Abstract
The aim of this study is to develop an automated deep-learning-based whole heart segmentation of ECG-gated computed tomography data. After 21 exclusions, CT acquired before transcatheter aortic valve implantation in 71 patients were reviewed and randomly split in a training (n = 55 patients), validation (n = 8 patients), and a test set (n = 8 patients). A fully automatic deep-learning method combining two convolutional neural networks performed segmentation of 10 cardiovascular structures, which was compared with the manually segmented reference by the Dice index. Correlations and agreement between myocardial volumes and mass were assessed. The algorithm demonstrated high accuracy (Dice score = 0.920; interquartile range: 0.906–0.925) and a low computing time (13.4 s, range 11.9–14.9). Correlations and agreement of volumes and mass were satisfactory for most structures. Six of ten structures were well segmented. Deep-learning-based method allowed automated WHS from ECG-gated CT data with a high accuracy. Challenges remain to improve right-sided structures segmentation and achieve daily clinical application.
Graphical abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Tridimensional (3D) cardiac imaging has emerged as a cornerstone in the evaluation and the management of patients in contemporary cardiology. As the number of procedures dedicated to the treatment of a variety of arrhythmias or structural heart diseases is exponentially growing [1,2,3,4,5,6,7], 3D cardiac imaging, providing a precise anatomic description of cardiothoracic structures, now plays a crucial role in patients selection and procedural planning [8,9,10,11]. Manually obtaining an accurate anatomic description of the heart and surrounding structures may be a tedious task and may suffer from reproducibility issues [12], which highlights the challenges posed by whole heart segmentation (WHS).
Recently, artificial intelligence (AI) has also established itself as an appealing tool in the analysis of multiple functional and anatomic parameters from imaging data [13,14,15,16]. Allowing user-free processes and high reproducibility, it appears promising for procedural planning, when a description of each cardiac chamber and structure is required. Several works evaluating WHS from computed tomography (CT) imaging have already been performed [17,18,19,20,21]. However, some of these studies were based on multi-atlas segmentation, which seems to be less accurate and more time consuming than deep-learning-based methods [20]. Furthermore, owing to the computing power required for the algorithms, to date none of the previously published work integrated the automatic segmentation process in a readily useable and standalone software, which may impede the use of these methods in daily practice, despite the good results reported in these studies.
In the present work, we developed and validated a deep-learning-based WHS segmentation process from a dedicated CT database, which was fully integrated in a routinely used procedural planning software.
Methods
Data Acquisition
Imaging data used in the present work were ECG-gated CT performed during the pre-procedural work-up of patients undergoing transcatheter aortic valve implantation (TAVI) at our institution. CT of consecutive TAVI recipients between May and September 2019 were analyzed. Exclusion criteria were image quality not allowing precise manual segmentation of the structures of interest, either because of artifacts or poor image contrast, which was assessed by the expert physician performing manual segmentation, and patients with prior surgical aortic valve replacement or TAVI. All patients gave written informed consent for the procedures and anonymous collection of their data, which were prospectively gathered in an electronic database as part of a national registry [3]. The present study was not pre-specified, observational and retrospective. Thus, the institutional review board waived specific consent for this study.
CT Acquisition Protocol
All patients underwent a cardiac CT scan for the procedural planning of TAVI according to established consensus [8]. CT scan was performed on a third generation dual-source CT scanner (SOMATOM Force, Siemens Healthcare, Forchheim, Germany). No systematic intravenous beta-blocker was used before CT scan. A prospective ECG-triggered high-pitch CT angiography extending from the carotid to the femoral arteries was performed during a single breath-hold. ECG gating was set on end-systole (30% interval of the cardiac cycle) and activated to trigger the acquisition on cardiac volume only. Acquisition parameters were as follows: collimation 192 × 0.6 mm, gantry rotation time 250 ms, fixed tube voltage 100 kV, current–time product ranging from 342–604 mAs, and spiral pitch factor 3.2. A bolus of 90 mL of iobitridol (Xenetix 300, Guerbet, Roissy, France) was injected at 4 mL/s, followed by a 40-mL saline chaser bolus. An automated bolus tracking system was used to synchronize the arrival of the contrast material with the initiation of the scan. Cardiac dataset was reconstructed using a 181-mm FOV, a 512 × 512 matrix, a Bv40 kernel and iterative reconstruction technique (Admire level 3, Siemens).
CT Manual Segmentation Protocol
For each CT, an expert interventional cardiologist with a cardiac imaging degree and extensive experience in the field of cardiac CT and TAVR performed a manual segmentation of the heart and surroundings large vessels using the Endosize© software (Therenva, Rennes, France), a CE- and FDA-marked medical device for planning and sizing of endovascular procedures [22]. The following ten structures were segmented: pulmonary veins (PV), left atrium (LA), left ventricular cavity (LVC), left ventricular myocardium (LVM), aorta (Ao), coronary sinus (CS), superior vena cava (SVC), right atrium (RA), right ventricular cavity (RVC), and pulmonary artery (PA). The segmentation method of the different elements has been previously described [20, 23,24,25]. LA and RA segmentation included the appendage. For the left ventricular myocardium, considering the end-systolic phase acquired, the frequent LV hypertrophy among TAVI recipients, and the procedural planning perspective of our work, we decided to include the main papillary muscles, i.e., exclude them from the cavity. The different cavities were delineated by identifying the endocardial border [21]. LVM mass was calculated as the left ventricular myocardial volume derived by the delineation of its endocardial and epicardial borders and multiplied with the specific gravity of myocardial tissue (assuming a tissue density of 1.05 g/mL) [26]. These segmentations were considered as the reference ground truth for our deep-learning model.
CT Automatic Segmentation Protocol
The automatic deep-learning-based WHS segmentation process was divided into two distinct stages: a localization step, which automatically selects the aortic valve to generate the region of interest (ROI) in the CT volume, and the segmentation step.
In the present study, the localization step was performed by using a regression convolutional neural network (CNN [27]) based on the SqueezeNet architecture. In contrast with classification approaches, during which CNNs detect the presence or absence of anatomical target structures in each of the orthogonal viewing planes independently and then combine them to obtain the coordinates of each landmark, here the distance (in mm) between the current slice and the slice belonging to the anatomical structure is used. After parsing the entire volume, distances can be converted to parabolic curves using polynomial regression, where the minimum is searched for each axis. The center of the aortic valve (defined as the intersections of the three commissures) was defined as the anatomical landmark to be found. 3D images were converted into three sets of 2D images for the axial, sagittal, and coronal axis, respectively, and preprocessed with cropping and padding operations. For each axis, a regression CNN was trained with the 2D image slice as input. Specifically, the VGG-16 CNN architecture [28] was adapted to output quantitative values by modifying the softmax loss layer with a Euclidean loss layer. All other weights were initialized from pre-training on the ImageNet database (accessed at http://www.image-net.org). The three networks were trained separately and did not share weights. Convergence was obtained after 100 epochs. Following this approach, the exact position of the aortic valve was estimated. After this automatic detection of the aortic valve, in the segmentation step, 3D data were resized to a 320 × 320 × 320 ROI and resampled to 0.7 × 0.7 × 1 mm by voxel to include all structures of interest. These preprocessed volumes were then used as the input dataset of the Dense V-Net 3D segmentation CNN [29]. Briefly, the volume is cropped into voxels batches and a succession of convolutional 3D 3 × 3 × 3 kernels, dense connections, and batches normalizations are applied to extract 3D spatial information from each batch. The resultant feature maps are then down-sampled by 2 and the operation is repeated 3 times. At the end, the feature maps are up-sampled to the original resolution and a softmax layer gives the result class for each voxel. Adam optimizer was preferred for training with an initial learning rate of 0.001, along with a loss combining the dice and cross-entropy coefficients. The output of the network is directly a 3D mask with one label for each structure. The network was trained for 300 epochs during 6 days on a Nvidia RTX 2070 GPU. The Niftynet open-source framework (https://niftynet.io/) was used for the training and the validation of the network. The complete workflow of the segmentation process is illustrated in the Fig. 1.
Deep-learning-based whole heart segmentation workflow. A. Native sagittal plane; B. Native coronal plane; C. Native axial plane. D. SqueezeNet ROI detection regression CNN localizes the center of the aortic valve in the entire CT volume allowing its resizing to a 320 × 320 × 320 ROI including all structures of interest. E. Dense V-Net 3D segmentation CNN performs the multilabel automatic segmentation. CNN: convolutional neural network; CT: computed tomography; ROI: region of interest
CT Automatic Segmentation Testing
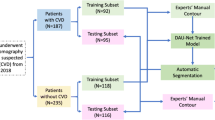
The dataset was randomly split in a training set (n = 55 patients, 17,600 slices), a validation set (n = 8 patients, 2560 slices), and a test set (n = 8 patients, 2560 slices). For testing purposes, the segmentation method was integrated in the Endosize® software (Therenva, Rennes, France). The automatic segmentations in the test set were obtained directly from this routinely used software using a standard 2-GHz workstation with 2 GB of RAM to evaluate the clinical applicability of our method.
WHS obtained using the deep-learning approach were compared with the manually segmented reference from the test set. The image-based performance metric was the Dice index [30]. Dice similarity score quantifies the voxel-wise degree of similarity between the model predicted segmentation mask and the ground truth and ranges from 0 (no similarity) to 1 (identical). Mathematically, it can be expressed as follows:
Statistical Analysis
Continuous variables are presented as mean ± standard deviation or median (interquartile [IQR] or full range) depending on their distribution, which was assessed using the Shapiro–Wilk test. Categorical variables were summarized as numbers (percentages). Dice scores were summarized as medians and quartiles. Comparisons between groups were performed with the use of the Kruskal–Wallis and the Fisher exact test for continuous and categorical variables, respectively. Levels of agreement between the automatic and manual segmentations were assessed on the test set with the Bland–Altman difference against mean plot. Pearson correlation coefficients of volumes between the manual reference and automatic prediction were also evaluated. Volumes measured by the automatic segmentation were compared with the manual segmentation results using the Wilcoxon’s signed rank test. Statistical analyses were conducted using the Statistical Package for Social Sciences version 25 (SPSS Inc., IBM, Armonk, NY).
Results
Population
Between May 15 and September 4, 2019, 107 consecutives patients underwent TAVI at our institution. Among them, nine had a history of aortic valve replacement whereas 21 had poor-image quality on their pre-procedural CT and were excluded (Supplemental Fig. 1). Moreover, the CT of six patients were successfully manually segmented but presented technical issues (mainly important motion artifacts in five patients), which precluded their analysis by the deep-learning-based algorithm. Therefore, these patients were excluded from the study population leaving 71 patients for analysis. Baseline characteristics of included patients are described in Table 1. Patients from the training and validation sets were comparable to patients from the test set at the exception of a lower body surface area in the test set. The 3D imaging dataset consisted of 2.064 billion voxels, i.e., 32.768 million voxels per volume.
Manual and Automatic Deep-Learning-Based Segmentations
Manual segmentations of the ten labels took a median of 90 min/patient (range: 57 to 153 min). The performance and results of a manual segmentation are illustrated in Fig. 2.
The aortic valve position was detected in less than 2 s in a standard workstation thanks to the multi-resolution search scheme for each axis. Automatic segmentations of the ten labels took a median of 13.4 s (range: 11.9 to 14.9 s) on a standard 2-GHz computer with 2 GB of RAM in the test set.
Validation of the Automatic Deep-Learning-Based Segmentation
The combined overall Dice index for the 10 labels was 0.920 (IQR: 0.906–0.925). The median Dice scores for Ao, CS, LA, LVC, LVM, PA, PV, RA, RVC, and SVC were 0.915 (IQR: 0.902–0.930), 0.604 (IQR: 0.516–0.652), 0.939 (IQR: 0.933–0.941), 0.852 (IQR: 0.793–0.867), 0.927 (IQR: 0.923–0.940), 0.878 (IQR: 0.865–0.888), 0.657 (IQR: 0.594–0.712), 0.877 (IQR: 0.816–0.901), 0.819 (IQR:0.763–0.862), and 0.627 (0.408–0.659), respectively (Table 2, Fig. 3). Bland–Altman and linear regression plots are shown (Fig. 4), with the mean difference and limits of agreement between the manual reference and automatic prediction for Ao, CS, LA, LVC, LVM, PA, PV, RA, RVC, and SVC being − 0.41 mL (95% confidence interval [CI]: − 20.6 to 19.7), − 0.40 mL (95%CI: − 1.56 to 0.76), − 1.07 mL (95%CI: − 10.4 to 8.2), − 6.63 mL (96%CI: − 16.2 to 2.9), − 1.76 g (95%CI: − 9.5 to 6.0), 0.53 mL (95%CI: − 9.0 to 10.1), − 4.67 mL (95%CI: − 9.68 to 0.35), − 0.76 mL (95%CI: − 14.6 to 13.1), − 8.17 mL (95%CI: − 18.0 to 1.6), and 2.51 mL (95%CI: − 9.9 to 14.9), respectively. Table 3 summarizes manual and automatic volumes and mass measurements. The automatic segmentation predictions correlated poorly with the manual reference for SVC (r = 0.49, p = 0.27), marginally better for CS (r = 0.77, p = 0.02), and significantly better for all other structures. Correlation coefficients were 0.97 (p < 0.001) for Ao, 0.98 (p < 0.001) for LA, 0.99 (p < 0.001) for LVC, 0.99 (p < 0.001) for LVM, 0.96 (p < 0.001) for PA, 0.90 (p = 0.002) for PV, 0.95 (p < 0.001) for RA, and 0.98 (p < 0.001) for RVC (Fig. 4).
Linear regression (A) and Bland–Altman (B) plots of model correlation and agreement with manual annotation. Ln, natural logarithm; other abbreviations as in Fig. 3
Discussion
In the present study, we proposed a deep-learning-based method allowing fast and automated WHS from ECG-gated CT data of TAVI candidates. The chief findings of the present study are as follows: (1) the proposed deep-learning-based model displayed an overall high level of accuracy with a Dice score of 0.92. (2) There were discrepancies in the model’s accuracy according to the considered structure. Especially automatic segmentation of CS, PV, and SVC were less accurate. (3) The automatic segmentation and manually obtained reference of volumes and mass correlated and agreed well for most structures. (4) The computing time of the model, fully integrated in a standalone routinely used software, was very limited (median: 13.4 s) which represents a first step towards a potential implementation in routine practice.
WHS remains a challenging task for which emerging deep-learning methods appear as innovative and appealing tools, especially from a computational cost standpoint, compared with previously described methods [12, 17, 20]. Zhuang et al. reported the results of a worldwide challenge of multimodality WHS [20]. In this work, twelve algorithms from twelve different teams were evaluated for the automatic segmentation of seven cardiac structures (Ao, LA, LVC, LVM, PA, RA, and RVC) from CT and magnetic resonance imaging data. For the CT dataset, composed of 60 cardiac CT volumes with only 20 for training, the best Dice score was 0.908 ± 0.086 and was obtained with a mean 104-s computing time on an Intel i7-4820 K 32-GB CPU with a Nvidia GTX TITAN X 12-GB GPU. Similarly to the proposed model in the present work, the best algorithm in this challenge used two separate CNNs to first localize the ROI in the volume and then perform the pixel-wise segmentation using a volumetric kernels equipped 3D CNN. Another interesting contribution to the field recently came from Baskaran et al. who trained, validated, and tested in a 70:20:10 split dataset of 166 CT, a U-Net-inspired, deep-learning model [21]. The authors identified five cardiac structures: LA, LVC, LVM, RA, and RV. They reported an overall Dice score of 0.925 (IQR: 0.887 to 0.948) for the identification of these structures by their model with Dice score for LA, LVC, LVM, RA, and RV being 0.934, 0.938, 0.920, 0.915, and 0.927, respectively. They demonstrated a good correlation and agreement between volumes and mass predicted by the model compared with their manual ground truth in a test set encompassing 17 patients with 1477 images. The mean computing time was 13.13 s/patient but the authors did not report the characteristics of the workstation they used for this work. In the present study, we attempted to increase the number of substructures segmented by adding surroundings vessels (Ao, CS, PA, PV, and SVC) considering their potential usefulness for the procedural planning of structural interventions. Our overall Dice score of 0.920 (IQR: 0.906–0.925) is comparable with values reported in these previously published state-of-the-art works. Nonetheless, we demonstrated significant discrepancies according to the segmented structures, i.e., the model was not sufficiently accurate for small structures such as CS, PV, and SVC and was marginally less accurate for right-sided structures and the LVC. Several reasons may explain these observations. First, we used CT data obtained during the pre-procedural work-up of TAVI recipients, for which acquisition parameters intend to optimize the contrast in the aorta and peripheral vascular structures. This may explain the sub-optimal results obtained for right-sided structures. Similarly, mixing of the non-contrasted blood from the inferior vena cava and the contrast-saturated blood from the SVC results in an inhomogeneous enhancement of the RA and beam hardening artifacts, which contribute to a decreased visualization of surrounding structures (CS, SVC, RVC). Second, our TAVI recipients population was older and likely sicker than the population of Baskaran et al., which was in average 20 years younger. Interestingly, Baskaran et al. reported worse prediction for the LVC in patients older than 65 years [21]. As we wanted to evaluate the feasibility of automatic WHS in routine practice, in contrast with this previous study, we did not exclude patients with elevated heart rate or atrial fibrillation. As expected among TAVI candidates, more than one-fourth of our population suffered from atrial fibrillation, which negatively affects CT images quality and is a known contributor to sub-optimal results of deep-learning-based WHS [20]. Furthermore, one-fifth of our population harbored chronic pulmonary diseases, which may also affect image quality, especially among patients who cannot sufficiently stand apnea. Despite these limitations, we believe that the population of the present study accurately represents current structural heart interventions candidates therefore allowing a precise evaluation of the potential clinical impact of our model. Third, regarding LVC, we elected to include the papillary muscles into the LVM label in contrast with previous studies [20, 21] and usual echocardiography guidelines [23], yet in line with magnetic resonance imaging measurements guidelines [24]. The interventional perspective we have set our work in motivated this choice. Indeed, accurate knowledge of any obstacle operators could meet when maneuvering or deploying a device into a cardiac chamber may be crucial to the procedural success. Thus, it makes sense to consider the main papillary muscles as myocardium to provide an appropriate description of the LVM shape and mass. From a segmentation standpoint, it likely complicated the automatic delineation of the LVC border, which was far less predictable than when papillary muscles are included in the LVC. In keeping with this point, CT data of the present study were acquired in systole, in patients with varying degrees of cardiac remodeling induced by their aortic stenosis, which may have resulted in different patterns of left ventricular hypertrophy, heterogeneously affecting the global geometry of LVC. These elements might have significantly participated in degrading the results of the automatic WHS explaining the lower Dice score values observed for LVC in the present study. However, on the contrary, a segmentation based on image density as the present one may be easier when the papillary muscles are not considered as a part of the ventricular cavity. Moreover, the systolic acquisition resulted in a lower LVC volume involving a reduced number of voxels. Therefore, any small difference between the manual and deep-learning-based segmentation has larger consequences upon the Dice score measurement than those expected from the measurement of LVC in a diastolic phase. This size consideration also applies for other small structures such as CS or PV. Fourth, regarding the PV, they usually exhibit a large degree of anatomical variation from one patient to another [31], which may explain the sub-optimal performance of our model to identify these structures.
Nevertheless, we reported excellent correlations between manually obtained and deep-learning predicted volumes for most structures. Although statistically significant absolute differences in volume measurement for the LVC, PV, and RVC were observed, the mean differences of measurement for all structures were low and would likely be clinically irrelevant. It is noticeable that the small size of our test set makes it vulnerable to the presence of outliers. However, the width of the 95%CI of the limits of agreement in the present study is in the range of those reported by Baskaran et al., which were themselves comparable or markedly lower than previously reported limits of agreement in deep-learning studies [21].
Clinical applicability of these deep-learning-based segmentation methods is crucial to whether they are to ultimately achieve widespread use. To the best of our knowledge, no recently published papers have mentioned the integration of such work in a readily useable system. Indeed, to provide optimal results, most of the published algorithms require powerful computing hardwares [20], which may not represent the majority of workstations available in the current daily medical environment. Before the advent of deep-learning approaches, computation times were never below 10 min. Using a dedicated hardware, Baskaran et al. reported an impressive processing time of 13 s/patient [21]. Furthermore, the mean computing time of the ten algorithms from the work by Zhuang et al. was also rather low at 312 s (range: 0.22 s to 21 min, 104 s for the most accurate model), with the use of dedicated workstations with such powerful GPUs [20]. The median computing time of our algorithm was only 13.4 s on a routinely used workstation, i.e., not equipped with a powerful hardware dedicated to research purposes. This point is of paramount importance for future clinical integration of the method. The current quickness of the algorithm also suggests that further work may easily achieve a refinement of the accuracy-computing time tradeoff, which would maximize the former while keeping the latter in a range compatible with a minimal disruption of clinical workflows. This good tradeoff was achieved thanks to our two-stage segmentation process, which had the benefit to keep relevant structures into a limited region of interest. With the prior aortic valve localization, a high resolution can be kept for precise 3D segmentation while restraining overall computation time into an acceptable range. The choice of the Dense V-Net architecture was also driven by this objective, while other segmentation network architectures (e.g., V-Net architecture) are recognized to be more precise but much more time consuming.
Limitations
A number of this study’s limitations have been discussed above. First, CT were acquired during the pre-procedural work-up of TAVI recipients using a dedicated protocol in accordance with an international expert consensus [8]. Whether the feasibility and performance of our algorithm, especially for the identification of right-sided structures, significantly differ according to the CT acquisition protocol or the underlying pathology will be addressed by our future works on a larger, more diverse database. In keeping with this point, this analysis was performed at a systolic phase in accordance with current guidelines for the measurement of aortic annulus. Future works will also have to determine the performances of our algorithm at a diastolic phase. Second, the training and validation sets of 63 patients encompassed a largely sufficient number of images to train a medical image deep-learning system to reach high accuracy [32]. However, this choice of keeping a large amount of data for model training limited the test set to 8 patients, which makes it vulnerable to outliers and likely resulted in larger 95%CI for the limits of agreement between the automatic and manual segmentations. Third, the manual segmentation was performed by a single expert. Fourth, a significant proportion of patients were excluded from this “pilot” study because of image quality, which precluded either manual or automatic segmentation, potentially raising generalizability issues. Finally, the localization step uses the intersection of the three aortic commissures to detect the center of the aortic valve, which may represent a limitation in case of bicuspid aortic valves. However, it should be emphasized that the localization step of the present algorithm is somewhat coarse, essentially used to crop an area of interest within the entire volume. Thus, it is unlikely that this aspect of our algorithm played a significant role in the results. Overall, this work is only the first step towards clinical application of our model. Aside from improving the accuracy-computing time tradeoff, further identification of structures such as cardiac valves remains a challenging task and an unmet need, which should be overcome to increase our model applicability in this transcatheter therapies era.
Conclusion
We developed a deep-learning-based segmentation method, which was fully integrated in a routinely used software supporting its potential clinical application. The method allowed fast, automated WHS from ECG-gated CT data with an overall high accuracy on a voxel level and demonstrated excellent correlations and adequate agreements compared with manual measurements for most segmented structures. However, further work is needed to improve right-sided and small structures segmentation, as well as to include other structures of interest (e.g., valves).
Abbreviations
- 3D:
-
Tridimensional
- WHS:
-
Whole heart segmentation
- CT:
-
Computed tomography
- AI:
-
Artificial intelligence
- TAVI:
-
Transcatheter aortic valve implantation
- PV:
-
Pulmonary veins
- LA:
-
Left atrium
- LVC:
-
Left ventricular cavity
- LVM:
-
Left ventricular myocardium
- Ao:
-
Aorta
- CS:
-
Coronary sinus
- SVC:
-
Superior vena cava
- RA:
-
Right atrium
- RVC:
-
Right ventricular cavity
- PA:
-
Pulmonary artery
- ROI:
-
Region of interest
- CNN:
-
Convolutional neural network
- IQR:
-
Interquartile
References
Siontis, G. C. M., Praz, F., Pilgrim, T., et al. (2016). Transcatheter aortic valve implantation vs. surgical aortic valve replacement for treatment of severe aortic stenosis: A meta-analysis of randomized trials. European Heart Journal, 37(47), 3503–3512. https://doi.org/10.1093/eurheartj/ehw225
Siontis, G. C. M., Overtchouk, P., Cahill, T. J., et al. (2019). Transcatheter aortic valve implantation vs. surgical aortic valve replacement for treatment of symptomatic severe aortic stenosis: An updated meta-analysis. European Heart Journal, 40(38), 3143–3153. https://doi.org/10.1093/eurheartj/ehz275
Auffret, V., Lefevre, T., Van Belle, E., et al. (2017). Temporal trends in transcatheter aortic valve replacement in France: FRANCE 2 to FRANCE TAVI. Journal of the American College of Cardiology, 70(1), 42–55. https://doi.org/10.1016/j.jacc.2017.04.053
Muller, D. W. M., Farivar, R. S., Jansz, P., et al. (2017). Transcatheter mitral valve replacement for patients with symptomatic mitral regurgitation: A global feasibility trial. Journal of the American College of Cardiology, 69(4), 381–391. https://doi.org/10.1016/j.jacc.2016.10.068 [published correction appears in J Am Coll Cardiol 2017;69(9):1213].
Guerrero, M., Urena, M., Himbert, D., et al. (2018). 1-Year outcomes of transcatheter mitral valve replacement in patients with severe mitral annular calcification. Journal of the American College of Cardiology, 71(17), 1841–1853. https://doi.org/10.1016/j.jacc.2018.02.054
Mas, J. L., Mas, J. L., Derumeaux, G., Guillon, B., et al. (2017). Patent foramen ovale closure or anticoagulation vs. antiplatelets after stroke. New England Journal of Medicine, 377(11), 1011–1021. https://doi.org/10.1056/NEJMoa1705915
Bishop, M., Rajani, R., Plank, G., et al. (2016). Three-dimensional atrial wall thickness maps to inform catheter ablation procedures for atrial fibrillation. Europace, 18(3), 376–383. https://doi.org/10.1093/europace/euv073
Blanke, P., Weir-McCall, J. R., Achenbach, S., et al. (2019). Computed tomography imaging in the context of transcatheter aortic valve implantation (TAVI)/transcatheter aortic valve replacement (TAVR): An expert consensus document of the Society of Cardiovascular Computed Tomography. JACC: Cardiovascular Imaging, 12(1), 1–24. https://doi.org/10.1016/j.jcmg.2018.12.003
Blanke, P., Dvir, D., Cheung, A., et al. (2015). Mitral annular evaluation with CT in the context of transcatheter mitral valve replacement. JACC: Cardiovascular Imaging, 8(5), 612–615. https://doi.org/10.1016/j.jcmg.2014.07.028
Blanke, P., Naoum, C., Dvir, D., et al. (2017). Predicting LVOT obstruction in transcatheter mitral valve implantation: Concept of the neo-LVOT. JACC: Cardiovascular Imaging, 10(4), 482–485. https://doi.org/10.1016/j.jcmg.2016.01.005
Weir-McCall, J. R., Blanke, P., Naoum, C., Delgado, V., Bax, J. J., & Leipsic, J. (2018). Mitral valve imaging with CT: Relationship with transcatheter mitral valve interventions. Radiology, 288(3), 638–655. https://doi.org/10.1148/radiol.2018172758
Zhuang, X., & Shen, J. (2016). Multi-scale patch and multi-modality atlases for whole heart segmentation of MRI. Medical Image Analysis, 31, 77–87. https://doi.org/10.1016/j.media.2016.02.006
Commandeur, F., Goeller, M., Betancur, J., Cadet, S., Doris, M., Chen, X., et al. (2018). Deep learning for quantification of epicardial and thoracic adipose tissue from non-contrast CT. IEEE Transactions on Medical Imaging, 37, 1835–1846.
Zreik, M., Lessmann, N., van Hamersvelt, R. W., et al. (2018). Deep learning analysis of the myocardium in coronary CT angiography for identification of patients with functionally significant coronary artery stenosis. Medical Image Analysis, 44, 72–85. https://doi.org/10.1016/j.media.2017.11.008
Itu, L., Rapaka, S., Passerini, T., et al. (2016). A machine-learning approach for computation of fractional flow reserve from coronary computed tomography. Journal of Applied Physiology1985, 121(1), 42–52. https://doi.org/10.1152/japplphysiol.00752.2015
Coenen, A., Kim, Y. H., Kruk, M., et al. (2018). Diagnostic accuracy of a machine-learning approach to coronary computed tomographic angiography-based fractional flow reserve: Result from the MACHINE consortium. Circulation: Cardiovascular Imaging, 11(6), e007217. https://doi.org/10.1161/CIRCIMAGING.117.007217
Zhuang, X., Bai, W., Song, J., et al. (2015). Multiatlas whole heart segmentation of CT data using conditional entropy for atlas ranking and selection. Medical Physics, 42(7), 3822–3833. https://doi.org/10.1118/1.4921366
Zhou, R., Liao, Z., Pan, T., et al. (2017). Cardiac atlas development and validation for automatic segmentation of cardiac substructures. Radiotherapy and Oncology, 122(1), 66–71. https://doi.org/10.1016/j.radonc.2016.11.016
Cai, K., Yang, R., Chen, H., et al. (2017). A framework combining window width-level adjustment and Gaussian filter-based multi-resolution for automatic whole heart segmentation. Neurocomputing, 220, 138–150. https://doi.org/10.1016/j.neucom.2016.03.106
Zhuang, X., Li, L., Payer, C., et al. (2019). Evaluation of algorithms for multi-modality whole heart segmentation: An open-access grand challenge. Medical Image Analysis, 58, 101537. https://doi.org/10.1016/j.media.2019.101537
Baskaran, L., Maliakal, G., Al’Aref, S. J., et al. (2020). Identification and quantification of cardiovascular structures from CCTA: An end-to-end, rapid, pixel-wise, deep-learning method. JACC: Cardiovascular Imaging, 13(5), 1163–1171. https://doi.org/10.1016/j.jcmg.2019.08.025
Kaladji, A., Lucas, A., Kervio, G., Haigron, P., & Cardon, A. (2010). Sizing for endovascular aneurysm repair: Clinical evaluation of a new automated three-dimensional software. Annals of Vascular Surgery, 24(7), 912–920. https://doi.org/10.1016/j.avsg.2010.03.018
Lang, R. M., Badano, L. P., Mor-Avi, V., et al. (2015). Recommendations for cardiac chamber quantification by echocardiography in adults: an update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. European Heart Journal – Cardiovascular Imaging, 16(3), 233–270. https://doi.org/10.1093/ehjci/jev014 [published correction appears in Eur Heart J Cardiovasc Imaging. 2016 Apr;17(4):412] [published correction appears in Eur Heart J Cardiovasc Imaging. 2016 Sep;17 (9):969].
Petersen, S. E., Khanji, M. Y., Plein, S., Lancellotti, P., & Bucciarelli-Ducci, C. (2019). European Association of Cardiovascular Imaging expert consensus paper: A comprehensive review of cardiovascular magnetic resonance normal values of cardiac chamber size and aortic root in adults and recommendations for grading severity. European Heart Journal - Cardiovascular Imaging, 20(12), 1321–1331. https://doi.org/10.1093/ehjci/jez232 [published correction appears in Eur Heart J Cardiovasc Imaging. 2019 Dec 1;20(12):1331].
Tobon-Gomez, C., Geers, A. J., Peters, J., et al. (2015). Benchmark for algorithms segmenting the left atrium from 3D CT and MRI datasets. IEEE Transactions on Medical Imaging, 34(7), 1460–1473. https://doi.org/10.1109/TMI.2015.2398818
Fuchs, A., Mejdahl, M. R., Kühl, J. T., et al. (2016). Normal values of left ventricular mass and cardiac chamber volumes assessed by 320-detector computed tomography angiography in the Copenhagen general population study. European Heart Journal Cardiovascular Imaging, 17(9), 1009–1017. https://doi.org/10.1093/ehjci/jev337
Forrest N. Landola, Song Han, Matthew W. Moskewicz, Khalid Ashraf, William J. Dally, Kurt Keutzer. SqueezeNet: AlexNet-level accuracy with 50× fewer parameters and < 0.50 MB model size. https://arxiv.org/abs/1602.07360. Accessed 18 April 2021
Karen Simonyan, Andrew Zisserman. Very deep convolutional network for large-scale image recognition. https://arxiv.org/abs/1409.1556. Accessed 18 April 2021
Gibson, E., Giganti, F., Hu, Y., et al. (2018). Automatic multi-organ segmentation on abdominal CT with dense V-networks. IEEE Transactions on Medical Imaging, 37(8), 1822–1834. https://doi.org/10.1109/TMI.2018.2806309
Eelbode, T., Bertels, J., Berman, M., et al. (2020). Optimization for medical image segmentation: Theory and practice when evaluating with dice score or Jaccard index. IEEE Transactions on Medical Imaging, 39(11), 3679–3690. https://doi.org/10.1109/TMI.2020.3002417
Chen, J., Yang, Z. G., Xu, H. Y., Shi, K., Long, Q. H., & Guo, Y. K. (2017). Assessments of pulmonary vein and left atrial anatomical variants in atrial fibrillation patients for catheter ablation with cardiac CT. European Radiology, 27(2), 660–670. https://doi.org/10.1007/s00330-016-4411-6
Cho J, Lee K, Shin E, Choy G, Do S. (2015) How much data is needed to train a medical image deep learning system to achieve necessary high accuracy? http://arxiv.org/abs/151106348. Accessed 18 April 2021
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Animal Studies
No animal studies were carried out by the authors for this article.
Conflict of Interest
Dr. Vincent Auffret and Dr. Le Breton received lecture fees from Edwards Lifescience. Mr. Florent Lalys is one of the employees of Therenva®. The other authors have nothing to disclose. All the authors take responsibility for all aspects of the reliability and freedom from bias of the data presented and their discussed interpretation.
Additional information
Associate Editor Jozine ter Maaten oversaw the review of this article
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sharobeem, S., Le Breton, H., Lalys, F. et al. Validation of a Whole Heart Segmentation from Computed Tomography Imaging Using a Deep-Learning Approach. J. of Cardiovasc. Trans. Res. 15, 427–437 (2022). https://doi.org/10.1007/s12265-021-10166-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12265-021-10166-0