Abstract
Recently, genome-wide association studies on coronary artery disease (CAD) identified a series of associated single-nucleotide polymorphisms (SNPs) in an intergenic region of chromosome 9p21.3, near the CDKN2A and CDKN2B genes. We investigated the association of this locus with CAD in 12 case–control studies of East Asians and undertook a meta-analysis for effect size, heterogeneity, publication bias, and strength of evidence. English and Chinese language articles were tested for 9p21.3 SNPs with coronary heart/artery disease or myocardial infarction as primary outcomes. Included articles also provided race, numbers of participants, and data to compute an odds ratio (OR). Articles were excluded if reporting other outcomes (e.g., stroke). Thirty-five articles were initially identified and 12 were included. Independent extraction was performed by two reviewers and consensus was reached. SNP rs1333049, rs2383206 and rs10757278 representing the 9p21.3 locus, were genotyped in 12 case–control studies involving a total of 9,813 patients and 10,710 controls. For rs1333049 (8 data sets), using a fixed-effects model, the summary OR was 1.29 (95 % CI, 1.23–1.36, P = 0.001). For rs2383206 (6 data sets), using a fixed-effects model, the summary OR was 1.24 (95 % CI, 1.18–1.31, P = 0.001). For rs10757278 (6 data sets), using a random-effects model, the summary OR was 1.34 (95 % CI, 1.21–1.50, P = 0.001). In addition, we defined the haploblock structure of SNPs within the region of 9p21.3 in China population in one study. This broad replication provides unprecedented evidence for association between genetic variants at chromosome 9p21.3 and risk of CAD in East Asians.
Similar content being viewed by others

Avoid common mistakes on your manuscript.
Introduction
Cardiovascular disease (CVD) is the leading cause of death and disability-adjusted life-years world wide, with increasing incidence and prevalence in low- and middle-income countries [1]. By 2020, more than 80 % of global CVD will be in these countries, with the largest burden occurring in the two largest countries, China and India, as they rapidly urbanise [1]. Nonmodifiable risk factors include increasing age, male sex, and heredity. Modifiable risk factors include smoking, hypertension, dyslipidemia, obesity, physical inactivity, and diabetes [2–4]. Biomarkers (e.g., C-reactive protein) have been combined with traditional risk factors to predict CVD events, and molecular markers hold further promise [5]. In 2007, genome-wide association studies on CVD identified a series of associated single-nucleotide polymorphisms (SNPs) in an intergenic region of chromosome 9p21, near the CDKN2A and CDKN2B genes [6, 7]. Recent success in identifying genes involved in complex diseases such as coronary artery disease (CAD) and myocardial infarction (MI) has been largely brought about by two major developments. First, modern array technology now enables simultaneous measurement of hundreds of thousands of single nucleotide polymorphisms (SNPs) across the human genome. Second, collaborations have been formed, bringing together large collections of well-phenotyped individuals.
However, only a small proportion of population of CAD has been explained in East Asia. In fact, for a typical association study with hundreds of cases and controls, the power to detect any effects at stringent significance levels is low. To increase the power, we formed a consortium to pool data across all published and multiple unpublished articles of East Asia population for CAD and MI. Here, we aim to provide a detailed description of the structure and functioning of our consortium.
Methods
Study populations
We systematically searched PubMed, Blackwell, Biosis Previews, Cochrane Library, China National Knowledge Infrastructure (CNKI), Chinese Biomedical (CBM) and Wan Fang (Chinese) databases for relevant articles published in different languages up to February 2012. We used search term “9p21 AND (cardiovascular disease OR coronary artery disease OR acute myocardial infarction OR myocardial infarction OR structural heart disease OR coronary atherosclerosis) AND (China OR Chinese OR Japanese OR Korean OR Mongol OR Asian population)” in this study. Other eligible studies were identified by a search of references cited in published articles.
The inclusion criteria of this meta-analysis were showed as follows: (a) including patients with a diagnosis of contained primary outcomes of coronary heart disease (CHD), myocardial infarction (MI), or coronary artery disease (CAD) and without other relate diseases; (b) describing the association of SNPs in 9p21 polymorphisms with CAD risk; (c) reported race and numbers of affected and unaffected participants, and race was East Asia population; (d) describing the genotyping method; (e) provided frequency genotype and the odds ratio (OR) with confidence intervals (CIs) or data sufficient to compute it. If more than 1 outcome was reported, the best-described phenotype was chosen. Several included articles reported consortium results with multiple independent populations. These populations were listed as separate data sets; all data were extracted independently by two reviewers according to the inclusion criteria. Discrepancies were documented and resolved by discussion with a third reviewer. The following information was extracted from all the eligible studies: patients with a diagnosis of disease not related with CAD, other race, SNPs not in the 9p21.
We studied subjects collected in eight different studies across China, Japan and Korea. All subjects were of East Asian origin. The large majority of cases had CAD as defined by China study (n = 3,413). The remaining cases had evidence of CAD based on either a revascularization procedure or anginal symptoms with a positive stress test. Details of each study are given in Table 1 and the online-only Data Supplement.
Not all researchers use the same 9p21 SNPs, and most articles reported results for multiple SNPs (uniquely identified by their rs number). As a proof-of-principle analysis, a number of SNPs in the 9p21 region with known association to CAD and MI were analyzed upfront. Specifically, 3 SNPs were reported as lead SNPs in the first publications by McPherson et al. (rs2383206), Helgadottir et al. (rs10757278), and Samani et al. (rs1333049). In addition, the SNP rs10757274 and rs2383207 in the same chromosomal region were also associated with CAD. We report here in three common SNPs (rs1333049, rs10757278, and rs2383206) that were included in more than six articles [8–19]. These SNPs are in high linkage disequilibrium (D′ > 0.9; r 2 > 0.85) [19]. The remaining four articles used two additional SNPs, rs2383207 and rs10757278 [8–19]. These were also in high linkage disequilibrium. Information about age at diagnosis and race/ethnicity was also collected.
Genotyping
Different genotyping platforms have been used across the studies. An analysis restricted only to SNPs genotyped on all platforms would have been severely limited. For instance, the estimated overlap between the Affymetrix Genome Wide Human SNP Array 6.0 and the Illumina Human-1 mol/L chip is only about 250,000 SNPs. To allow for combined analyses across different platforms, missing SNPs were imputed by each study.
Haplotype analysis
Haplotype association analysis was performed with the Haploview software and SAS software (version 9.1.3, SAS Institute Inc) implementing the Stochastic-EM algorithm. Using this approach, we were able to simultaneously estimate haplotype frequencies for each haplotype pair consistent with the observed data. When ambiguous haplotypes were encountered, multiple, probability-weighted haplotypes were created.
Statistical analysis methods
For each data set, the observed genotype frequencies in controls were compared with expected frequencies based on Hardy–Weinberg equilibrium (χ 2 test with 2 degrees of freedom). All P values are 2-sided at the P = 0.05 level. After upload of the summary data and centralized quality control, a meta-analysis across all studies is performed for every SNP separately. Here, depending on the heterogeneity between studies, fixed or random effects models are calculated, and outlying studies excluded. Summary ORs and corresponding 95 % CIs were derived (by reanalysis when possible) and summarized using random-effects modeling weighted by each data set’s total variance (STATA) [20]. Subgroup differences were compared using the Q test for heterogeneity for each covariate separately. Publication bias was examined by performing a cumulative effects analysis [20, 21]. Wider ranges of these summary ORs indicate potential for publication bias.
Results
Literature search and meta-analysis databases
A total of 35 articles were identified in the initial search with the medical subject heading terms. After screening the abstracts or full contents, 23 articles were excluded (twelve were not related to CAD, five were not about polymorphism of SNPs in 9p21, four reported other races, two were republished studies). Finally, twelve studies (9,813 cases and 10,710 controls) were included in this meta-analysis. Eight studies of them were performed in China [8–13, 15, 18, 19], two in Japan [14, 16], and two in Korea (Table 1; Fig. 1). [15, 17] All genotype frequencies of studies were conformed to the HWE in control group.
Quantitative synthesis
The results of all the East Asia population for our proof-of-principle analysis are shown in Figs. 2, 3, 4 for rs1333049 (8 data sets), using a fixed-effects model, the summary OR was 1.29 (95 % CI, 1.23–1.36, P = 0.001). Heterogeneity was checked separately using Q statistic (Q = 10.14; I 2 = 31 %; P = 0.181). For rs2383206 (6 data sets), using a fixed-effects model, the summary OR was 1.24 (95 % CI, 1.18–1.31, P = 0.001). Heterogeneity was low (Q = 8.24; I 2 = 39.3 %; P = 0.144). For rs10757278 (6 data sets), using a random-effects model, the summary OR was 1.34 (95 % CI, 1.21–1.50, P = 0.001). Heterogeneity was checked separately using Q statistic (Q = 13.20; I 2 = 62.1 %; P = 0.022).
In subgroup analyses, the G allele was also significantly associated with CAD in every race (Table 2). But there were little differences in ORs found between China population (5 data sets) and other races (Japan 2 data sets, Korea 1 data sets) (ORs of rs1333049, 1.27, 1.38, and 1.19).
Sensitivity analysis
Sensitivity analyses were performed by excluding the studies that were not in HWE, and the pooled OR were not materially altered in recessive genetic model (data not shown), indicating that the model results were statistically robust. Sensitivity analyses could not be done because of the small sample size in the subgroup analyses.
Publication bias
Begg rank correlation method and Egger weighted regression method were performed to assess the publication bias in recessive genetic model. No publication bias evidence was found (Table 2).
Discussion
Based on previous studies, evidence for the association between heart disease and chromosome 9p21 SNP markers had strong credibility. The 9p21 SNP markers have been identified though genome wide association studies and appear independent of traditional risk factors or family history [20, 22, 23]. The associated SNPs were rs1333049 (18 researches, OR = 1.23), rs10757274 (17 researches, OR = 1.24), rs2383207 (6 researches, OR = 1.28), rs2891168 (4 researches, OR = 1.29), rs10757278 (2 researches, OR = 1.27) [2]. In Asian race, rs1333049 (Japan, China), rs10757274 (China), rs10757278 (China) showed an association with the MI [2].
Most data sets are limited to white populations, usually of European descent, so the results for other racial groups might differ. We aim to investigate the association of this locus with CAD in 12 case–control studies of East Asians and undertook a meta-analysis for effect size, heterogeneity, publication bias, and strength of evidence. The SNP rs1333049 was also part of the previous meta-analysis by Schunkert et al. [24, 25]. In that report, the overall OR was 1.29 with a 95 % CI of 1.22–1.37, virtually identical to our current result. In our research, for rs1333049 (8 data sets), we received the similar result that the summary OR was 1.29 (95 % CI, 1.23–1.36, P = 0.001). The OR associated with rs2383206 was 1.24 (95 % CI, 1.18–1.31, P = 0.001). It was lower than rs2383206 analyzed in Europe population (OR = 1.28 (95 % CI, 1.22–1.35, P = 1.64 × 10−20), by Michael Preuss et al.). The OR associated with rs10757278 was 1.34 (95 % CI, 1.21–1.50, P = 0.001). It was higher than rs10757278 analyzed in Europe population (OR = 1.28 (95 % CI, 1.21–1.35, P = 1.64 × 10−20), by Michael Preuss et al.).
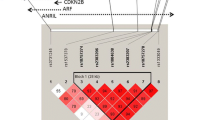
HapMap data suggests that rs10757274 and rs2383206 were located within 20 kb of each other on chromosome 9p21 and were in strong linkage disequilibrium (r 2 = 0.89) in North American population [5], and the five SNPs (rs2383206, rs2383207, rs10757274, rs10757278, and rs1333049) are in 1 block, and the D’ between each of the 5 SNPs was 1.0 in Chinese Han origin [13]. In our study, we identified one haploblock in the 9p21 region (rs10757274, rs2383206, rs10757278, and rs1333049) in Chinese population, and it’s similar to previous North American population study. On the basis of the stringent analysis of these SNPs and replication in our research, we are confident that 9p21 region is a strong candidate locus for CAD susceptibility in the Chinese population. Figure 5 shows the results of haplotype analysis for the SNPs examined. And then we calculated possible haplotype frequency of 8 loci in 9p21, using haploview and SAS software. Using the genotypes of 1,185 persons, we defined the haploblock structure of SNPs within the region of 9p21 in the Chinese population. By defining a solid spine of LD as D′ > 0.90, we identified one haploblock in the 9p21 region.
The region of chromosome 9p21 contained the coding sequences of gene for 2 cyclin-dependent kinase inhibitors, CDKN2A and CDKN2B, which played an important role in the regulation of the cell cycle and would be implicated, through their role in transforming growth factor (TGF)-β-induced growth inhibition, in the pathogenesis of atherosclerosis [13, 26–28]. Given the strong implication of this genomic region in CHD, the exact molecular mechanism of its involvement remains to be confirmed. McPherson et al. [7] resequenced the coding regions of the 2 genes most proximal to the risk locus and found no explanation of the CHD risk associated with this locus. However, the same region has recently been associated with increased susceptibility to type 2 diabetes [29, 30]. These results suggest that the region of 9p21 plays a role in multiple complex diseases. Further studies should focus on the identification of the underlying mechanism at this locus of CHD. In addition, the interactions between gender, smoking, and overweight with these SNPs need to be replicated in other populations.
References
Teo KK, Liu L, Chow CK et al (2009) Potentially modifiable risk factors associated with myocardial infarction in China: the INTERHEART China study. Heart 95:1857–1864
Glenn E, Palomaki, Stephanie Melillo, Linda A (2010) Bradley association between 9p21 genomic markers and heart disease: a meta-analysis. JAMA 303:605–656
Hobbs FD (2004) Cardiovascular disease: different strategies for primary and secondary prevention. Heart 90:1217–1223
van Wyk JT, van Wijk MA, Sturkenboom MC, Moorman PW, van der Lei J (2005) Identification of the four conventional cardiovascular disease risk factors by Dutch general practitioners. Chest 128:2521–2527
Wang TJ, Gona P, Larson MG et al (2006) Multiple biomarkers for the prediction of first major cardiovascular events and death. N Engl J Med 355:2631–2639
Helgadottir A, Thorleifsson G, Manolescu A et al (2007) A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 316:1491–1493
McPherson R, Pertsemlidis A, Kavaslar N et al (2007) A common allele on chromosome 9 associated with coronary heart disease. Science 316:1488–1491
Xie F, Chu X, Wu H et al (2011) Replication of putative susceptibility loci from genome-wide association studies associated with coronary atherosclerosis in Chinese Han population. Plos One 6:e20833
Wang W, Peng W, Lu L et al (2011) Polymorphism on chromosome 9p21.3 contributes to early-onset and severity of coronary artery disease in non-diabetic and type 2 diabetic patients. Chin Med J 124:66–71
Ding H, Xu Y, Wang X et al (2009) 9p21 is a shared susceptibility locus strongly for coronary artery disease and weakly for ischemic stroke in Chinese Han population. Circ Cardiovasc Genet 2:338–346
Peng W, Lu L, Zhang Q et al (2009) Chromosome 9p21 polymorphism is associated with myocardial infarction but not with clinical outcome in Han Chinese. Clin Chem Lab Med 47:917–922
Zhang Q, Wang X, Cheng S et al (2009) Three SNPs on chromosome 9p21 confer increased risk of myocardial infarction in Chinese subjects. Atherosclerosis 207:26–28
Zhou L, Zhang X, He M et al (2008) Associations between single nucleotide polymorphisms on chromosome 9p21 and risk of coronary heart disease in Chinese Han population. Arterioscler Thromb Vasc Biol 28:2085–2089
Hiura Y, Fukushima Y, Yuno M et al (2008) Validation of the association of genetic variants on chromosome 9p21 and 1q41 with myocardial infarction in a Japanese population. Circ J 72:1213–1217
Chen Z, Qian Q, Ma G et al (2009) A common variant on chromosome 9p21 affects the risk of early-onset coronary artery disease. Mol Biol Rep 36:889–893
Hinohara K, Nakajima T, Takahashi M et al (2008) Replication of the association between a chromosome 9p21 polymorphism and coronary artery disease in Japanese and Korean populations. J Hum Genet 53:357–359
Shen G, Li L, Rao S et al (2008) Four SNPs on chromosome 9p21 in a South Korean population implicate a genetic locus that confers high cross-race risk for development of coronary artery disease. Arterioscler Thromb Vasc Biol 28:360–365
Qiu Z, Zhang Y, Gao S et al (2011) SusceptibiHty study of 9p21 polymophism in patients with coronary heart disease in Chinese Han population. Geriatr Health Care 17:21–24
Guo J, Li W, Liu X et al (2011) Association study of single-nucleotide polymorphisms on chromosome 1p13 and 9p21 with Acute myocardial infarction (AMI) in a Chinese population: the Interheart China Study. Acad J Second Mil Med University 32:822–829
Palomaki G, Melillo S, Bradley L (2010) Association between 9p21 genomic markers and heart disease. JAMA 303:7
Borenstein M, Hedges LV, Higgins JPT, Rothstein HR (2009) Introduction to meta-analysis. Wiley, West Sussex
Anderson JL, Horne BD, Kolek MJ et al (2008) Genetic variation at the 9p21 locus predicts angiographic coronary artery disease prevalence but not extent and has clinical utility. Am Heart J 156:1155–1162 e2
Brautbar A, Ballantyne CM, Lawson K et al (2009) Impact of adding a single allele in the 9p21 locus to traditional risk factors on reclassification of coronary heart disease risk and implications for lipid-modifying therapy in the atherosclerosis risk in communities study. Circ Cardiovasc Genet 2:279–285
Preuss M, König R, Thompson R et al (2010) Design of the coronary artery disease genome-wide replication and meta-analysis (CARDIoGRAM) study : a genome-wide association meta-analysis involving more than 22,000 cases and 60,000 controls. Circ Cardiovasc Genet 3:475–483
Schunkert H, Go¨tz A, Braund P et al (2008) Repeated replication and a prospective meta-analysis of the association between chromosome 9p21.3 and coronary artery disease. Circulation 117:1675–1684
Lowe SW, Sherr CJ (2003) Tumor suppression by Ink4a-Arf: progress and puzzles. Curr Opin Genet Dev 13:77–83
Hannon GJ, Beach D (1994) p15INK4B is a potential effector of TGF-betainduced cell cycle arrest. Nature 371:257–261
Kalinina N, Agrotis A, Antropova Y et al (2004) Smad expression in human atherosclerotic lesions: evidence for impaired TGF-beta/Smad signaling in smooth muscle cells of fibrofatty lesions. Arterioscler Thromb Vasc Biol 24:1391–1396
Saxena R, Voight BF, Lyssenko V et al (2007) Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 316:1336–1341
Scott LJ, Mohlke KL, Bonnycastle LL et al (2007) A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316:1341–1345
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Guo, J., Li, W., Wu, Z. et al. Association between 9p21.3 genomic markers and coronary artery disease in East Asians: a meta-analysis involving 9,813 cases and 10,710 controls. Mol Biol Rep 40, 337–343 (2013). https://doi.org/10.1007/s11033-012-2066-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-012-2066-1