Abstract
In this study, we verify the association between the rs1333049 single nucleotide polymorphism (9p21.3) within CDKN2A-CDKN2B and coronary artery disease (CAD) in an Italian population. We replicated rs1333049_G allele association with a significantly reduced risk of CAD (OR = 0.816; 95% confidence interval [0.705–0.945]; p = 0.0065) in 711 CAD patients and 755 normal healthy individuals. This effect is maintained even stratifying patients by gender and by risk factors. A significant association was found with age of CAD onset. Interestingly, we found a protective trend of association between the rs1333049_G allele and peripheral artery disease, a progressive atherosclerotic condition in which plaque builds up in the arteries that carry blood to the head, organs, and limbs (OR = 0.724; 95% CI [0.520–1.007]; p = 0.054). No genotype-phenotype association was found with more severe CAD clinical parameters. If certain genetic factors predispose individuals to adverse outcomes, the knowledge of a patient’s genotype may influence clinical management.
Similar content being viewed by others

Avoid common mistakes on your manuscript.
Coronary artery disease (CAD), the most common form of heart disease, is a group of pathology including stable and unstable angina, myocardial infarction, and sudden coronary death [1]. CAD is one of the leading causes of morbidity and mortality in both developed and developing countries. According to estimates, 30–60% interindividual variations in the risk of CAD are due to heritability, suggesting that genetic risk factors play a critical role in the pathogenesis of CAD. Genome-wide association studies (GWASs) have identified several independent CAD susceptibility loci to date [2], some of which seem to confer risk associated with ethnic differences, while others are thought to be ethnicity-specific [3]. In these loci, there are many genetic variants that were reported to be associated to the risk of CAD, but the exact number of candidate genes, as well as the effect they have on the development and progression of the disease, is not fully understood. Locus 9p21.3 is known for presenting the strongest association with CAD and acute myocardial infarction, and it is associated with other important diseases such as abdominal aortic and intracranial aneurysms, type 2 diabetes, metabolic syndrome, and stroke [4].
The association between the single nucleotide polymorphism (SNP) rs1333049 in CDKN2A-CDKN2B and CAD has already been replicated in various studies and in different ethnicities. Nevertheless, the linkage disequilibrium (LD) structure of the 9p21.3 region is different in populations of European and Asian ancestry (Supplementary Fig.1) [5]. For instance, four SNPs (rs9632884, rs10757274, rs1333042, and rs1333049) showing significant association with CAD are in almost complete LD in individuals of European descent (r 2 = 0.84–0.90). In the Chinese population, however, two of them (rs10757274 and rs1333049) were in strong LD with each other (r 2 = 0.78) but were in only moderate LD with the other two SNPs rs9632884 and rs1333042 (r 2 = 0.27–0.43) [3]. Thus, the risk of CAD related to the chromosome 9p21.3 locus may vary among different ethnic groups (Supplementary Fig.1 and Supplementary Table 1). Hence, the role of this locus in other ethnic groups remains to be investigated. Sometimes, replication studies fail to confirm initial findings because of substantial differences between study populations and population specificity that may consist in differences in LD block, population-specific interactions between genes, and epigenetic modifications.
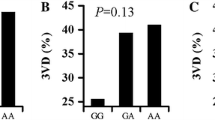
In this case-control study, we sought to evaluate a correlation between rs1333049 C/G and CAD in an Italian population. Here, we confirm that G allele has a protective effect against CAD (Table 1; p = 0.0065) using 711 Italian CAD patients and 755 controls, free of CAD by familiar history, clinical examination, and electrocardiography. This association was confirmed both in male (OR = 0.82; CI = [0.677–0.994]; p = 0.043) and in female subjects (OR = 0.64; CI = [0.471–0.877]; p = 0.005). Subgroup analyses performed in patients with CAD showed no difference in both the genotype and allele distribution of rs1333049 according to presence of risk factors (obesity, hypertension, hypercholesterolemia, diabetes mellitus) and markers of CAD severity (acute coronary syndrome, carotid plaques). These findings are in line with current literature, in which no association between rs1333049 and mortality or adverse cardiovascular events was shown. On the other hand, we observed a lower rate of G allele presence in patients with peripheral artery disease (PAD) (Table 1). Moreover, a trend towards a lower rate of rs1333049 was also observed in CAD patients with carotid atherosclerotic disease (Table 1). This evidence might suggest a role of rs1333049 as a protective factor in systemic atherosclerosis, rather than being responsible of the coronary localization of the atherosclerotic disease.
Since CAD is a late-onset disease, we investigated whether the polymorphism could be associated with age at onset. For this purpose, we divided our CAD population by age groups and analyzed the possible association with the genotype (Table 1 and Supplementary Table 2). We found a significantly lower rate of G allele in patients whose onset of CAD occurred before turning 50 years old (OR = 0.68; CI = [0.464–0.888]; p = 0.007). In a multivariate logistic regression analysis, G allele was found to be an independent protective factor (Supplementary Table 3) of early onset of CAD (OR = 0.507; p = 0.003). These findings suggest that the allele G might have a role in deferring the clinical manifestation of CAD. Indeed, among CAD patients, the clinical variables “normal arterial blood pressure” and “normal blood sugar levels” were more frequent in young people when compared with the old ones (Supplementary Table 2) whereas the genetic protective allele G was less frequent in the same young patients (Table 1). Therefore, genetic factors might have a relevant role in the development of CAD among young people which are not affected from risk diseases such as hypertension and diabetes mellitus.
In order to utilize new genetic information for treatment and prevention of CAD, it will be necessary to understand the functions of the gene(s) at the disease-associated loci and the mechanisms through which they affect coronary risk. Therefore, the present study replicated a significant evidence of association in an independent cohort of Italians clarifying the genetic component’s contribution in different populations for a multifactorial highly widespread disease like CAD. Finally, the protective effect of the rs1333049 G allele in patients with different localization of atherosclerotic disease (like the limbs or carotid arteries) and its potential role in deferring the onset of CAD, shown in our analysis, might be hypothesis generating for future studies.
Change history
12 July 2017
An erratum to this article has been published.
References
O'Donnell, C. J., & Nabel, E. G. (2011). Genomics of cardiovascular disease. The New England Journal of Medicine, 365, 2098–2109. doi:10.1056/NEJMra1105239.
CARDIoGRAMplusC4D Consortium. (2015). A comprehensive 1000 genomes–based genome-wide association meta-analysis of coronary artery disease. Nature Genetics, 47, 1121–1130. doi:10.1038/ng.3396.
Lu, X., Wang, L., Chen, S., He, L., Yang, X., Shi, Y., Cheng, J., Liang, Z., Gu, C. C., Huang, J., Wu, T., Ma, Y., Li, J., Cao, J., Chen, J., Ge, D., Fan, Z., Li, Y., Zhao, L., Li, H., Zhou, X., Chen, L., Liu, D., Chen, J., Duan, X., Hao, Y., Wang, L., Lu, F., Liu, Z., Yao, C., Shen, C., Xiaodong, P., Yu, L., Fang, X., Xu, L., Mu, J., Wu, X., Zheng, R., Wu, N., Zhao, Q., Li, Y., Liu, X., Wang, M., Yu, D., Hu, D., Ji, X., Guo, D., Sun, D., Wang, Q., Yang, Y., Liu, F., Mao, Q., Liang, X., Ji, J., Chen, P., Mo, X., Li, D., Chai, G., Tang, Y., Li, X., Du, Z., Liu, X., Dou, C., Yang, Z., Meng, Q., Wang, D., Wang, R., Yang, J., Schunkert, H., Samani, N. J., Kathiresan, S., Reilly, M. P., Erdmann, J., The Coronary ARteryDIsease Genome-Wide Replication And Meta-Analysis (CARDIoGRAM) Consortium, Peng, X., Wu, X., Liu, D., Yang, Y., Chen, R., Qiang, B., & Gu, D. (2012). Genome-wide association study in Han Chinese identifies four new susceptibility loci for coronary artery disease. Nature Genetics, 44(8), 890–894. doi:10.1038/ng.2337.
Helgadottir, A., Thorleifsson, G., Magnusson, K. P., Gretarsdottir, S., Steinthorsdottir, V., Manolescu, A., Jones, G. T., Rinkel, G. J., Blankensteijn, J. D., Ronkainen, A., et al. (2008). The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nature Genetics, 40, 217–224. doi:10.1038/ng.72.
Guo, J., Li, W., Wu, Z., Cheng, X., Wang, Y., & Chen, T. (2013). Association between 9p21.3 genomic markers and coronary artery disease in east Asians: A meta-analysis involving 9,813 cases and 10,710 controls. Molecular Biology Reports, 40(1), 337–343. doi:10.1007/s11033-012-2066-1.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This study was approved by the Ethics Committee of the Medical University of Naples.
Conflict of Interest
The authors declare that they have no conflicts of interest.
Additional information
Associate Editor Paul J. R. Barton oversaw the review of this article
Rights and permissions
About this article
Cite this article
Pignataro, P., Pezone, L., Di Gioia, G. et al. Association Study Between Coronary Artery Disease and rs1333049 Polymorphism at 9p21.3 Locus in Italian Population. J. of Cardiovasc. Trans. Res. 10, 455–458 (2017). https://doi.org/10.1007/s12265-017-9758-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12265-017-9758-9