Abstract
Both NRIP1 and DOK1 genes are considered candidate tumor suppressor genes (TSGs). Also, cell polarity-related genes PARD3, PRKCI and DLGAP3, and autophagy-related genes ULK1 and ULK2 genes are considered to play crucial roles in tumorigenesis. The aim of our study was to find whether these genes were mutated in colorectal cancer (CRC). In a genome database, we observed that each of these genes harbored mononucleotide repeats in the coding sequences, which could be mutated in cancers with high microsatellite instability (MSI-H). For this, we studied 124 CRCs for the frameshift mutations of these genes and their intratumoral heterogeneity (ITH). NRIP1, DOK1, PARD3, PRKCI, DLGAP3, ULK1 and ULK2 harbored 18 (22.8%), 2 (2.5%), 2 (2.5%), 2 (2.5%), 5 (6.3%), 2 (2.5%) and 2 (2.5%) of 79 CRCs with MSI-H, respectively. However, we found no such mutations in microsatellite stable (MSS) cancers in the nucleotide repeats. We also studied ITH for the frameshift mutations in 16 cases of CRCs and detected that the frameshift mutations of NRIP1, DOK1, PARD3, PRKCI, DLGAP3, ULK1 and ULK2 showed regional ITH in 5 (31.3%), 2 (12.5%), 0 (0%), 0 (0%), 1 (6.3%), 1 (6.3%) and 3 (18.8%) cases, respectively. Our data exhibit that candidate cancer-related genes NRIP1, DOK1, PARD3, PRKCI, DLGAP3, ULK1 and ULK2 harbor mutational ITH as well as the frameshift mutations in CRC with MSI-H. Also, the results suggest that frameshift mutations of these genes might play a role in tumorigenesis through their inactivation in CRC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Transcription cofactor NRIP1 (nuclear receptor–interacting protein 1) represses E2F1 activity and inhibits E2F1 target gene expression such as CCNE1 and CCNB2, subsequently inhibiting cell cycle progression [1]. Decreased expression of NRIP1 has been reported in many tumors including breast and colorectal cancers (CRC) [1, 2]. Docking proteins (DOK1, DOK2 and DOK3) belong to a tyrosine-phosphorylated adaptor protein family and inhibit tyrosine kinase activation [3]. DOK proteins are frequently downregulated in cancers [4]. Evidence exists that aberrant expression and localization of the polarity proteins are common in human tumors [5]. Atypical PKCs, PARD3 and PARD6 bind together and constitute the Pard complex that is central to the development of junctional structures and apical-basolateral polarization in epithelial cells [6]. PARD3 and PRKCI encode PARD3 and protein kinase C iota (an atypical PKC), respectively [7, 8]. DLGAP3 is an interacting protein with DLG4, a Dlg family protein in the Scribble complex [9]. Autophagy is implicated in many human diseases, including neurodegenerative disorders, microbial infections and cancers [10]. ULK1 and ULK2 form Unc-51 like autophagy activating kinase 1 (ULK1) protein kinase complex that is located at the most upstream position during autophagy process [10].
About one third of CRCs are classified as high microsatellite instability (MSI-H) cancers [11]. Many TSGs harbor frameshift mutations at monocleotide repeats in MSI-H cancers [11]. In the human genome database, we observed that NRIP1, DOK1, PARD3, PRKCI, DLGAP3, ULK1 and ULK2 genes possess nucleotide repeats in coding sequences that might be mutated in MSI-H cancers. However, frameshift mutations of the repeats in these genes in CRC have not been reported. Intratumoral heterogeneity (ITH) is a common phenomenon in cancers, which may result in cancer evolution and influence on clinical outcomes [12,13,14]. Thus, identification of genetic ITH is important in understanding biological and clinicopathologic features of the cancers. The current study aimed to find whether NRIP1, DOK1, PARD3, PRKCI, DLGAP3, ULK1 and ULK2 frameshift mutations are common and harbor ITH in MSI-H CRC.
Materials and Methods
Tissue Samples and Microdissection
In this study, we used 124 CRCs that consisted of 45 CRCs with microsatellite stable (MSS) and 79 CRCs with MSI-H. These samples overrepresent MSI-H cases due to our collection method (MSI-H and MSS cases were separately collected in different times). We adopted an MSI evaluation system using five mononucleotide repeats [15]. For 16 MSI-H CRCs, we picked 4–7 tumor areas and one normal area per CRC for the ITH analysis. Each ITH area was histologically confirmed under light microscope. These ITH areas were studied for detecting mutational ITH of NRIP1, DOK1, PARD3, PRKCI, DLGAP3, ULK1 and ULK2. Pathologic features of the cancers were evaluated in all blocks of all cases by a pathologist and are summarized in Table 1. Tumor and normal cells were microdissected as described previously [16]. Approval of this study was obtained from the institutional review board of the Catholic University of Korea.
Single Strand Conformation Polymorphism (SSCP) Analysis
We analyzed DNA sequences in NRIP1 (A8 and A7), DOK1 (T), DLGAP3 (G7), PARD3 (A7), PRKCI (A8), ULK1 (C7 and C5) and ULK2 (A7). Genomic DNA was amplified using polymerase chain reaction (PCR). [32P]dCTP was incorporated to the PCR products for visualization in autoradiogram. We determined aberrant gel motility in the SSCP (FMC Mutation Detection Enhancement system; Intermountain Scientific, Kaysville, UT, USA) using visual inspection, which subsequently sequenced by Sanger DNA sequencing (3730 DNA Analyzer, Applied Biosystem, Carlsbad, CA, USA). Other procedures in detail were described in our earlier studies [17, 18].
Results
Mutational Analysis
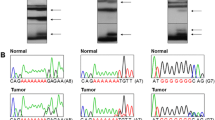
Genomic DNAs of the 124 CRCs (79 with MSI-H and 45 with MSS) were studied to detect frameshift mutations in the mononucleotide repeats by the PCR-SSCP analyses. On SSCP and Sanger sequencing, we observed that NRIP1, DOK1, PARD3, PRKCI, DLGAP3, ULK1 and ULK2 harbored 18 (22.8%), 2 (2.5%), 2 (2.5%), 2 (2.5%), 5 (6.3%), 2 (2.5%) and 2 (2.5%) of 79 CRCs with MSI-H, respectively. Normal tissues of the corresponding patients from the slides were microdissected and analyzed, but there was no evidence of the mutations on SSCP and Sanger sequencing, which indicated that the mutations had risen somatically (Fig. 1 and Table 2). All detected mutations exhibited both wild-type and mutant signals in SSCP and Sanger sequencing, indicating that they were heterozygous mutations (Fig. 1).
Representative DNA sequencings of the repeats in NRIP1 and DOK1 in colon cancers. DNA sequencing analyses of A8 and A7 repeats in NRIP1 and T7 repeat in DOK1 from tumor (T) and normal tissues (N). a-c Direct DNA sequencing analyses show a heterozygous A deletion or a heterozygous A duplication for the NRIP1 repeats in the tumor tissues as compared to normal tissues. D-E. Direct DNA sequencing analyses show a heterozygous T deletion or a heterozygous T duplication for the DOK1 repeats in the tumor tissues as compared to normal tissues
The frameshift mutations detected in this study were found in MSI-H CRCs, but there was none in MSS CRCs (Table 2) (Fisher’s exact test, p < 0.001). There was no significant association of these mutations with the grade, differentiation and tumor stage. In those with MSI-H, no correlation was observed between histological features (medullary pattern, mucinous histology and tumor-infiltrating lymphocytes) and the mutations.
Intratumoral Heterogeneity of the Frameshift Mutations
Ninety-six areas from 16 CRCs with MSI-H were analyzed to find ITH in the frameshift mutations. We observed that the frameshift mutations of NRIP1, DOK1, PARD3, PRKCI, DLGAP3, ULK1 and ULK2 showed regional ITH in 5 (31.3%), 2 (12.5%), 0 (0%), 0 (0%), 1 (6.3%), 1 (6.3%) and 3 (18.8%) cases, respectively (Table 3 and Fig. 2). There was no significant histological difference between mutation (+) and mutation (−) ITH areas.
Discussion
Tumor suppressor genes (TSGs) or anti-oncogene is a gene that protects a cell from one step on the path to cancer [19]. When a TSG is mutated, the cells can progress to cancer in combination with other genetic or epigenetic changes. Earlier studies identified that both NRIP1 and DOK1 possessed TSG activities in cells [1,2,3,4]. Loss of cell polarity is a typical hallmark of cancer development and progression in epithelial cells [5]. DLGAP3, PARD3 and PRKCI are polarity signaling-related genes that are related to TSG functions [5]. Loss of autophagy contributes to tumorigenesis [10]. Both ULK1 and ULK2 play crucial roles in autophagy formation [10]. The present study here identified that MSI-H CRC exhibited frameshift mutations within the repeats of NRIP1, DOK1, PARD3, PRKCI, DLGAP3, ULK1 and ULK2 in CRC. These results indicate that the functions of these genes are altered by frameshift mutations identified in this study and suggest that these inactivating mutations might inhibit their functions in MSI-H CRC as well. The NRIP1, DOK1, PARD3, PRKCI, DLGAP3 frameshift mutations might play a role in the pathogenesis of MSI-H CRC by inhibiting their TSG activities. Also, the inactivating mutations of ULK1 and ULK2 might play a role in tumorigenesis of CRC by inhibiting autophagy activities of ULK1 and ULK2.
The frameshift mutations identified in the present study may truncate and disable their encoded proteins and hence may be considered loss-of-function mutations. It is also possible that both of these would be expected to lead to premature stop codons in the mRNAs but not necessarily the production of a truncated protein as these transcripts are more likely to be degraded by nonsense mediated decay [11]. Since cancers with MSI show extremely high mutator phenotype with frameshift mutation in microsatellite sequences, the frameshift mutations in this study could be passenger mutations. By contrast, many TSGs have been found to harbor mutations at nucleotide repeats in the coding sequences in the cancers with MSI (type II transforming growth factor β receptor, BAX, caspase-5, hMSH3, hMSH6 and TCF4) [20]. It remains undetermined whether the frameshift mutations detected in this study are driver or passenger mutations for the development of MSI-H cancers. We were not able to find any significant difference in clinocopathologic parameters between those with and without the mutations probably due to the small number of mutated cases. Analysis of a larger cohort with the mutations is required in future studies.
In this study, we identified mutational ITH of NRIP1, DOK1, DLGAP3, ULK1 and ULK2 in CRCs, which is consistent with previous studies that had reported frequent mutational ITH in CRCs with MSI-H [21]. Genomic instability in a cancer may result in an elevated level of somatic mutations and contribute to ITH development by providing a pool of mutations upon which selection can act in a given microenvironment [12,13,14]. The cancer ITH of driver mutations could result in poor clinical outcomes in patients as well. For instance, single or a group of mutations with a metastasis potential might redirect clinical outcomes since such rare clones may accomplish dominance during tumor progression [12,13,14]. Based on the cancer-related properties of the genes, our results suggest that loss of their properties may have regional heterogeneity in CRC that could be further selected and influence the clinical outcomes. However, presence of ITH of the frameshift mutations might suggest a possibility that there could be a mixed or ameliorated effect of the frameshift mutations in MSI-H cancers.
References
Docquier A, Harmand PO, Fritsch S, Chanrion M, Darbon JM, Cavaillès V (2010) The transcriptional coregulator RIP140 represses E2F1 activity and discriminates breast cancer subtypes. Clin Cancer Res 16:2959–2970
Lapierre M, Bonnet S, Bascoul-Mollevi C, Ait-Arsa I, Jalaguier S, Del Rio M, Plateroti M, Roepman P, Ychou M, Pannequin J, Hollande F, Parker M, Cavailles V (2014) RIP140 increases APC expression and controls intestinal homeostasis and tumorigenesis. J Clin Invest 124:1899–1913
Di Cristofano A, Carpino N, Dunant N, Friedland G, Kobayashi R, Strife A, Wisniewski D, Clarkson B, Pandolfi PP, Resh MD (1998) Molecular cloning and characterization of p56dok-2 defines a new family of RasGAP-binding proteins. J Biol Chem 273:4827–4830
Berger AH, Niki M, Morotti A, Taylor BS, Socci ND, Viale A, Brennan C, Szoke J, Motoi N, Rothman PB, Teruya-Feldstein J, Gerald WL, Ladanyi M, Pandolfi PP (2010) Identification of DOK genes as lung tumor suppressors. Nat Genet 42:216–223
Zen K, Yasui K, Gen Y, Dohi O, Wakabayashi N, Mitsufuji S, Itoh Y, Zen Y, Nakanuma Y, Taniwaki M, Okanoue T, Yoshikawa T (2009) Defective expression of polarity protein PAR-3 gene (PARD3) in esophageal squamous cell carcinoma. Oncogene 28:2910–2918
Suzuki A, Yamanaka T, Hirose T, Manabe N, Mizuno K, Shimizu M, Akimoto K, Izumi Y, Ohnishi T, Ohno S (2001) Atypical protein kinase C is involved in the evolutionarily conserved par protein complex and plays a critical role in establishing epithelia-specific junctional structures. J Cell Biol 152:1183–1196
Mazzarella R, Ciccodicola A, Esposito T, Arcucci A, Migliaccio C, Jones C, Schlessinger D, D'Urso M, D'Esposito M (1995) Human protein kinase C iota gene (PRKCI) is closely linked to the BTK gene in Xq21.3. Genomics 26:629–631
Ebnet K, Suzuki A, Horikoshi Y, Hirose T, Meyer Zu Brickwedde MK, Ohno S, Vestweber D (2001) The cell polarity protein ASIP/PAR-3 directly associates with junctional adhesion molecule (JAM). EMBO J 20:3738–3748
Welch JM, Wang D, Feng G (2004) Differential mRNA expression and protein localization of the SAP90/PSD-95-associated proteins (SAPAPs) in the nervous system of the mouse. J Comp Neurol 472:24–39
Jiang X, Overholtzer M, Thompson CB (2015) Autophagy in cellular metabolism and cancer. J Clin Invest 125:47–54
Imai K, Yamamoto H (2008) Carcinogenesis and microsatellite instability: the interrelationship between genetics and epigenetics. Carcinogenesis 29:673–680
Marusyk A, Almendro V, Polyak K (2012) Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer 12:323–334
McGranahan N, Swanton C (2015) Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 27:15–26
Choi YJ, Rhee JK, Hur SY, Kim MS, Lee SH, Chung YJ, Kim TM, Lee SH (2017) Intraindividual genomic heterogeneity of high-grade serous carcinoma of the ovary and clinical utility of ascitic cancer cells for mutation profiling. J Pathol 241:57–66
Murphy K, Zhang S, Geiger T, Hafez MJ, Bacher J, Berg KD, Eshleman JR (2006) Comparison of the microsatellite instability analysis system and the Bethesda panel for the determination of microsatellite instability in colorectal cancers. J Mol Diagn 8:305–311
Jung SH, Kim MS, Jung CK, Park HC, Choi HJ, Maeng L, Min KO, Kim J, Park TI, Shin OR, Kim TJ, Xu H, Lee KY, Kim TM, Song SY, Lee C, Chung YJ, Lee SH (2016) Whole-exome sequencing identifies recurrent AKT1 mutations in sclerosing hemangioma of lung. Proc Natl Acad Sci U S A 113:10672–10677
Jo YS, Choi MR, Song SY, Kim MS, Yoo NJ, Lee SH (2016) Frameshift mutations of HSPA4 and MED13 in gastric and colorectal cancers. Pathol Oncol Res 22:769–772
Choi EJ, Kim MS, Song SY, Yoo NJ, Lee SH (2017) Intratumoral heterogeneity of frameshift mutations in MECOM gene is frequent in colorectal cancers with high microsatellite instability. Pathol Oncol Res 23:145–149
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Nature Reviews Cell 144:646–674
Calin GA, Gafà R, Tibiletti MG, Herlea V, Becheanu G, Cavazzini L, Barbanti-Brodano G, Nenci I, Negrini M, Lanza G (2000) Genetic progression in microsatellite instability high (MSI-H) colon cancers correlates with clinico-pathological parameters: a study of the TGRbetaRII, BAX, hMSH3, hMSH6, IGFIIR and BLM genes. Int J Cancer 89:230–235
Choi YJ, Kim MS, An CH, Yoo NJ, Lee SH (2014) Regional bias of intratumoral genetic heterogeneity of nucleotide repeats in colon cancers with microsatellite instability. Pathol Oncol Res 20:965–971
Acknowledgements
This study was supported by grants from National Research Foundation of Korea (2012R1A5A2047939 and 2016R1D1A1B01007490).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of Interest and Financial Sponsorship and Support
None.
Rights and permissions
About this article
Cite this article
Choi, E.J., Lee, J.H., Kim, M.S. et al. Intratumoral Heterogeneity of Somatic Mutations for NRIP1, DOK1, ULK1, ULK2, DLGAP3, PARD3 and PRKCI in Colon Cancers. Pathol. Oncol. Res. 24, 827–832 (2018). https://doi.org/10.1007/s12253-017-0297-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12253-017-0297-0