Summary
Background
Acute lymphoblastic leukemia (ALL), characterized by overproduction and accumulation of immature lymphoid cells in bone marrow and peripheral blood, is the most common malignancy in children. NOTCH signaling is suggested to be a key event in hematological malignancies and appears to be a major oncogenic trigger in leukemia. Several studies on NOTCH target gene (HES‑1, p21 and c‑Myc) expression evaluated the correlation between these genes in AML (acute myeloid leukemia), but this relationship has not yet been clarified in ALL. Therefore, we aimed to study the expression of these genes in our Egyptian patients with ALL to obtain more information.
Patients and methods
RNA was extracted from peripheral blood mononuclear cells (PBMNCs) of 91 pediatric ALL patients (49 B-cell acute lymphoblastic leukemia [B-ALL] and 42 T-cell acute lymphoblastic leukemia [T-ALL]) and 52 healthy controls. The expression levels were determined by quantitative real-time polymerase chain reaction (qRT-PCR).
Results
Median p21 and HES1 expressions were down regulated, while c‑Myc expression was up-regulated in B‑ALL cases (p < 0.001, p = 0.008, p < 0.001, respectively) and in T‑ALL cases (p = 0.049, p = 0.015, p < 0.001, respectively) when compared to the control group. Median HES1 expression was down regulated, in B‑ALL cases compared to T‑ALL cases (p = 0.002), while P21 and c‑Myc did not differ significantly between B‑ALL and T‑ALL cases.
Conclusion
P21 expression showed a significant positive correlation with HES1 expression and c‑Myc showed nonsignificant negative correlations with p21 and HES1, thus, suggesting that HES1 may affect ALL cells through the HES1–p21 pathway. Patients with over expressed c‑Myc had worse survival than patients with low expression which suggested it is a risk predictor.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acute lymphoblastic leukemia (ALL) is a malignant disease of the bone marrow in which early lymphoid precursors proliferate and replace the normal hematopoietic cells, resulting in a marked decrease in normal blood cell production [1]. ALL constitutes 30% of all pediatric malignancies and 70% of pediatric leukemia; cases show a male to female ratio of 2.3:1. The 2–10 year age group constitutes 68.5% [2].
NOTCH can act as both an oncogene and a tumor suppressor [3]. A prime example of oncogenic NOTCH signaling is provided by T‑cell acute lymphoblastic leukemia (T-ALL), which is an aggressive neoplasm of immature T cells. In this disease, gain-of-function mutations give rise to constitutively active forms of NOTCH [4]. In addition, NOTCH signaling is involved in the pathogenesis of certain colon cancers [5].
The best known NOTCH target genes include the following: HES (Hairy enhancer of split) which is a helix–loop–helix transcription factor that functions as a transcriptional repressor [6], c‑Myc (MYC proto-oncogene) which drives cell cycle progression and regulates the expression of key enzymes that control cellular metabolism and stimulates ribosome biogenesis and protein synthesis through interactions with RNA polymerase III and RNA polymerase I [7,8,9,10,11,12], and finally p21 gene (also known as cyclin-dependent kinase inhibitor 1) which is involved in replicative senescence and terminal differentiation and proliferation in nonhemopoietic and hemopoietic cells [13]. Expression of p21 is induced by wild-type p53 in the presence of DNA damage, leading to apoptosis or cell cycle arrest at the G1 checkpoint [14].
Due to the important role of NOTCH target gene expression in tumorigenesis and because this relationship in ALL is not clarified yet due to the small number of studies in ALL, our goal was to evaluate the expression of these genes in pediatric Egyptian ALL patients and correlate the results with clinical and laboratory data of the patients.
Patients and methods
Patients’ samples
This study involved 91 newly diagnosed pediatric ALL patients treated at the Oncology Center Mansoura University. Diagnosis of ALL was performed according to standard cytomorphology and immunophenotypic criteria. All samples were obtained in accordance with the Declaration of Helsinki, with informed consent obtained from the patients, parents or guardians and approval from the faculty of medicine Mansoura University institutional review board. Peripheral blood (PB) samples were obtained from the 91 ALL patients (40 [44%] male and 51 [56%] female with a median age of 6 years); healthy control PB samples were obtained from 52 healthy donors (median age 6 years).
Methods
PB mononuclear cells isolation
Mononuclear cells were isolated by density gradient centrifugation using lymphocyte separation medium (Lonza, Walkersville, MD, USA); RNA was isolated from mononuclear cells using miRNeasy Mini kits (Qiagen, Germantown, MD, USA). RNA concentration and purity were determined using NanoDrop (Maestrogen, Hsinchu, Taiwan).
cDNA and quantitative real-time PCR (qRT-PCR)
cDNA was synthesized from 2 μg RNA using high capacity reverse transcription kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. Quantitative real-time PCR was performed on the StepOne™ by using TaqMan gene expression assays for P21, HES1 and c‑MYC genes (Life Technologies, Grand Island, NY, USA). The house-keeping gene Glyceraldehyde 3‑phosphate dehydrogenase (GAPDH) used as internal controls. The relative gene expression level was calculated as 2−∆∆Ct.
Statistical analysis
The collected data were revised, coded and tabulated using Statistical package for Social Science (Released 2011, SPSS Statistics for Windows, Version 20.0., IBM Corp., Armonk, NY, USA). Data were compiled and suitable analyses were done according to the type of data obtained for each parameter. The Kolmogrov–Smirnov test was done to test the normality of data distribution. Significant data were considered to be nonparametric.
Results
Expression levels of HES1, P21, and c-Myc genes
To determine the expression pattern of HES1, P21, and c‑Myc genes, respectively, in T‑ALL and B‑ALL patients, 91 samples of PB mononuclear cells from pediatric ALL patients and 52 from normal subjects were analyzed using qRT-PCR. In all, 91 patients (40 males, 51 females) with T‑ALL (n = 42) and B‑ALL (n = 49) at a median age of 6 years (range 1–18 years) were analyzed.
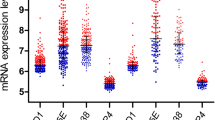
Median p21 and HES1 expressions were down regulated, while c‑Myc expression was up regulated in ALL cases when compared to the control group (p < 0.001 each). Median p21 and HES1 expressions were down regulated, while c‑Myc expression was up regulated in B‑ALL cases when compared to the control group (p < 0.001, p = 0.008, p < 0.001, respectively). Median p21 and HES1 expressions were down regulated, while c‑Myc expression was up regulated in T‑ALL cases when compared to the control group (p = 0.049, p = 0.015, p < 0.001, respectively). Median HES1 expression was down regulated in B‑ALL cases compared to T‑ALL cases (p = 0.002). P21 and c‑Myc did not differ significantly between B‑ALL and T‑ALL cases.
Relationship between HES1, p21, and c-Myc expression and the clinicopathological characteristics of pediatric ALL patients
To determine whether HES1, p21, and c‑Myc expression levels correlate with the clinicopathological characteristics of pediatric ALL patients, we divided patients into high and low groups based on the median expression value of HES1, p21, and c‑Myc expression.
B‑ALL patients with low HES1 gene expression had higher peripheral blasts (75% vs. 90%, p = 0.003), higher frequency of hepatomegaly (54.5% vs. 88.9%, p = 0.007), and high risk rate (13.6% vs. 81.5%, p = <0.001) compared to those with high HES1 expression as shown in Table 1. Moreover, low p21 gene expression was significantly associated with higher TLC (5.3 × 109/L vs. 11.25 × 109/L, p = 0.003), higher frequency of hepatomegaly (48% vs. 100%, p ≤ 0.001), high risk rate (24% vs. 79.2%, p ≤ 0.001), nonremission rate (8% vs. 33.3%, p = 0.037), and lower platelet count (56 × 109/L vs. 39.5 × 109/L, p = 0.016) compared to high p21 gene expression (Table 2). High c‑Myc gene expression was significantly associated with lower frequency of hepatomegaly (100% vs. 50%, p ≤ 0.001) and splenomegaly (65.2% vs. 34.6%, p = 0.032) compared to patients with low c‑Myc gene expression (Table 3).
In T‑ALL patients, low HES1 gene expression was significantly associated with higher frequency of high-risk rate (33.3% vs. 95.2%, p = <0.001) and nonremission rate (0% vs. 52.4%, p ≤ 0.001; Table 1). Low p21 gene expression was significantly associated with higher TLC (8.1 × 109/L vs. 66.5 × 109/L, p ≤ 0.001) and higher frequency of high risk (33.3% vs. 95.2% p ≤ 0.001) than those with high p21 gene expression (Table 2). High c‑Myc gene expression was significantly associated with higher frequency of nonremission rate (9.5% vs. 42.9%, p = 0.032) compared to patients with low c‑Myc gene expression (Table 3).
When we compared the expression of the genes in the different studied groups, we found that median p21 and HES1 expression was down regulated, while c‑Myc expression was up regulated in B‑ALL cases when compared to the control group (p < 0.001, p = 0.008, p < 0.001, respectively). Median p21 and HES1 expressions were down regulated, while c‑Myc expression was up regulated in T‑ALL cases when compared to the control group (p = 0.049, p = 0.015, p < 0.001, respectively; Fig. 1).
Gene expression in the control, B‑ALL, and T‑ALL groups: a HES1 expression, b P21 expression, c c-Myc expression. B-ALL B‑cell acute lymphoblastic leukemia, T-ALL T‑cell acute lymphoblastic leukemia, cMyc MYC proto-oncogene, P21 cyclin-dependent kinase inhibitor 1, HES1 hairy enhancer of split 1, OS overall survival, DFS Disease-free survival
P21 expression showed a significant positive correlation with HES1 expression in total ALL, B‑ALL, and T‑ALL cases (rs = 0.667, p < 0.001; rs = 0.673, p < 0.001; rs = 741, p < 0.001, respectively). But c‑Myc showed nonsignificant negative correlations with p21 and HES1 in total ALL, B‑ALL, and T‑ALL cases.
Shorter OS was significantly associated with low p21, low HES1, and high c‑Myc expressions in B‑ALL and T‑ALL cases (Fig. 2). Also, patients with high c‑Myc expression levels in B‑ALL and T‑ALL patients showed shorter DFS than patients with low c‑Myc expression levels (17.4 months vs. 8 months, p = 0.12; 11.5 months vs. 3.6 months, p = 0.001) as shown in Fig. 3.
Association of HES1, p21, and c‑Myc expression with OS in B‑ALL and T‑ALL cases. a OS according to HES1 expression in B‑ALL cases. b OS according to HES1 expression in T‑ALL cases. c OS according to p21 expression in B‑ALL cases. d OS according to p21 expression in T‑ALL cases. e OS according to c‑Myc expression in B‑ALL cases. f OS according to c‑Myc expression in T‑ALL cases
Discussion
In this study, our goal was to identify the role of NOTCH target genes such as HES1, P21 and c‑Myc genes to clarify their role in ALL pathogenesis and correlate the results with clinical and laboratory data of the patients.
The role of HES1 in the prognosis of ALL has not been well demonstrated. We found that the expression level of HES1 gene was low in PBMNC samples of pediatric ALL compared to normal PB control (p value < 0.001). This is consistent with another study reporting that the average expression of HES1 in AML (acute myeloid leukemia) BMNCs was lower than that in normal BMNCs [15]. In our study, HES‑1 expression in pediatric B‑ALL was significantly decreased in comparison to pediatric T‑ALL. (p = 0.002), which is consistent with another study reporting that B‑ALL samples showed lower levels of HES1 expression than T‑ALL samples, thus, suggesting lower levels of NOTCH activation in B‑ALL samples [16]. Low HES1 gene expression in B‑ALL was significantly associated with higher peripheral blasts, higher hepatomegaly, and high risk rate compared to those with high HES1 gene expression. However, in the T‑ALL group, low HES1 gene expression was significantly associated with high risk and nonremission rate. In the evaluation of the expression of HES1 as a prognostic factor for T‑ALL and B‑ALL patients, our study showed that the low-expression group had a shorter OS time compared with those of the high-expression group and this is consistent with another study on AML reporting that the OS of the high-expression group is significantly longer than that of the low-expression group (37.6 ± 1.6 months versus 54.0 ± 1.3 months, p < 0.05) [15].
In this study median p21 expression were down regulated in the entire ALL group, B‑ALL cases, and T‑ALL cases when compared to the control group (p < 0.001, p < 0.001, p = 0.049, respectively). Furthermore, low p21 gene expression was significantly associated with higher TLC, higher hepatomegaly, high risk rate, nonremission rate, and lower platelet count in the B‑ALL group and in the T‑ALL group. This might give an important role to study the expression of p21 as it is considering as a strong candidate for participation in tumor progression and over expression of p21 might suppress tumor growth in different experimental models which might justify the importance of target therapy usage [17, 18]. Our finding in ALL was consistent with a previous study done on P21 in AML patients that pointed out the importance of p21 in human leukemias and indicated that the lower p21 expression might be prognostic factor in acute myeloid leukemia patients [19]. Another study showed that p21 was frequently down regulated in adult T‑cell leukemia/lymphoma (ATLL) cells [20], while others reported an absence of p21 expression has been noted in T‑ALL cells [21]. In our study, shorter OS was significantly associated with low p21expression group in B‑ALL and T‑ALL and this was suggested to be poor prognostic factor. DFS was not significantly associated with p21 expression in the B‑ALL or T‑ALL group.
On the other hand, c‑Myc expression was up regulated in the entire ALL group, B‑ALL cases, and T‑ALL cases when compared to the control group (p < 0.001), and did not differ significantly between B‑ALL and T‑ALL cases. This is compatible with other studies reporting that c‑Myc is frequently overexpressed in human acute lymphoblastic and myeloid leukemia [22,23,24,25]. Up regulation of c‑Myc in human ALL has been reported through chromosome translocations, aberrant c‑Myc stability, and genetic gene fusion [26,27,28]. No significant differences in c‑Myc expression were observed with age, sex, or peripheral blood blasts.
Conclusion
In our study, we found a positive correlation between HES1 expression and p21 expression in pediatric ALL which suggests that HES1 may affect ALL cells through the HES1–p21 pathway. Down regulation of p21 and HES1 expression and up regulation of c‑Myc expression were significantly associated with high-risk leukemia, nonremission rate, shorter OS, and shorter DFS which suggest that these are poor prognostic factors in ALL. Therefore, we recommend further studies of these genes in ALL in order to assess the importance of these genes in ALL.
References
Maloney KW, Carroll WL, Carroll AJ, Devidas M, Borowitz MJ, Martin PL, et al. Down syndrome childhood acute lymphoblastic leukemia has a unique spectrum of sentinel cytogenetic lesions that influences treatment outcome: a report from the Children’s Oncology Group. Blood. 2010;116:1045–50. https://doi.org/10.3324/haematol.2010.024968.
Shalaby H, Ashaat A, El-Wahab A, El-Hamid M, El-Wakeel S. Bcl‑2 expression and chromosomal abnormalities in childhood acute lymphoblastic leukemia. Acad J Cancer Res. 2010;3:34–43.
Koch U, Radtke F. Notch and cancer: a double-edged sword. Cell Mol Life Sci. 2007;64:2746–62. https://doi.org/10.1007/s00018-007-7164-1.
Borggrefe T, Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol Life Sci. 2009;66:1631–46.
Qiao L, Wong BC. Role of Notch signaling in colorectal cancer. Carcinogenesis. 2009;30:1979–86. https://doi.org/10.1093/carcin/bgp236.
Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194:237–55. https://doi.org/10.1002/jcp.10208.
Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–99. https://doi.org/10.1146/annurev.cellbio.16.1.653.
Levens DL. Reconstructing Myc. Genes Dev. 2003;17:1071–7. https://doi.org/10.1101/gad.1095203.
Felton-Edkins ZA, Kenneth NS, Brown TR, Daly NL, Gomez-Roman N, Grandori C, et al. Direct regulation of RNA polymerase III transcription by RB,p53 and c‑Myc. Cell Cycle. 2003;2:180–3.
Arabi A, Wu S, Ridderstråle K, Bierhoff H, Shiue C, Fatyol K, et al. c‑Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol. 2005;7:303. https://doi.org/10.1038/ncb1225.
Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, et al. c‑Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol. 2005;7:311. https://doi.org/10.1038/ncb1224.
Grewal SS, Li L, Orian A, Eisenman RN, Edgar BA. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat Cell Biol. 2005;7:295. https://doi.org/10.1038/ncb1223.
Taniguchi T, Endo H, Chikatsu N, Uchimaru K, Asano S, Fujita T, et al. Expression of p21Cip1/Waf1/Sdi1 and p27Kip1cyclin-dependent kinase inhibitors during human hematopoiesis. Blood. 1999;93:4167–78.
El-Deiry WS, Harper JW, O’Connor PM, Velculescu VE, Canman CE, Jackman J, et al. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54:1169–74.
Tian C, Tang Y, Wang T, Yu Y, Wang X, Wang Y, et al. HES1 is an independent prognostic factor for acute myeloid leukemia. OTT. 2015;8:899. https://doi.org/10.1007/s00277-015-2413-0.
Kannan S, Fang W, Song G, Mullighan CG, Hammitt R, McMurray J, et al. Notch/HES1-mediated PARP1 activation: a cell type–specific mechanism for tumor suppression. Blood. 2011;117:2891–900. https://doi.org/10.1182/blood-2009-12-253419.
Chen YQ, Cipriano SC, Arenkiel JM, Miller FR. Tumor suppression by p21WAF1. Cancer Res. 1995;55:4536–9.
Bae I, Fan S, Bhatia K, Kohn KW, Fornace AJ, O’Connor PM. Relationships between G1 arrest and stability of the p53 and p21Cip1/Waf1 proteins following γ‑irradiation of human lymphoma cells. Cancer Res. 1995;55:2387–93.
Polak J, Pekova S, Schwarz J, Kozak T, Haskovec C. Expression of cyclin-dependent kinase inhibitors in leukemia. Cas Lek Ceskych. 2003;142:25–8.
Watanabe M, Nakahata S, Hamasaki M, Saito Y, Kawano Y, Hidaka T, et al. Downregulation of CDKN1A in adult T‑cell leukemia/lymphoma despite overexpression of CDKN1A in human T‑lymphotropic virus 1‑infected cell lines. J Virol. 2010;84:6966–77. https://doi.org/10.1128/JVI.00073-10.
Scott SA, Kimura T, Dong W‑F, Ichinohasama R, Bergen S, Kerviche A, et al. Methylation status of cyclin-dependent kinase inhibitor genes within the transforming growth factor beta pathway in human T‑cell lymphoblastic lymphoma/leukemia. Leuk Res. 2004;28:1293–301.
Grabher C, von Boehmer H, Look AT. Notch 1 activation in the molecular pathogenesis of T‑cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2006;6:347. https://doi.org/10.1038/nrc1880.
Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, et al. c‑Myc is an important direct target of Notch1 in T‑cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–109.
Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, et al. NOTCH1 directly regulates c‑MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci. 2006;103:18261–6. https://doi.org/10.1073/pnas.0606108103.
Gutierrez A, Sanda T, Ma W, Zhang J, Grebliunaite R, Dahlberg S, et al. Inactivation of LEF1 in T‑cell acute lymphoblastic leukemia. Blood. 2010;115:2845–51. https://doi.org/10.1182/blood-2009-07-234377.
Faderl S, O’Brien S, Pui CH, Stock W, Wetzler M, Hoelzer D, et al. Adult acute lymphoblastic leukemia: concepts and strategies. Cancer. 2010;116:1165–76. https://doi.org/10.1002/cncr.24862.
Malempati S, Tibbitts D, Cunningham M, Akkari Y, Olson S, Fan G, et al. Aberrant stabilization of c‑Myc protein in some lymphoblastic leukemias. Leukemia. 2006;20:1572.
Rice KL, Hormaeche I, Doulatov S, Flatow JM, Grimwade D, Mills KI, et al. Comprehensive genomic screens identify a role for PLZF-RARα as a positive regulator of cell proliferation via direct regulation of c‑MYC. Blood. 2009;114:5499–511. https://doi.org/10.1182/blood-2009-03-206524.
Acknowledgements
The authors like to thank the patients and healthy volunteers who participated in this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
M. Reyad, S. Abdel-Aziz, L.M. Saleh, S. El-Ghlban, I. El Tantawy El Sayed and H. Abdel-ghaffar declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Reyad, M., Abdel-Aziz, S., Saleh, L.M. et al. The emerging role of NOTCH target genes in Egyptian childhood acute lymphoblastic leukemia. memo 14, 119–126 (2021). https://doi.org/10.1007/s12254-020-00665-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12254-020-00665-2