyran class of selective COX-2 inhibitors (Wang et al. 2010a, b, c) was collected from the Binding Database server (https://www.bindingdb.org). The bioactivities of the inhibitors selected were reported in the form of IC50 values. The IC50 (nM) values were converted into their negative logarithmic form (pIC50) (M) with the resulting pIC50 values ranged from 3.523 to 8.523. The 3D coordinates of the inhibitors were generated using the LigPrep module in Maestro 10.2 (Schrödinger, USA) using the following conditions: (i) the force field used was OPLS3 (Harder et al. 2016), (ii) all possible ionization states at pH (7.0 ± 2.0) were derived using the Epik method (Shelley et al. 2007) (iii) all possible tautomers and stereoisomers of the compounds were generated based on the number of chiral centers, and (iv) one low energy ring conformation per ligand was generated (Brooks et al. 2008). The energy-minimized molecules with chirality consistent with that reported in the literature were selected and the resulting dataset was divided into training (45) and test sets (13) considering that both sets contain high, medium and low active compounds with chemical diversity. The experimental and predicted pIC50 values of all molecules in the training and test sets are listed in Table 1.
Alignment based on template using Surflex-sim
The determination of the active conformation and alignment of molecules is the most important step for the generation of molecular field-based 3D-QSAR models. The success of any 3D-QSAR method relies on the appropriate relative positioning of ligands in a fixed lattice prior to the generation of molecular field values. In this study, the Surflex-Sim module of SYBYL-X was used to generate the alignment of all the molecules. This method uses a surface-based morphological similarity function to align molecules and considers molecular shape, hydrogen bonding and electrostatic properties (Jain 2000). The co-crystal bound ligand (5c-S) in protein complex (PDB: 3LN0) was chosen for the first phase of alignment. As the co-crystal ligand in 3LN0 is a representative molecule of dataset, the co-crystal ligand was extracted (Wang et al. 2010b) and 29c_2 molecule of the dataset were aligned using the mutual alignment mode. This generates multiple hypotheses and ranks according to a similarity score value. The top similarity score hypothesis was selected as template for aligning the rest of the molecules using a flexible superposition approach. Figure 1 shows the alignment of all the inhibitors from the dataset.
Molecular docking using GLIDE
Molecular docking was carried out using the extra-precision (XP) mode of Glide 6.7 (Schrödinger). Glide (Grid-based Ligand Docking with Energetics) (Friesner et al. 2004) uses a hierarchical series of filters to search for possible locations of the ligand in the active-site region of a receptor. The shape and properties of the receptor are represented on a grid by several different sets of molecular fields that provide progressively more accurate scoring of the ligand poses. The protein structure (PDB ID: 3LN0) was prepared using the protein preparation wizard in Maestro 10.2 (Schrödinger) using its standard protocol and restrained protein minimization was performed using the OPLS3 force field until the energy converges with a RMSD of 0.3 Å. The prepared protein structure complex was then used to generate a receptor grid using a grid box size of 64 Å × 64 Å × 64 Å with its centroid located on the bound ligand (5c-S). The OPLS-AA force field (Siu et al. 2012) was used for grid-based energy evaluation and refinement of docking solutions including torsional and rigid-body movements of ligands. A small number of docking solutions were then subjected to Monte Carlo energy minimization (Friesner et al. 2006). The final energy evaluation was done with Glide score (GScore):
where vdW is the van der Waals energy, Coul is the Coulomb energy, Lipo is the lipophilic contact energy, Hbond is the hydrogen-bonding energy, Metal is the metal binding energy, Rewards is the rewards and penalties for buried polar groups, hydrophobic enclosures, correlated hydrogen bonds, etc., RotB is the penalty for freezing rotatable bonds, and Site is the polar interactions in the active site. A single best pose was generated as the output for each ligand and taken for further analysis.
Validation of docking protocol
Before docking the dataset of molecules, the use of Glide was validated by both a redocking approach and an enrichment study. In the redocking approach, the bound ligand found in the X-ray crystal structure was extracted and docked back into the binding pocket of the enzyme. This was done to ensure that Glide could reproduce the orientation and position of the inhibitors as observed in the X-ray structure. In the enrichment study, the performance of Glide was tested to determine whether it can discriminate active from inactive molecules. For this purpose, Glide decoys (532) were mixed with the active molecules (activity >7.167) of the benzopyran dataset and docked into the binding pocket. A receiver operating characteristic (ROC) curve and enrichment factor (EF) were calculated in order to determine the performance of Glide. The performance in a ROC curve can be measured as the area under the curve (AUC), which should have values between 0 and 1. A value of AUC = 1 indicates complete discrimination between active and inactive molecules. Similarly, EF is defined as the fraction of active compounds found divided by the fraction of the screened library (Kumar et al. Kumar and Tiwari 2013; Poongavanam and Kongsted 2013). Enrichment calculations were performed using the following equation as available in the program:
Gaussian-based 3D-QSAR model
Gaussian-based 3D-QSAR analysis was performed using the molecular field-based QSAR tool in Maestro 10.2 (Schrödinger) (Cappel et al. 2015). Generally, any 3D-QSAR method constructs a model by correlating the activities and the 3D molecular field values of a set of aligned molecules. In molecular field-based QSAR methods, two kinds of field values are used. The first molecular field type is similar to CoMFA, which consists of a steric field based on Lennard-Jones and electrostatic potentials, with the latter having a distance-dependent dielectric with a 1/r2 dependence. The second molecular field type is based on Gaussian-based potential similar to CoMSIA, consisting of steric, electrostatic, hydrophobic, H-bond acceptor, and H-bond donor fields. The dataset of benzopyrans was divided into training and test sets for the development of a 3D-QSAR model and its validation, respectively. The Gaussian-based potential function was selected to build the 3D-QSAR model using Partial Least Squares (PLS) regression. During model building, the following parameters were used: grid spacing: 1.0 Å; Extend grid by 3 Å beyond the training set limits; Ignore force fields within 2.0 Å of any training set atom; Truncate steric and electrostatic force fields at 30 kcal/mol; Eliminate variables with SD < 0.01; Eliminate variables with |t-value| <2.0; Number of ligands to leave out for cross-validation: 1 (leave-one-out).
The QSAR models developed for each PLS factor were evaluated by internal and external validation parameters: squared correlation coefficient (R2), cross-validated correlation coefficient (\({\text{Q}}^{ 2}_{\text{training}}\)), \({\text{R}}^{ 2}\) scramble, Stability, Fisher test (the variance ratio, which indicates the statistical significance of model), P value (level of variance ratio, with small P values usually indicating a greater degree of significance), SD (standard deviation of the regression), \({\text{RMSE}}_{\text{test}}\) (root-mean-square error of the test set), \({\text{Q}}^{ 2}_{\text{test}}\) (value for the predicted activities of the test set), and Pearson-r (correlation coefficient between observed and predicted activities for the test set).
Gaussian-based 3D-QSAR model generation workflow
Successful generation of a 3D-QSAR model requires the correct alignment in 3D of the optimized conformations of all the molecules. As numerous bioactive conformations of inhibitors of COX-2 are available, an alignment could be done based on these bioactive conformations. For this purpose, PDB structure 3LN0 containing bound ligand (5c-S), one of the dataset inhibitors, was considered for alignment using Surflex-Sim. The aligned dataset of molecules was imported into Maestro 10.2 and Gaussian-based 3D-QSAR models were generated using PLS statistics.
Protein–ligand interaction fingerprint analysis
A detailed understanding of protein–ligand interactions is an important part of drug design. To aid the visualization of protein–ligand complexes, the protein–ligand interaction fingerprint (PLIF) approach available in MOE2009.10 (Molecular Operating Environment) was applied to better understand the ligand–protein interactions. The PLIF approach was applied to the 23 known X-ray crystal structures of inhibitors of COX-2 enzyme obtained from the Protein Data Bank to reveal the similarity and diversity of their binding interactions. Since the structure 3LN0 was chosen as the reference receptor, the protein–ligand interaction fingerprints of its cognate ligand and those of all other ligands were generated from their docked poses (Table S1; Figure S1 in the Supporting Information). The protein–ligand interaction fingerprints of the cognate ligand and the dataset molecules were compared and contrasted to investigate the similarity and diversity of interactions.
Results and discussion
Molecular docking
The aim of the molecular docking simulations was to predict the binding affinity and interactions of benzopyran derivatives known to modulate the activity of human COX-2 (Wang et al. 2010b). Validation of the docking program Glide by predicting the binding interactions of inhibitor 5c-S, as shown in Fig. 2, with the best binding pose exhibiting a low RMSD of 0.1031 Å and revealing excellent agreement between the predicted and experimental binding pose. All the benzopyran analogs and the standard drug Celecoxib were then docked into the binding site of COX-2 to predict their binding affinity and interactions (Table S2; Figures S2, S3 in the Supporting Information). Docking of Celecoxib resulted in a prediction of high binding affinity, with a docking score of −11.241 and the formation of strong H-bonds to important binding pocket polar amino acid residues Tyr-371 and Ser-516. On the other hand, docking of the benzopyran analogs predicted somewhat lower binding affinities with docking scores between −10.924 to −2.342, with all molecules forming between 1 and 2 H-bonds to the same polar amino acid residues Tyr-371 and Ser-516, and occasionally a H-bond to Tyr 341. A common binding mode was observed for benzopyran molecules and were predicted to be within a radius of 3 Å with diverse properties of amino acid residues in the binding site: hydrophobic, for example, Val-335, Leu-338, Met-508, Val-509, Ala-513, Leu-517, and Gly-512; aromatic (hydrophobic), for example, Tyr-341, Tyr-371, Trp-373, and Phe-504; polar uncharged, for example, Ser-339, and Ser-516. It was further noted that, since benzopyran molecules possess a stereocenter at C-2 position, they may exist in two possible conformations. The molecular docking simulation reveal both conformations, where the trifluoromethyl group at C-2 position is opposite in both cases in the binding pocket comprised of Val-335, Leu-338, Met-508 and Val-509 amino acid residues. Since the majority of interactions between ligands and protein were hydrophobic in nature, this suggests that strong hydrophobic interactions with COX-2 play a key role in determining the binding affinity and interactions of this class of molecules.
Enrichment study
Since the molecular docking scores for dataset molecules poorly correlated with experimental activity (pIC50 (M)), (data not shown here) thus it is worthwhile to perform enrichment study in order to assess, whether, current molecular docking protocol is able to discriminate the actives from inactives/decoys. Thus in enrichment study, a total 25 known active ligands and 532 decoys were docked into the binding pocket of COX-2 (PDB structure 3LN0) and the top ranked pose for each ligand (based on the Glide score) was considered. As shown in Figure S4 (A), the ROC curve shows that the Glide docking protocol followed effectively discriminates between the active and inactive molecules, with an AUC of 0.742. In Figure S4 (B), the blue stepped line displays a standard enrichment curve of the fraction of active molecules found against the fraction of database screened. The count and percentage of active molecules in the top N% of decoys were 1% (12 and 48%), 5% (17 and 68%) and 20% (19 and 76%). Similarly, the enrichment factor in the top 1, 5 and 20% were 22, 14, and 3.8, respectively. The enrichment study clearly indicates the docking program is able to decrease the number of false positive and false negatives in virtual screening. The ROC curve and screen results support the reliability of Glide for further use in virtual screening for hit identification.
Gaussian-based 3D-QSAR analysis
The aligned dataset of molecules obtained from Surflex-Sim was subjected to Gaussian-based 3D-QSAR modeling using PLS statistics with different field contributions values. In order to select the best field combination model and avoid the over-fitting problem, the stability statistic was considered. As a rule of thumb, the optimum number of PLS factor for a robust model should have higher stability and lower \({\text{R}}^{ 2}_{\text{Scramble}}\) values (<0.5). Based on this criterion, two models with field contribution of (Steric + Hydrophobic + Hydrogen bond acceptor) and (Steric + Electrostatic + Hydrophobic + Hydrogen bond acceptor) have higher stability values of 0.873, 0.867 and lower \({\text{R}}^{ 2}_{\text{Scramble}}\) of 0.465 and 0.489 at PLS factor 3, moreover their internal (\({\text{Q}}^{ 2}_{\text{training}}\)) validation parameters have values of 0.638 and 0.660 respectively. Comparative statistical analysis afforded a robust model with \({\text{r}}^{ 2}\) = 0.866, \({\text{Q}}^{ 2}_{\text{training}}\) = 0.660, \({\text{Q}}^{ 2}_{\text{test}}\) = 0.846, and \({\text{RMSE}}_{\text{test}}\) = 0.41, exhibiting good correlation with gaussian steric, electrostatic, hydrophobic and hydrogen bond acceptor (HBA) field values, along with their potential field contributions being 0.34, 0.125, 0.327, 0.204, respectively. The statistical summary of the model is listed in Table 2 and in supplementary Table S3. The molecular field contribution values indicate that the steric and hydrophobic features of the molecules play an important role in governing the biological activity, which further supports the molecular docking findings, which revealed that the hydrophobic interaction is a major contributor in modulating the binding affinity. Moreover, the contour maps were superimposed in the protein binding pocket to further aid in rationalizing the structural features responsible for activity. The contribution of different molecular field intensities is given in Table 2.
Gaussian steric contour maps
In this work, the steric field is represented by green and yellow contour maps, in which green coloured regions indicate areas where increased bulk is associated with enhanced activity and yellow regions suggest areas where increased steric bulk is unfavourable to activity. Figure 3a shows only two green contour areas. In order to aid the visualization, the potent compound (5a) is overlaid on the map of contours along with the protein binding pocket. A large green contour is located over the region containing C-2 substituents. It is noteworthy to mention here that C-2 is a chiral center and many molecules exist as either R or S stereoisomers. The green contour suggests that the R-stereoisomer of benzopyran molecules are better inhibitors of COX-2 compared to their S-stereoisomer counterparts. Moreover, a small green contour at a distance from C-8 position suggests linear alkyl substituents like in compounds 29c, 29d, 32a favours the activity.
Contour maps in protein binding pocket for Gaussian-based 3D-QSAR model. a Steric favored regions are shown in green and disfavored regions are shown in yellow. (presented as 29d_2: ball-stick; 5a: tube; 29c_2: thin-tube). b Electrostatic favorable electropositive regions are shown in blue and favorable electronegative regions are shown in red (presented as 29c_2: ball-stick; 19: tube; 23b: thin-tube)
Gaussian electrostatic contour maps
In this work, the electrostatic field is represented by blue and red contour maps, where blue areas are the regions where electronegative substituents are unfavourable to activity, while red areas are the regions where electronegative substituents are favourable to activity (Fig. 3b). All benzopyran molecules have a carboxylic group at the C-3 position and are supported by red contour map at that position. Similarly, a red contour at C-6 position suggests that electronegative substituents like (–Cl, –CCH, –OCF3) are favourable for activity. The electrostatic contour map also shows two blue regions around the C-5 and C-7 positions, which reflect substituents of compounds (7, 34a, 34b) and (23b, 23c, 23d, 23e, 29c_2, 29d, 29e, 32a, 32b), respectively, and which are potent molecules in the dataset. However, low activity compounds (19, 20) with -Ph and -MePh substituents at the C-5 position suggests that electropositive substituents are favourable for activity. Moreover, a large red contour was observed over the C-7 position, suggest that substituents with electronegative atom (–O–) as a linker is favourable for activity (compounds 23b, 23c, 23d).
Gaussian hydrophobic contour maps
In this work, the hydrophobic fields are represented as yellow and white contour maps, where yellow regions are favourable and white regions are unfavourable for activity, respectively. In Fig. 4a, a large yellow contour region at the C-5 and C-6 positions indicates that compounds 7, 7_2, 5a, 5 g, 34a, and 34b have hydrophobic groups that are well tolerated at that position. As discussed previously in relation to steric contour maps, substituents at the C-2 position in the R-stereoisomers of the molecules show good activity compared to the S-stereoisomers. Similarly, S-stereoisomers of the molecules exhibit a white contour region at the C-2 position (–CF3). Similarly, four white contour regions occupying the space at certain distance for substituents at C-5 and C-7 positions suggest that extended network of linear or branched substituents are unfavourable for activity as evidenced by compounds (12, 19, 20, 26a, 26b, 26c, 26d, 26e). It further suggests that careful selection of hydrophobic groups for substitutions is needed. Furthermore, a large yellow contour map in between C-7 and C-8 further support that, hydrophobic groups with not too much bulky substituent is favouring the activity (23b, 23c, 23d, 29c_2, 29d_2, 32a). It is worth mentioning here that many structural features of the hydrophobic contour map pattern follow the steric contour map pattern, as expected.
Contour maps in protein binding pocket for Gaussian-based 3D-QSAR model. a Hydrophobic favorable regions are shown in yellow and unfavorable regions are shown in grey (presented as 29c_2: ball-stick; 5a: tube; 26d_2: thin-tube). b Hydrogen bond acceptor favorable groups are shown in red and unfavorable groups are shown in magenta (presented as 8e: ball-stick; 23b: tube; 33: thin-tube)
Gaussian hydrogen bond acceptor contour maps
In this work, hydrogen-bond acceptor (HBA) fields are represented as red and magenta contour maps, where red regions correspond to favourable groups and magenta regions correspond to unfavourable groups. In Fig. 4b, a large red contour region near the C-7 position in compounds 23b, 23c, 23d, 23e, 32a and 33 suggests that the presence of oxygen or sulphur as a HBA linker between terminal substituents and the C-7 carbon enhance activity. In the dataset, the majority of molecules (R-stereoisomers) contain –CF3, substituent at the C-2 position. These groups are not good hydrogen-bond acceptors, which is associated with the presence of a magenta contour map at the neighbouring C-2 position. Similarly, a large magenta contour map is found at a distance near the C-8 position. On mapping of compounds 8e and 8f, the magenta contour is found over the oxygen functionality of –SO2NH2 group and comparing the activity, it revealed that the extension of substituents at C-8 position (8f) is unfavourable for activity.
The structural requirements in terms of contour maps corroborate with the protein binding amino acid residues and further supports the field based 3D-QSAR hypothesis.
Protein–ligand interaction fingerprint analysis
The PLIF analysis is used to study the interaction between the residues in the protein and the ligands on a superimposed structure of complexes. The derived fingerprint bits are used to investigate the common residues that interact with the ligand. In this study, protein–ligand interaction fingerprints (PLIF) was used to analyze the similarity and diversity of interactions with COX-2 between inhibitors taken from known X-ray crystal structures and the dataset molecules. The fingerprint bits created from the PLIF analysis for the known X-ray crystal structures and the dataset molecules are displayed in Table S4, S5 and Figure S5. The fingerprint bits in Table S4 and S5, showed that the ligand in both kind of complexes (X-ray crystal structures and dataset molecules) have interaction commonly with amino acids such as Arg-106, Leu-338, Ser-339, Tyr-341, Tyr-371, Arg-499, Phe-504 and Ser-516 amino acid residues through side chain H-bond acceptor, donor, and Ionic interaction. The dataset molecules additionally, showed backbone H-bond acceptor interaction with Gln-178, Met-508 amino acid residues. Such interactions are uncommon in known inhibitors, however, these residues are in vicinity of the binding pocket, further revealed the structural diversity of dataset molecules, and capable of making additional interaction, which stabilizing the inhibitors in COX-2 binding pocket. Figure S5 shows a histogram of the frequencies of the different interaction made by the known inhibitors and dataset molecules respectively. The interaction matrices for known inhibitors and dataset molecules revealed the similarity of the binding interaction profiles for both sets of molecules and suggest the existence of a common binding mode and interaction pattern.
Validation of Gaussian-based 3D-QSAR models
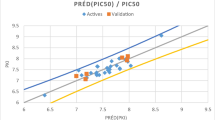
Experimental versus predicted activities for both the training and test set inhibitors are shown in Fig. 5, which represents the above described Gaussian-based 3D-QSAR model. The correlation coefficients for the selected model at PLS factor 3 for the training and test sets are 0.866 and 0.949 respectively. The statistical validation parameters as recommended by Tropsha et al. (Golbraikh and Tropsha 2002) were calculated to determine the robustness and reliability of the model. The predictive power of the model was further accessed by calculating additional validation parameters such as the modified \({\text{r}}^{ 2}\) term (\({\text{r}}^{ 2}_{\text{m}}\)), and \({\text{R}}^{ 2}_{\text{p}}\) as reported by Pratim Roy et al. (2009). The values of all of these parameters are shown in Table 3, which suggest a good level of robustness and reliability of the selected model.
The Gaussian-based 3D-QSAR model shows strong predictive power and provides detailed information regarding the structural features of the molecules that govern their inhibitory activity. Figure 6 summarises the positive and negative COX-2 inhibition activity coefficients for the core benzopyran structure: (i) an electronegative group (pink color) at position C-6 of the benzopyran ring favours activity, (ii) an electropositive group (blue colour) at the C-7 position favours activity, iii) a hydrophobic and bulky group at the C-6 position (yellow and green colour) favours activity, and (iii) HBA (magenta) in C-6 substituents favours activity.
Conclusions
A molecular modelling study was carried out to gain insight into the structural basis of the inhibitory properties of the novel benzopyran class of COX-2 inhibitors. A Gaussian-based 3D-QSAR model was developed which not only exhibited good predictive powers in both the internal and external validations, but also identified critical structural features influencing the inhibitory activity. The statistically robust QSAR model showed \({\text{R}}^{ 2}_{\text{training}}\) = 0.866, \({\text{Q}}^{ 2}_{\text{training}}\) = 0.66, \({\text{RMSE}}_{\text{test}}\) = 0.41, \({\text{Q}}^{ 2}_{\text{test}}\) = 0.846 and \({\text{r}}^{ 2}_{\text{m}}\) = 0.866, based on steric, electrostatic, hydrophobic, and HBA, respectively. The analysis of contour field maps revealed the suitable size and shape of substituents at the C-5, C-6 and C-7 positions enhance activity. The stereochemistry at the C-2 position also modulates activity, such that R-stereoisomers are favourable for activity. Similarly, electronegative and electropositive groups at the C-6 and C-7 positions, respectively, modulate activity. Molecular docking simulations predicted the binding modes of these molecules and their key interaction features. The carboxylic group at the C-3 position appears to be responsible for establishing strong hydrogen bonds with Tyr-371 and Ser-516 in the binding site. Analysis of protein–ligand interaction fingerprints further revealed the existence of a common set of residues (Arg-106, Ser-339, Tyr-341, Tyr-371 and Ser-516) that interact with all known inhibitors as well as the benzopyran molecules. These findings provide useful insights into the binding mechanism between COX-2 and inhibitors that may be of value for the future design of novel potent COX-2 inhibitors.
References
Brooks WH, Daniel KG, Sung SS, Guida WC (2008) Computational validation of the importance of absolute stereochemistry in virtual screening. J Chem Inf Model 48:639–645
Cappel D, Dixon SL, Sherman W, Duan J (2015) Exploring conformational search protocols for ligand-based virtual screening and 3-D QSAR modeling. J Comput Aided Mol Des 29:165–182
Cramer RD, Patterson DE, Bunce JD (1988) Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J Am Chem Soc 110:5959–5967
Crofford LJ (1997) COX-1 and COX-2 tissue expression: implications and predictions. J Rheumatol Suppl 49:15–19
Deng Z, Chuaqui C, Singh J (2004) Structural interaction fingerprint (SIFt): a novel method for analyzing three-dimensional protein–ligand binding interactions. J Med Chem 47:337–344
Fiorucci S, Meli R, Bucci M, Cirino G (2001) Dual inhibitors of cyclooxygenase and 5-lipoxygenase. A new avenue in anti-inflammatory therapy? Biochem Pharmacol 62:1433–1438
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47:1739–1749
Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT (2006) Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem 49:6177–6196
Fu JY, Masferrer JL, Seibert K, Raz A, Needleman P (1990) The induction and suppression of prostaglandin H2 synthase (cyclooxygenase) in human monocytes. J Biol Chem 265:16737–16740
Golbraikh A, Tropsha A (2002) Beware of q2! J Mol Gr Model 20:269–276
Harder E, Damm W, Maple J, Wu C, Reboul M, Xiang JY, Wang L, Lupyan D, Dahlgren MK, Knight JL, Kaus JW, Cerutti DS, Krilov G, Jorgensen WL, Abel R, Friesner RA (2016) OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J Chem Theory Comput 12:281–296
Jain AN (2000) Morphological similarity: a 3D molecular similarity method correlated with protein-ligand recognition. J Comput Aided Mol Des 14:199–213
Kelly MD, Mancera RL (2004) Expanded interaction fingerprint method for analyzing ligand binding modes in docking and structure-based drug design. J Chem Inf Comput Sci 44:1942–1951
Kujubu DA, Fletcher BS, Varnum BC, Lim RW, Herschman HR (1991) TIS10, a phorbol ester tumor promoter-inducible mRNA from Swiss 3T3 cells, encodes a novel prostaglandin synthase/cyclooxygenase homologue. J Biol Chem 266:12866–12872
Kumar S, Tiwari M (2013) Variable selection based QSAR modeling on Bisphenylbenzimidazole as inhibitor of HIV-1 reverse transcriptase. Med Chem 9:955–967
Masferrer JL, Seibert K, Zweifel B, Needleman P (1992) Endogenous glucocorticoids regulate an inducible cyclooxygenase enzyme. Proc Natl Acad Sci USA 89:3917–3921
O’Banion MK, Winn VD, Young DA (1992) cDNA cloning and functional activity of a glucocorticoid-regulated inflammatory cyclooxygenase. Proc Natl Acad Sci USA 89:4888–4892
O’Neill GP, Ford-Hutchinson AW (1993) Expression of mRNA for cyclooxygenase-1 and cyclooxygenase-2 in human tissues. FEBS Lett 330:156–160
Otto JC, Smith WL (1995) Prostaglandin endoperoxide synthases-1 and -2. J Lipid Mediat Cell Signal 12:139–156
Poongavanam V, Kongsted J (2013) Virtual screening models for prediction of HIV-1 RT associated RNase H inhibition. PLoS ONE 8:e73478
Pratim Roy P, Paul S, Mitra I, Roy K (2009) On two novel parameters for validation of predictive QSAR models. Molecules 14:1660–1701
Seibert K, Zhang Y, Leahy K, Hauser S, Masferrer J, Perkins W, Lee L, Isakson P (1994) Pharmacological and biochemical demonstration of the role of cyclooxygenase 2 in inflammation and pain. Proc Natl Acad Sci USA 91:12013–12017
Shelley JC, Cholleti A, Frye LL, Greenwood JR, Timlin MR, Uchimaya M (2007) Epik: a software program for pK(a) prediction and protonation state generation for drug-like molecules. J Comput Aided Mol Des 21:681–691
Siu SW, Pluhackova K, Bockmann RA (2012) Optimization of the OPLS-AA force field for long hydrocarbons. J Chem Theory Comput 8:1459–1470
Vane JR (1971) Inhibition of prostaglandin synthesis as a mechanism of action for aspirin-like drugs. Nat New Biol 231:232–235
Wang JL, Aston K, Limburg D, Ludwig C, Hallinan AE, Koszyk F, Hamper B, Brown D, Graneto M, Talley J, Maziasz T, Masferrer J, Carter J (2010a) The novel benzopyran class of selective cyclooxygenase-2 inhibitors. Part III: the three microdose candidates. Bioorg Med Chem Lett 20:7164–7168
Wang JL, Carter J, Kiefer JR, Kurumbail RG, Pawlitz JL, Brown D, Hartmann SJ, Graneto MJ, Seibert K, Talley JJ (2010b) The novel benzopyran class of selective cyclooxygenase-2 inhibitors-part I: the first clinical candidate. Bioorg Med Chem Lett 20:7155–7158
Wang JL, Limburg D, Graneto MJ, Springer J, Hamper JR, Liao S, Pawlitz JL, Kurumbail RG, Maziasz T, Talley JJ, Kiefer JR, Carter J (2010c) The novel benzopyran class of selective cyclooxygenase-2 inhibitors. Part 2: the second clinical candidate having a shorter and favorable human half-life. Bioorg Med Chem Lett 20:7159–7163
Xie WL, Chipman JG, Robertson DL, Erikson RL, Simmons DL (1991) Expression of a mitogen-responsive gene encoding prostaglandin synthase is regulated by mRNA splicing. Proc Natl Acad Sci USA 88:2692–2696
Acknowledegements
We acknowledge the Science & Engineering Research Board (A Statutory body under the Department of Science & Technology, Government of India), New Delhi for financial support through the Young Scientist Project SB/YS/LS-130/2014 at the All India Institute of Medical Sciences (AIIMS), Jodhpur, India and National Research Foundation of Korea (NRF), which is funded by the Ministry of Education, Science, and Technology (No: 2012R1A6A3A04038302 and 2017R1C1B2003380).
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: DKY, SK. Performed the experiments: DKY, Saloni, SK, HS. Analyzed the data: DKY SK, Saloni. Contributed reagents/materials/analysis tools: DKY, SK, SM, HS, PS, SM, KK, JC, MHK and HPS. Wrote the paper: DKY, SK and RLM.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have declared that no competing interests exist.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yadav, D.K., Saloni, Sharma, P. et al. Studies of the benzopyran class of selective COX-2 inhibitors using 3D-QSAR and molecular docking. Arch. Pharm. Res. 41, 1178–1189 (2018). https://doi.org/10.1007/s12272-017-0945-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12272-017-0945-7