Abstract
Few studies have investigated the biodegradation of microplastics in marine environments. Microorganisms that can degrade microplastics in high-salinity conditions are sought after. Therefore, we aimed to isolate a halotolerant poly(ε-caprolactone) (PCL)-degrading bacterium for applications in biotechnology. The bacterium isolated from seaside soil was identified as Bacillus gibsonii via phylogenetic analysis based on 16S rRNA gene sequences and designated as KRICT-1. We tested whether the KRICT-1 strain showed halotolerance by determining the sodium chloride (NaCl) tolerance at various concentrations. The KRICT-1 strain showed growth at up to 10% NaCl on Luria–Bertani (LB) medium agar plates and 10% NaCl in liquid LB medium, indicating that KRICT-1 can grow and produce a PCL-depolymerizing enzyme under high-salt conditions. The KRICT-1 strain could depolymerize PCL with a PCL film weight loss of 2.82% at up to 10% NaCl concentration after cultivation of 7 weeks. KRICT-1 is the first strain of B. gibsonii which shows PCL-depolymerizing activity. Scanning electron microscopy and water contact angle results confirmed the degradation of PCL by the KRICT-1 strain. The extracellular enzyme produced by the KRICT-1 strain was stable over a wide range of temperatures (15–40 °C) and pH (7.0–9.5). This halotolerant PCL-degrading bacterium can be used in the degradation of biodegradable plastics present in saline soils, saline water, and wastewater.
Graphical abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
The accumulation of plastic on earth leads to serious environmental and economic problems [1]. As plastic becomes more prevalent in daily life and technology, the accumulation of non-biodegradable waste also increases year after year. Many researchers have focused a lot of effort on reducing terrestrial white pollution; however, they were relatively less concerned about plastic pollution in the sea. The extensive use of plastics has also led to an increase in frequency of marine pollution [2]. Pollution in marine environments negatively impacts marine ecosystems and human health as follows: marine organisms can ingest microplastics and people consume these marine organisms through the food chain. The ingested microplastics accumulate in the body, and eventually, they can cause inflammation, physiological stress, and behavioral disorders in people [3, 4]. As this seriousness has recently been emphasized, various studies are being attempted to address the problem of marine plastic pollution [5, 6].
In the marine environment, plastic can be degraded in various ways including photo-oxidative degradation, thermal degradation, ozone-induced degradation, mechanochemical degradation, catalytic degradation, and biodegradation [7]. Compared to physicochemical methods, microbial plastic degradation in marine environments offers several advantages such as higher environmental compatibility, natural breakdown processes, enhanced cost-effectiveness, broader plastic types degraded, and promotion of sustainability [8]. Numerous microbial species of algae, fungi, bacteria, and actinomycetes have demonstrated degradation activity against various plastics such as polyethylene (PE) [9, 10], polyethylene terephthalate (PET) [11,12,13], polystyrene [14, 15], polypropylene (PP) [16, 17], polyurethane [18, 19], and polyvinyl chloride [20, 21] by secreting plastic-degrading enzymes. However, researches on the biodegradation of plastic by these microorganisms have mainly focused on terrestrial and freshwater environments, with limited studies in seawater conditions.
The biodegradation of plastics in seawater is affected by several factors such as temperature, pH, and salinity [22, 23]. Among these factors, salinity significantly reduces plastic degradation efficiency by inhibiting growth of plastic-degrading bacteria [24, 25]. For this reason, it is essential to secure halotolerant microorganisms with plastic-degrading activity to biodegrade the plastics in seawater. We are interested in microplastics contained in food, especially microplastics mixed with salt. Currently, about 40% of the world's salt is produced in salt fields. Microplastic pollution in seawater is becoming more serious, and the salt produced in salt fields inevitably contains a large amount of microplastics. After salt is formed, it becomes very difficult to remove microplastics present with salt. Hence, it is important to remove microplastics present in seawater during salt formation. However, since seawater collected in salt fields is evaporated by sunlight, the salt concentration is higher than that (3.5%) of general seawater. Few studies have been conducted to biodegrade plastics at higher salt concentration than typical seawater. To obtain proof for plastic biodegradation in high salt concentration (10% sodium chloride [NaCl]), we selected poly(ε-caprolactone) (PCL), a relatively well-studied plastic in biodegradation, as the target plastic for degradation.
Thus, in this study, we attempted to isolate new PCL-degrading bacteria with high salt tolerance from costal soils. Using a PCL emulsion agar plate with 10% NaCl, we isolated a new PCL-degrading bacterium with high NaCl tolerance. We compared the NaCl tolerance and PCL degradation activity of the isolated strain to those of Pseudomonas putida KRICT-1, which was an efficient PCL-degrading bacterium isolated from environment soil in our previous study. This is the first report identifying a new PCL-degrading bacterium with high salt tolerance up to 10% NaCl, and this strain could help degrade microplastics in sea water located in salt fields.
2 Materials and methods
2.1 Materials and sample collection
PCL with a low molecular weight (average Mn: approximately 14,000) for the screening plate and PCL with a high molecular weight (average Mn: 80,000) for the PCL film were purchased from Sigma–Aldrich. NaCl and calcium carbonate (CaCO3) were obtained from Junsei. Luria–Bertani (LB) broth medium (5.0 g/L yeast extract; 10.0 g/L peptone; 10.0 g/L NaCl) was used for cell cultivation. All other chemicals were commercial products of analytical grade. Seaside soil samples were collected at depths of 10–12 inches from Jeju Island, Korea (33°31′05.9′′ N and 126°29′13.3′′ E). The samples were stored in a sterile PP tube at room temperature.
2.2 Screening of PCL agar plates to isolate halotolerant PCL-depolymerizing microorganisms
Briefly, 1 g of each soil sample was used to prepare a suspension in distilled water and then serially diluted (10–3, 10–5, and 10–7). Each diluted sample was spread on emulsified PCL-LB agar plates containing 2%, 4%, 6%, 8%, and 10% (w/v) NaCl for 2 days at 30 °C. The screening plate of emulsified PCL-LB agar was prepared by homogenizing 10 g/L PCL in LB medium (10 g/L bactotryptone, 5 g/L yeast extract, and 5 g/L NaCl) using a sonicator (Vibra-cell VCX-750; Sonics). Following the addition of 15 g/L agar and NaCl, the emulsified PCL-LB agar medium was autoclaved. The medium was poured into Petri plates and allowed to solidify. After spreading, the plates were evaluated for the zone of clearance around the colonies. Only one clear zone was observed on 8% and 10% NaCl plates, and the zone of clearance indicated the biodegradation capacity of the halophilic bacteria.
2.3 Identification of the isolated microorganism via 16S rRNA nucleotide sequence analysis
To identify the isolated halotolerant PCL-depolymerizing bacterium using 16S rRNA nucleotide sequence analysis, the 16S rRNA gene was amplified via PCR using the universal primer set 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′). The sequences were aligned and compared with sequences from the NCBI GenBank database to identify the most similar strains using the Basic Local Alignment and Search Tool (BLAST) algorithm [26].
2.4 NaCl tolerance of the PCL-depolymerizing isolate
To determine the influence of different salt concentrations on the growth and PCL depolymerization activity of the isolate obtained in this study, the previously isolated P. putida strain showing PCL-depolymerizing activity was cultured to compare the activity of the two strains under salt stress. Both strains were grown in LB medium containing various concentrations of NaCl (0.5%, 2.0%, 4.0%, 6.0%, 8.0%, and 10% (w/v)) in a shaking incubator at 200 rpm at 30 °C for 49 days. A piece of PCL film (60 mg), which was cut into a round shape with a diameter of 15 mm and a thickness of 0.7 mm, was added to a culture flask containing culture medium. The PCL film was sterilized using 70% ethanol followed by UV treatment for 2 h. Cell growth was analyzed by successively measuring the optical density at 600 nm every 24 h for 5 days using a NanoDrop spectrophotometer. To determine the amount of PCL film weight loss, PCL films were recovered from the culture medium and thoroughly washed with distilled water to remove the adherent biomass and components of the medium. The weights of the PCL films were measured after oven drying at 45 °C for 12 h.
2.5 Scanning electron microscopic analysis of PCL film
The surface morphological changes in the PCL films were observed using a scanning electron microscope (SEM, JSM-5910; JEOL). After 7 weeks of incubation, the bacterium-treated and untreated PCL films in 10% NaCl-containing culture medium were collected and gently washed with distilled water and 70% ethanol solution to remove any surface-attached cells. The film samples were coated with a thin layer of gold (JEOL-JSM-420) and observed under a SEM to detect any changes in the PCL surface.
2.6 Hydrophilicity measurement of PCL film
Water contact angles (WCAs) of the PCL films after incubation of the isolate and P. putida strain were measured using a SEO Phoenix-300 analyzer (SEO). The films were removed from the culture medium, washed with 2% sodium dodecyl sulfate solution, rinsed with distilled water, and then oven-dried overnight at 50 °C. Contact angle measurements were performed in air with water as the probe liquid at room temperature. The WCAs were calculated using the average of values from three different sites on the films.
2.7 PCL-depolymerizing activity assay
The production of extracellular PCL-depolymerizing enzymes by the isolate was evaluated. After the isolate was cultivated in LB medium at 30 °C under shaking conditions (200 rpm) for 3 days, the culture broth was centrifuged at 12,000×g for 15 min to remove the cells. The cell-free supernatant containing the PCL-depolymerizing enzyme was concentrated via ultrafiltration with a 10 kDa cutoff size (Amicon Ultra-15; Millipore).
An activity assay for PCL depolymerization was performed based on an esterase activity assay with minor modifications using p-nitrophenyl butyrate (pNPB) as a substrate [27]. The reaction mixture (1 mL) contained 0.8 M potassium phosphate buffer (125 μL; pH 8.0), 10 mM pNPB solution dissolved in dimethyl sulfoxide (5 μL), and appropriately diluted enzyme solution (5 μL). After 10 min of reaction at 50 °C, the released pNP was quantified by measuring the absorbance at 410 nm using a spectrophotometer (Shimadzu UV-2600; Shimadzu). One unit of enzyme activity was defined as the amount of enzyme that produced 1 μmol pNP per min under the aforementioned conditions. The protein concentration of the enzyme preparation was determined using the Bradford method [28], and bovine serum albumin was used as a standard.
2.8 Biochemical characterization of extracellular PCL-depolymerizing enzyme
The effects of temperature and pH on the activity of the PCL-polymerizing enzyme and stability of the isolate were examined by measuring the enzyme activity at various temperatures and pH ranges. The optimal temperatures for the hydrolysis reaction facilitated by the isolate were determined by assaying the hydrolysis activity at various temperatures (15–65 °C) at pH 8.0. To examine the effect of temperature on enzyme stability, the cell-free lysate containing enzyme was pre-incubated at different temperatures for 30 min before the assay.
The optimal pH for the hydrolysis reaction facilitated by the isolate was determined by assaying the hydrolysis activity in various pH conditions (pH 3.5–10.0). After the enzyme solution was added to the reaction solutions at each reaction temperature and incubated for 10 min, enzyme activity was calculated based on absorbance. In this study, different buffers were used to maintain various pH conditions, such as 100 mM acetate buffer for the pH range 3.5–5.5, 100 mM phosphate buffer for pH range 5.5–8.0, and 100 mM Tris–HCl buffer for pH range 8.0–10.0 [29]. The stability of the enzyme at various pH values was characterized by determining the residual activity after incubating the enzyme in various pH buffers for 30 min before the assay. Each experiment was performed in triplicate.
2.9 Nucleotide sequence
The 16S rRNA sequence of the isolate was submitted to the GenBank database of NCBI under accession number OQ390097.
3 Results
3.1 Isolation and identification of the halotolerant PCL-depolymerizing strain
To isolate halotolerant PCL-degrading bacteria, soil samples were collected from the seaside and samples were spread on a PCL-emulsified LB agar plate containing > 10% NaCl. After culturing on the plate for 5 days, only one strain was successfully isolated, which formed a colony with a clear zone on the plate by hydrolyzing PCL. The isolate KRICT-1 was identified based on its 16S rRNA sequence using a BLAST search against sequences in the GenBank DNA database. The isolate KRICT-1 was identified as Bacillus gibsonii with a sequence similarity of 99%.
3.2 Effects of different salt concentrations on growth and PCL depolymerization activity
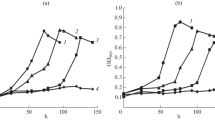
The effect of different NaCl concentrations on the growth and depolymerization of PCL by the KRICT-1 strain was studied by culturing the strain on PCL-emulsified LB agar plates containing different salt concentrations (0.5–10.0% (w/v)). A previously isolated P. putida strain showing high PCL-depolymerizing activity was used as a control strain to evaluate the activity of KRICT-1 under salt stress. The KRICT-1 strain showed growth in plates containing NaCl up to a concentration of 10% with formation of clear zones, while P. putida showed growth at a maximum NaCl concentration of 8% at 30 °C (Fig. 1). When both strains were cultured in LB liquid medium containing different NaCl concentrations, the growth of the KIRCT-1 strain was partially increased at relatively higher salt concentrations. In contrast, an increase in NaCl concentration suppressed the cell growth of the P. putida strain and decreased the maximum cell growth (Fig. 2). Under varying salt concentrations, KRICT-1 maintains growth or even shows enhanced growth in saline conditions, since KRICT-1 exhibits higher salt tolerance due to its adaptation to high-salinity environments. After 7 weeks of incubation, the KRICT-1 strain alone was capable of depolymerizing PCL with a 2.82% PCL film weight loss at up to 10% NaCl concentration, whereas P. putida lost its PCL depolymerization activity at the same NaCl concentration (Fig. 3).
Growth of a Pseudomonas putida and b KRICT-1 strain on media containing PCL and 0.5%, 2%, 4%, 6%, 8%, or 10% NaCl. The isolated KRICT-1 strain alone showed growth and formed a transparent halo around colonies on agar plates containing 10% NaCl after 5 days of incubation at 30 °C. PCL: poly(ε-caprolactone), NaCl: sodium chloride
Comparison of the NaCl tolerance of a Pseudomonas putida and b KRICT-1 strain using LB medium containing various NaCl concentrations of 0.5%, 2%, 4%, 6%, 8%, and 10%. The cell growth of P. Putida and KRICT-1 strain was monitored by measuring the optical density at 600 nm. Error bars represent standard deviation from three independent experiments. NaCl: sodium chloride, LB: Luria–Bertani
Time-course profile of PCL film degradation performed by a Pseudomonas putida and b KRICT-1 strain using LB medium containing various concentrations of 0.5%, 2%, 4%, 6%, 8%, and 10% NaCl. The strains were incubated in LB medium containing 60 mg of PCL film and grown in a shaking incubator at 30 °C and 200 rpm for 7 weeks. Error bars represent the standard deviation from three independent experiments. PCL: poly(ε-caprolactone), LB: Luria–Bertani, NaCl: sodium chloride
3.3 Physical properties of PCL after biodegradation
SEM analysis was performed to investigate the effects of PCL depolymerization by observing changes in surface morphology of PCL films after incubation with the KRICT-1 strain under 10% NaCl conditions. The PCL films incubated in the uncultured control and P. putida culture had a smooth surface (Fig. 4a, b). In contrast, the roughness of the PCL film increased after incubation with the KRICT-1 strain and hemispherical holes were formed (Fig. 4c). The WCA measurement was used to evaluate the hydrophilic/hydrophobic properties of the PCL films; the contact angle of the PCL film changed from initially hydrophobic to hydrophilic after biodegradation. The contact angle of the PCL film incubated with KRICT-1 strain culture medium containing 10% NaCl for 7 weeks was 74.74 ± 1.68°, while those of PCL films in uncultured medium as a control and P. putida culture medium were 84.41 ± 1.77° and 84.51 ± 0.33°, respectively (Fig. 5). PCL films were not affected by P. putida cultured under NaCl concentrations up to 10%. Therefore, the hydrophilicity of the PCL film cultured under 10% NaCl conditions only increased owing to the hydrolysis reaction facilitated by strain KRICT-1.
Scanning electron microscopy analysis of PCL film. a Abiotic control of PCL film (× 20,000 magnification) and b surface structures of PCL film (× 20,000 magnification) after 7 weeks of incubation with Pseudomonas putida in LB medium containing 10% NaCl, and the c surface structures of PCL film (× 20,000 magnification) after 7 weeks of incubation with the KRICT-1 strain in LB medium containing 10% NaCl. The bar represents 2 μm length. PCL: poly(ε-caprolactone), LB: Luria–Bertani, NaCl: sodium chloride
Water contact angles of the PCL films. The contact angle of the PCL films from three different culture conditions was measured using an SEO Phoenix-300 analyzer. The contact angles were analyzed by dropping water on the film surface at room temperature and it was measured at three points in triplicate. Error bars represent standard deviation from three independent experiments. PCL: poly(ε-caprolactone)
3.4 Biochemical characterization of extracellular PCL-depolymerizing enzyme
The extracellular PCL-depolymerizing enzyme derived from the KRICT-1 strain displayed high relative activity at 25–50 °C (Fig. 6a) and was stable during pre-incubation at up to 40 °C (Fig. 6b). An increase in temperature gradually decreased the activity and stability of the PCL-depolymerizing enzyme. The enzyme was characterized under various pH conditions (3.5–10.0). The optimum pH condition was found to be pH 8.0; enzyme activity gradually decreased in the acidic range below pH 7.0 (Fig. 6c). Although enzyme stability continued to increase with pre-incubation at up to pH 8.5, it declined rapidly beyond pH 9.0. The enzyme was stable with > 40% relative activity between pH 7.5 and 8.5 (Fig. 6d). The temperature and pH characteristics of the extracellular PCL-depolymerizing enzyme play crucial roles in determining its industrial applications. Characterization of the temperature range where the enzyme exhibits maximal activity and stability, along with the ideal pH conditions, ensures efficient PCL breakdown. Appropriate temperature and pH conditions directly impact the efficiency of the PCL depolymerization process. Optimal conditions enhance the enzyme's catalytic activity, leading to faster and more effective degradation of PCL. Furthermore, esterase is a key enzyme facilitating PCL depolymerization, breaking down ester bonds within the polymer. Their stability is crucial for efficient enzymatic degradation. Stable enzyme maintains their catalytic activity over time and harsh conditions, ensuring sustained PCL breakdown [30], and enhances bioremediation efficacy by prolonging their action on PCL, accelerating the polymer's degradation [31]. Stable esterases are pivotal for sustained and efficient PCL depolymerization, influencing the feasibility of enzymatic recycling strategies for managing plastic waste. From the results, the extracellular PCL-depolymerizing Enzymes of KRICT-1 with broader temperature and pH ranges of activity offer versatility in various industrial settings. Enzymes that perform well across a range of conditions can be more easily integrated into diverse manufacturing or waste treatment processes. Understanding and optimizing the temperature and pH characteristics of the extracellular PCL-depolymerizing enzyme are pivotal for its efficient and adaptable utilization across diverse industrial settings, waste management, and sustainable production processes.
Effects of temperature and pH on the activity and stability of extracellular PCL-depolymerizing enzyme derived from the KRICT-1 strain. a Effects of temperature on enzyme activity. The residue activities were measured at between 15 and 65 °C at pH 8.0. b Effects of temperature on enzyme stability. c Effects of pH on enzyme activity. Activity was measured after incubation in the following pH buffers: 100 mM acetate buffer for pH range 3.5–5.5, 100 mM phosphate buffer for pH range 5.5–8.0, and 100 mM Tris–HCl buffer for pH range 8.0–10.0 at 50 °C. d Effect of pH on enzyme stability. The stability of the enzyme at various pH values was characterized by determining the residual activity after the enzyme was incubated in various buffers for 30 min before assaying. Error bars represent standard deviation from three independent experiments. PCL: poly(ε-caprolactone)
4 Discussion
In this study, we screened and identified the halotolerant PCL-depolymerizing bacterium, B. gibsonii KRICT-1, from seaside soil using PCL-emulsified LB agar plates containing 10% NaCl. Although previous studies have reported that B. gibsonii shows halotolerance [32, 33], this is the first documented case that B. gibsonii can degrade PCL. Currently, we are looking for PCL depolymerization enzymes in the chromosome of this strain. The degradation of PCL in nature is attributed to microorganisms that secrete extracellular PCL depolymerases belong to the carboxylic hydrolase subfamily (EC 3.1.1), mainly including carboxylesterases (EC 3.1.1.1), cutinase (EC 3.1.1.74) and lipase (EC 3.1.1.3) which are active on aliphatic polyesters and thus can be employed in PCL hydrolysis [34, 35]. In the case of the mechanism for PCL biodegradation by esterase, esterase catalyzes PCL depolymerization via a surface erosion mechanism, initiating degradation at the polymer PCL surface [36]. Esterase breaks down PCL by hydrolyzing ester linkages, cleaving the polymer chains into smaller oligomers and monomers by the formation of hydroxyl and carboxyl end groups [37]. Unlike previously known PCL depolymerization enzymes, the enzymes of B. gibsonii KRICT-1 exhibited plastic depolymerization enzyme activity even at high salt concentrations. Therefore, if we study the enzymatic and structural characteristics of the enzymes, we will be able to gain insight into the mechanisms that exhibit structural stability and enzymatic activity at high salt concentrations.
Recently, researches on plastic degradation using recombinant microorganisms are also active. In particular, studies on degrading plastic by recombinant expression of the plastic degradation enzyme gene in microorganisms that can grow under extreme conditions (high temperature, low temperature, high acidity, and high alkalinity, etc.) are also being actively conducted [38]. If the B. gibsonii strain we screened recombinantly expresses enzymes that can degrade other plastics (PET, PP, and PE) in addition to PCL, other microplastics that can be found in salt field seawater with high salt concentration can be degraded. This study presented the basis for a microbial method to degrade microplastics present in seawater with high salt concentrations in salt fields. In future, through more researches, we would like to suggest an effective way to degrade microplastics from salt manufactured in the salt field via a biological method.
To study the plastic degradation activity of B. gibsonii KRICT-1 under high-salinity conditions, we compared its activity with that of P. putida KRICT-1, which has been found to show the highest PCL degradation activity in our previous study. In the present study, B. gibsonii KRICT-1 alone showed growth and degraded PCL when cultured in the presence of 10% NaCl (Figs. 2 and 3). We found that the enzyme was stable between pH 7.5 and 8.5. Sonkar and Singh [39] similarly reported that the maximum lipase activity of B. gibsonii is observed between pH 8.0 and 9.0. The PCL depolymerization activity was considerably lower than that of P. putida and other strains [40, 41]. We screened and identified a halotolerant PCL-depolymerizing bacterium from seaside soil. This strain, which was identified as B. gibsonii based on 16S rRNA sequence, demonstrated effective PCL depolymerization activity under high-salinity conditions. The isolated B. gibsonii KRICT-1 as a halotolerant bacterium has application prospects in bioremediation because it can potentially degrade plastic pollutants, especially in high-salinity environments. Given its potential application in biotechnology, B. gibsonii KRICT-1 represents a promising strain for the degradation of biodegradable plastics in seawater with high salt concentrations in salt fields.
However, further research into plastic biodegradation mediated by B. gibsonii is needed, and there are still many challenges to overcome before the biodegradation process mediated by B. gibsonii can be widely implemented. To implement B. gibsonii KRICT-1 for plastic degradation in saline environments, we need to solve these challenges: (1) to investigate and optimize the pathways B. gibsonii KRICT-1 employ to break down various types of plastics, especially low-density polyethylene and other commonly used polymers in saline environments, (2) to enhance the degradation efficiency of B. gibsonii KRICT-1 by understanding the interplay between salinity levels, environmental conditions, and other bacterial activity in saline environments, and (3) to assess the ecological impact of employing KRICT-1 in saline environments to ensure natural microbial balance or unintended environmental consequences.
5 Conclusion
In the present study, we screened and identified the halotolerant PCL-depolymerizing bacteria from the soil of seaside. This strain, classified according to 16S rRNA sequence, belonging to the genera B. gibsonii, demonstrated an effective PCL depolymerization activity under high saline conditions. In our study, the observed increase in hydrophilicity of PCL film due to the biodegradation of B. gibsonii KRICT-1 under high-salinity conditions. This suggests the capability of B. gibsonii KRICT-1 to break down PCL into more hydrophilic compounds, potentially aiding in its degradation and assimilation by microorganisms in saline ecosystems. It demonstrates its potential for use in bioremediation strategies targeting plastic pollution in salt-affected environments, addressing plastic waste concerns in such environments. Given their potential application in biotechnology, the B. gibsonii KRICT-1 is a promising strain for degradation of biodegradable plastics existing in saline soils, saline water, and wastewater condition.
References
Bahl S, Dolma J, Singh JJ et al (2021) Biodegradation of plastics: a state of the art review. Mater Today Proc 39:31–34. https://doi.org/10.1016/j.matpr.2020.06.096
Jambeck JR, Geyer R, Wilcox C et al (2015) Marine pollution. Plastic waste inputs from land into the ocean. Science 347:768–771. https://doi.org/10.1126/science.1260352
Alfaro-Núñez A, Astorga D, Cáceres-Farías L et al (2021) Microplastic pollution in seawater and marine organisms across the Tropical Eastern Pacific and Galápagos. Sci Rep 11:6424. https://doi.org/10.1038/s41598-021-85939-3
Varó I, Perini A, Torreblanca A et al (2019) Time-dependent effects of polystyrene nanoparticles in brine shrimp Artemia franciscana at physiological, biochemical and molecular levels. Sci Total Environ 675:570–580. https://doi.org/10.1016/j.scitotenv.2019.04.157
Chamas A, Moon H, Zheng J et al (2020) Degradation rates of plastics in the environment. ACS Sustain Chem Eng 8:3494–3511. https://doi.org/10.1021/acssuschemeng.9b06635
Thoden van Velzen EU, Santomasi G (2022) Tailor-made enzymes for plastic recycling. Nature 604:631–633. https://doi.org/10.1038/d41586-022-01075-6
Grassie N, Scott G (1985) Polymer degradation and stabilization. Cambridge University Press, Cambridge
Webb HK, Arnott J, Crawford RJ et al (2013) Plastic degradation and its environmental implications with special reference to poly(ethylene terephthalate). Polymers 5:1–18. https://doi.org/10.3390/polym5010001
Delacuvellerie A, Cyriaque V, Gobert S et al (2019) The plastisphere in marine ecosystem hosts potential specific microbial degraders including Alcanivorax borkumensis as a key player for the low-density polyethylene degradation. J Hazard Mater 380:120899. https://doi.org/10.1016/j.jhazmat.2019.120899
Bombelli P, Howe CJ, Bertocchini F (2017) Polyethylene bio-degradation by caterpillars of the wax moth Galleria mellonella. Curr Biol 27:R292–R293. https://doi.org/10.1016/j.cub.2017.02.060
Carr CM, Clarke DJ, Dobson ADW (2020) Microbial polyethylene terephthalate hydrolases: current and future perspectives. Front Microbiol 11:571265. https://doi.org/10.3389/fmicb.2020.571265
Yoshida S, Hiraga K, Takehana T et al (2016) A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 351:1196–1199. https://doi.org/10.1126/science.aad6359
Eugenio EQ, Campisano ISP, Dias AG et al (2022) Novel efficient enzymatic synthesis of the key-reaction intermediate of PET depolymerization, mono(2-hydroxyethyl terephthalate)—MHET. J Biotechnol 358:102–110. https://doi.org/10.1016/j.jbiotec.2022.08.019
Atiq N (2011) Biodegradability of synthetic plastics polystyrene and styrofoam by fungal isolates. Dissertation, Quaid-i-Azam University Islamabad
Peng BY, Su Y, Chen Z et al (2019) Biodegradation of polystyrene by dark (Tenebrio obscurus) and yellow (Tenebrio molitor) mealworms (Coleoptera: Tenebrionidae). Environ Sci Technol 53:5256–5265. https://doi.org/10.1021/acs.est.8b06963
Jeon HJ, Kim MN (2016) Isolation of mesophilic bacterium for biodegradation of polypropylene. Int Biodeterior Biodegrad 115:244–249. https://doi.org/10.1016/j.ibiod.2016.08.025
Mihreteab M, Stubblefield BA, Gilbert ES (2019) Microbial bioconversion of thermally depolymerized polypropylene by Yarrowia lipolytica for fatty acid production. Appl Microbiol Biotechnol 103:7729–7740. https://doi.org/10.1007/s00253-019-09999-2
Cregut M, Bedas M, Durand MJ et al (2013) New insights into polyurethane biodegradation and realistic prospects for the development of a sustainable waste recycling process. Biotechnol Adv 31:1634–1647. https://doi.org/10.1016/j.biotechadv.2013.08.011
Filip Z (1979) Polyurethane as the sole nutrient source for Aspergillus niger and Cladosporium herbarum. Eur J Appl Microbiol Biotechnol 7:277–280. https://doi.org/10.1007/BF00498022
Giacomucci L, Raddadi N, Soccio M et al (2019) Polyvinyl chloride biodegradation by Pseudomonas citronellolis and Bacillus flexus. N Biotechnol 52:35–41. https://doi.org/10.1016/j.nbt.2019.04.005
Webb JS, Nixon M, Eastwood IM et al (2000) Fungal colonization and biodeterioration of plasticized polyvinyl chloride. Appl Environ Microbiol 66:3194–3200. https://doi.org/10.1128/AEM.66.8.3194-3200.2000
Dimassi SN, Hahladakis JN, Yahia MND et al (2022) Degradation-fragmentation of marine plastic waste and their environmental implications: a critical review. Arab J Chem 15:104262. https://doi.org/10.1016/j.arabjc.2022.104262
Chen H (2022) Biodegradable plastics in the marine environment: a potential source of risk? Water Emerg Contam Nanoplast 1:16. https://doi.org/10.20517/wecn.2022.11
Wang GX, Huang D, Ji JH et al (2020) Seawater-degradable polymers-fighting the marine plastic pollution. Adv Sci (Weinh) 8:2001121. https://doi.org/10.1002/advs.202001121
Dong X, Zhu L, He Y et al (2023) Salinity significantly reduces plastic-degrading bacteria from rivers to oceans. J Hazard Mater 451:131125. https://doi.org/10.1016/j.jhazmat.2023.131125
Tamura K, Stecher G, Peterson D et al (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Eom GT, Lee SH, Song BK et al (2013) High-level extracellular production and characterization of Candida antarctica lipase B in Pichia pastoris. J Biosci Bioeng 116:165–170. https://doi.org/10.1016/j.jbiosc.2013.02.016
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. https://doi.org/10.1006/abio.1976.9999
Oh YR, Jang YA, Hong SH et al (2020) Purification and characterization of a malate:quinone oxidoreductase from Pseudomonas taetrolens capable of producing valuable lactobionic acid. J Agric Food Chem 68:13770–13778. https://doi.org/10.1021/acs.jafc.0c04094
Almeida BC, Figueiredo P, Carvalho ATP (2019) Polycaprolactone enzymatic hydrolysis: a mechanistic study. ACS Omega 4:6769–6774. https://doi.org/10.1021/acsomega.9b00345
Oh YR, Jang YA, Song JK et al (2022) Efficient enzymatic depolymerization of polycaprolactone into 6-hydroxyhexanoic acid by optimizing reaction conditions and microbial conversion of 6-hydroxyhexanoic acid into adipic acid for eco-friendly upcycling of polycaprolactone. Biochem Eng J 185:108504. https://doi.org/10.1016/j.bej.2022.108504
Orhan F (2016) Alleviation of salt stress by halotolerant and halophilic plant growth-promoting bacteria in wheat (Triticum aestivum). Braz J Microbiol 47:621–627. https://doi.org/10.1016/j.bjm.2016.04.001
Shameer S (2016) Haloalkaliphilic Bacillus species from solar salterns: an ideal prokaryote for bioprospecting studies. Ann Microbiol 66:1315–1327. https://doi.org/10.1007/s13213-016-1221-7
Murphy C, Cameron J, Huang S et al (1998) A second polycaprolactone depolymerase from Fusarium, a lipase distinct from cutinase. Appl Microbiol Biotechnol 50:692–696. https://doi.org/10.1007/s002530051352
Tseng M, Hoang KC, Yang MK et al (2007) Polyester-degrading thermophilic actinomycetes isolated from different environment in Taiwan. Biodegradation 18:579–583. https://doi.org/10.1007/s10532-006-9089-z
Blackwell CJ, Haernvall K, Guebitz GM et al (2018) Enzymatic degradation of star poly(ε-caprolactone) with different central units. Polymers 10:1266. https://doi.org/10.3390/polym10111266
Li L, Lin X, Bao J et al (2022) Two extracellular poly(ε-caprolactone)-degrading enzymes from Pseudomonas hydrolytica sp. DSWY01T: purification, characterization, and gene analysis. Front Bioeng Biotechnol 10:835847. https://doi.org/10.3389/fbioe.2022.835847
Atanasova N, Stoitsova S, Paunova-Krasteva T et al (2021) Plastic degradation by extremophilic bacteria. Int J Mol Sci 22:5610. https://doi.org/10.3390/ijms22115610
Sonkar K, Singh DP (2021) Kinetic and thermodynamic characterization of novel alkaline lipase from halotolerant Bacillus gibsonii. Arch Microbiol 203:2199–2209. https://doi.org/10.1007/s00203-021-02197-7
Urbanek AK, Rymowicz W, Strzelecki MC et al (2017) Isolation and characterization of Arctic microorganisms decomposing bioplastics. AMB Express 7:148. https://doi.org/10.1186/s13568-017-0448-4
Abdel-Motaal FF, El-Sayed MA, El-Zayat SA et al (2014) Biodegradation of poly (ε-caprolactone) (PCL) film and foam plastic by Pseudozyma japonica sp. Nov., a novel cutinolytic ustilaginomycetous yeast species. Biotech 4:507–512. https://doi.org/10.1007/s13205-013-0182-9
Acknowledgements
This work was supported in part by the R&D programs of MOTIE/KEIT (10077291), and KRICT (KS2442-10 and BSF23-505).
Author information
Authors and Affiliations
Contributions
Ki Ryeon Kim contributed to investigation, validation, data curation, and writing—original draft. Jin-Wan Park, Eun-Bi Cho, and Young-Ah Jang contributed to investigation, data curation, and methodology. Yu Ri Oh and Gyeong Tae Eom contributed to conceptualization, project administration, supervision, writing—original draft, writing—review and editing, funding acquisition, and resources.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Informed consent
Neither ethical approval nor informed consent was required for this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kim, KR., Park, JW., Cho, Eb. et al. Isolation of a halotolerant poly(ε-caprolactone)-depolymerizing strain of Bacillus gibsonii from seaside soil. Biotechnol Bioproc E (2024). https://doi.org/10.1007/s12257-024-00133-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12257-024-00133-2