Abstract
Breast cancer (BC) is the leading cause of cancer related death among women in 2014. The AURKA gene that encodes the protein called Aurora kinase A plays an important role in the progression of the cell cycle, by controlling and promoting the entry into the phase of mitosis. The single nucleotide polymorphism AURKA T91A (rs2273535) (Phe21Ile) has been identified as functional alternator of this kinase, the Ile allele is associated with the occurrence of chromosome segregation errors and tumor progression. Therefore, it is essential to know how BC risk is associated with histopathological characteristics, immunohistochemical characteristics, and genotype polymorphism in a high altitude Ecuadorian mestizo population. In this retrospective case-control study 200 individuals were analyzed. DNA was extracted from 100 healthy and 100 affected women. Genotypes were determined by genomic sequencing. We found significant association between the AURKA T91A (rs2273535) (Phe21Ile) genotype and an increased risk of BC development: Phe/Ile (odds ratio [OR] = 2.6; 95% confidence interval [CI] = 1.4–4.9; P = 0.004), Ile/Ile (OR = 3.8; 95% CI = 1.6–9.0; P = 0.002), and Phe/Ile + Ile/Ile (OR = 2.9; 95% CI = 1.6–5.2; P = 0.001). Additionally, the rs2273535 variant was associated with the tumor grade SBR III (OR = 9.6; 95% CI = 1.0–91.9; P = 0.048) and the Ki-67 ≥ 20 (OR = 16.5; 95% CI = 2.7–101.3; P = 0.002). In brief, this study provides the first evidence where the Ile allele of the AURKA gene could act as potentially predictive biomarker of BC in the high altitude Ecuadorian mestizo population that lives at 2800 m above sea level (masl).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer (BC) is the most commonly diagnosed cancer (1,676,633 cases) and the leading cause of cancer-related death among women (521,817 cases) in 2012 [1]. Worldwide, the areas with a higher incidence of BC per each 100,000 inhabitants are Western Europe (89.9), Oceania (85.5), and Northern Europe (76.7) while South America has a lower incidence (44.3) [2]. In Ecuador, the incidence rate of BC was 32.7 and the mortality rate was 10.3 in 2012 [3].
The histopathologic classification of BC (noninvasive or in situ carcinoma, and invasive or infiltrating ductal carcinoma) together with the molecular subtypification of the estrogen receptor (ER) status, progesterone receptor (PR) status, and HER2/neu status provides valuable information on tumoral behavior for an efficient treatment of BC [4,5,6,7].
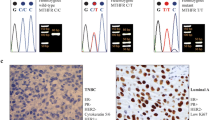
BC is driven by genetic alterations, including single nucleotide variants (SNVs), small insertions or deletions (indels), gene fusion, copy-number variations (CNVs) and large chromosome rearrangements. The next-generation sequencing (NGS) technologies have generated massive amounts of cancer genome data, providing somatic mutation landscapes to better understand breast cancer biology [8,9,10,11,12,13,14,15,16]. The driver mutations have selective growth advantage in tumor cells. These mutations have been considered prognostic markers and as therapeutic targets in the control of cell division and tumor growth inhibition [17]. AURKA (20q13.2) gene encodes the Aurora kinase A protein that has straight relation to the cell cycle since it performs both in the centrosome maturation and in the correct assembly of the spindle apparatus in mitosis (Fig. 1a) [18, 19].
Genetic analyses of T91A (rs2273535) (Phe21Ile) polymorphism in the AURKA gene. a Location of rs2273535 polymorphism in chromosome 20. b Genotyping of rs2273535 polymorphism by DNA sequencing. c AURKA pathway: The AURKA acts as a regulatory component for cell oncogenic transformation through the phosphorylation and stabilization of p53/TP53. It phosphorylates the protein phosphatase-1 (PP1) to inhibit its activity. AURKA is overexpressed in the late G2 phase to trigger mitosis. AURKA acts upon CDC25B, a direct regulator of the complex cyclin B1-CDK1, essential for mitosis. The rs2273535 polymorphism is associated with the overexpression of Aurora A and the loss of the anchorage site of ubiquitin 2E (UBE2N), related to tumor progression. P53: Inhibition of tumor suppression, CDK: Mitosis activation, BUBR1: Microtubules bonded to kinetochore, NPL: Assembly of mitotic use, PLK: Chromosome segregation, AURKA: Centrosome duplication
The Aurora kinase A has a high level of expression detected in human malignant neoplasm and it performs as a key regulatory component in critical control points of response for cell oncogenic transformation through the phosphorylation of p53/TP53. It phosphorylates the isoforms of the protein phosphatase 1 (PP1) in order to inhibit its activity. The Aurora kinase A is overexpressed in the late G2 phase to trigger the mitotic process, which turns it into a promoting factor of tumorigenesis [20]. The polymorphism T91A (rs2273535) (Phe21Ile) of the AURKA gene has been identified as functional alternator of this kinase, the Ile allele is associated with the occurrence of chromosome segregation errors and tumor progression (Fig. 1b, c) [21]. Recent studies by Xu et al., and Mendiola et al., have proposed the Aurora kinase A as a prognostic factor in cancer treatment [22, 23]. The results obtained so far do not report data on in the incidence and the behavior of the AURKA gene in mestizo populations in Latin America that live at 2800 m above sea level (masl).
The objective was to determine the risk of breast cancer associated with histopathological characteristics, immunohistochemical characteristics, and the genetic polymorphism AURKA T91A (rs2273535) (Phe21Ile) in a high altitude Ecuadorian mestizo population.
Materials and Methods
Study Subjects
The Bioethics Committee of the Universidad de las Americas approved this retrospective case-control study following the Declaration of Helsinki. A total of 200 Ecuadorian mestizo women who lived at 2800 masl were included into the analysis. Concerning the 100 individuals with BC, all samples were obtained from the Pathology Department at Solon Espinosa Ayala Oncologic Hospital. Affected individuals were diagnosed with BC between 2008 and 2011. Each case history conferred relevant information such as pTNM (tumor, nodule, metastasis) classification, tumor stage (T1-T4), Scharf Bloom-Richardson staging system (SRBIII: slightly-differentiated tumor, SRBII: mildly-differentiated tumor, SRBI: well-differentiated tumor), age, affected breast, histopathological classification, ER status, PR status, HER2/neu status, metastasis status, and Ki-67 levels (cut-off 20%) like prognostic factor. With regards to the control group, 100 peripheral blood samples from individuals of the mestizo population with no smoking history were selected at random from the sample bank at Centro de Investigación Genética y Genómica at Universidad Tecnológica Equinoccial. Thus, the matching of cases and controls presented similar age at menarche (14.2 vs. 14.6 years), age at menopause (47.4 vs. 46.7 years), age (54.0 vs. 52.3 years), age at first live birth (26.8 vs. 26.2 years), breast cancer in first-degree relative (3.1 vs. 2.1%), and the mean number of live births (2.5 vs. 2.5), respectively. Furthermore, informed consent was obtained from all individual participants included in the study.
DNA Extraction and Purification
The extraction and purification of the genomic DNA of the case and control individuals were obtained by means of the PureLink Genomic DNA Kit (Invitrogen, Carlsbad, CA). The tumor tissues were previously dissolved with proteinase K (Qiagen, Hilden, Germany). The DNA of the affected individuals, that presented an average concentration of 84 ng/μl, was extracted from ten sections (5 μm) of formalin-fixed paraffin-embedded breast tumor tissue previously cut with a microtome CUT 6062 (SLEE, Mainz, Germany). Meanwhile, the DNA of the healthy individuals was extracted from peripheral blood samples and presented an average concentration of 135 ng/μl. Both calculations were obtained, using a NanoDrop 2000 (Thermo Scientific, Waltham, MA).
Genotyping
Genotyping was performed using the polymerase chain reaction (PCR) and DNA sequencing analysis. A final volume of 25 μl was used for each PCR reaction for AURKA. Each reaction consisted of 4 μl of DNA template (20 ng/μl), 0.5 μM of forward (FW) 5′- CTGCTTGCTCTTTTGGGTGT -3′ and reverse (RV) 5′- CTCTTCCATTCTAGGCTACAGCTC -3′ primers, 0.4 μM of each deoxynucleotide triphosphate (dNTPs), 3 mM of MgCl2, 1.5 U of Platinum Taq DNA polymerase with 1X reaction buffer (500 mM of KCl, 200 mM of Tris-HCl, pH = 8.4) (Invitrogen, Madison, WI).
The polymorphism studied was AURKA T91A (rs2273535) (Phe21Ile). This polymorphism is located within two conserved motifs in the N-terminal region of AURKA gene [24]. For the analysis of the rs2273535 polymorphism, a fragment consisting of 272 base pairs (pb) carrying exon 3 was amplified. The PCR program started with an initial denaturation stage lasting 4 min at 94 °C, followed by 32 cycles of 30 s at 95 °C, 30 s at 54 °C, 45 s at 72 °C, and a final elongation for 3 min at 72 °C. Each run was completed using a Sure Cycler 8800 thermocycler (Agilent, Santa Clara, CA). The amplified fragment (272 pb) was analyzed by electrophoresis in 2% agarose gels stained with ethidium bromide and was observed in an ImageQuant 300 transilluminator (General Electric, Fairfield, SC).
Subsequently, the PCR amplicons were analyzed through sequence analysis, using the Genetic Analyzer 3130 (Applied Biosystems, Austin, TX). The final volume of the reaction was 12 μl and contained 2.8 of Milli-Q water, 2 μl of 5X buffer, 1 μl of primer FW (3.2 pmol), 1 μl of BigDye Terminator v3.1 sequencing standard (Applied Biosystems, Austin, TX), and 3 μl of PCR product (3 to 10 ng). Once the product was amplified, it was purified, using Agentcourt Cleanseq (Beckman Coulter, Miami, FL). The amplification program consisted of 3 min at 96 °C, followed by 30 cycles of 10 s at 96 °C, 5 s at 50 °C, and 4 min at 60 °C [25]. Finally, sequence analysis was performed using Sequencing Analysis Software 5.3.1 (Applied Biosystems, Austin, TX) and the alignment with sequence from GeneBank (AURKA NG_012133.1) was performed using Seq-Scape Software v2.6 (Applied Biosystems, Austin, TX).
Statistical Analysis
The information from the clinical records of the patients was collected in a database. Genotypic and allelic frequencies of the AURKA gene were calculated; also, Hardy-Weinberg equilibrium (HWE) was determined by using a web-tool (http://www.oege.org/software/hwe-mr-calc.shtml) [26]. With the use of IBM SPSS Statistics 22 software (SPSS Inc., Chicago, IL), chi-square (χ2) and odds ratio (OR) (with a 95% confidence interval [CI] and 2 × 2 contingency table) test were applied to determine the association between the risk of developing BC, the SNP rs2273535, and histopathological and immunohistochemical features. A value of P < 0.05 was considered statistically significant.
Results
The genotype distribution and allele frequency of the AURKA T91A (rs2273535) (Phe21Ile) polymorphism are shown in Table 1. The genotypic frequency of the Phe/Phe normal homozygous genotype was 0.3, of the Phe/Ile normal heterozygous was 0.5, and the Ile/Ile rare homozygous genotype was 0.2. The allele frequency of the Phe allele was 0.6 and the Ile allele was 0.4. The P value >0.05 demonstrated that there was no significant difference in the frequencies; consequently, there was HWE in all study population.
The distribution of baseline characteristics in all patients with BC is shown in Table 2. Regarding the studied population, the 46% of the healthy group had the Phe/Phe normal genotype, the 54% of the affected group had the Ile/Ile genotype (P = 0.002). The 57% of the studied population had right breast affected and the 43% left breast affected (P = 0.931). The 79.5% of the affected individuals had invasive ductal carcinoma, the 9% had lobular carcinoma in situ, the 7% mucinous carcinoma, and the 4.5% papillary carcinoma subtype (P = 0.398). In relation to Scarff-Bloom-Richardson tumor grade, the 29% of the individuals had SBR III (reference), the 45% had SBR II (P = 0.017), and the 26% SBR I (P = 0.007). As for membrane receptors, the 68% of individuals had ER+ status and the 32% had ER- status (P = 0.561). The 58.5% had PR+ status and the 41.5% PR- status (P = 0.972). The 22% had HER2+ status and the 78% HER2- status (P = 0.875). The 66% of individuals had Ki-67 < 20 and the 34% Ki-67 ≥ 20 (P = 0.000). Concerning immunohistotype, the combinations from most frequent to least frequent were: 47.4% with ER+ RP+ HER2- (P = 0.875), 20% with ER- RP- HER2- (P = 0.771), 8.4% with ER+ RP- HER2- (P = 0.922), 7.4% with ER- RP- HER2+ (P = 0.692), 6.3% with ER+ RP+ HER2+ (reference), 5.3% with ER+ RP- HER2+ (P = 0.632), 3.2% with ER- RP+ HER2- (P = 0.153), and 2.1% with ER- RP+ HER2+ (P = 0.587).
The association between the studied AURKA Phe31Ile polymorphism and the risk of developing BC is detailed in Table 3. Regarding variant rs2273535, the AURKA Phe/Ile genotype presented an OR of 2.6 (95% CI = 1.4–4.9; P = 0.004); the AURKA Ile/Ile genotype presented an OR of 3.8 (95% CI = 1.6–9.0; P = 0.002). Whereas the combination of AURKA Phe/Ile + Ile/Ile genotypes presented an OR of 2.9 (95% CI = 1.6–5.2; P = 0.001).
The association among the AURKA Phe31Ile polymorphism, breast cancer risk, and baseline characteristics is detailed in Table 4. Regarding the SBR tumor grade, the analysis of logistic regression between SBR III vs. SBR I resulted in an OR of 0.48 (95% CI = 0.1–2.5; P = 0.040) in individuals affected with the Phe/Ile genotype, and an OR of 12.0 (95% CI = 1.0–92.0; P = 0.048) with the Ile/Ile genotype. As for membrane receptors, the comparison between ER- vs. ER+ resulted in an OR of 0.7 (95% CI = 0.2–2.0; P = 0.523) with the Phe/Ile genotype and an OR of 1.2 (95% CI = 0.4–4.3; P = 0.724) with the Ile/Ile genotype. The comparison between PR- vs. PR+ resulted in an OR of 0.9 (95% CI = 0.3–2.5; P = 0.852) with the Phe/Ile genotype and an OR of 0.8 (95% CI = 0.3–2.9; P = 0.817) with the Ile/Ile genotype. The comparison between HER2+ vs. HER2- resulted in an OR of 0.9 (95% CI = 0.3–3.3; P = 0.884) with the Phe/Ile genotype and an OR of 0.7 (95% CI = 0.2–2.8; P = 0.679) with the Ile/Ile genotype. Lastly, the comparison of the cell proliferation marker Ki-67 between ≥20 vs. < 20 resulted in an OR of 0.8 (95% CI = 0.3–3.1; P = 0.829) with the Phe/Ile genotype and an OR of 16.5 (95% CI = 2.7–101.3; P = 0.002) with the Ile/Ile genotype.
Discussion
Identifying genetic alterations involved in the development of cancer is crucial to fully understand BC biology, as well as for the development of new therapeutics [27]. During the last decade, BC genome-wide association studies (GWAS) have identified ∼80 loci with small to moderate effects on OR ranging from 1.05 to 1.53, the most studied populations being European, North American, and Asian. However, Latin American populations have been poorly studied and the NGS analyses consider very few samples of mestizo individuals. Therefore, the genetic characterization of these populations is essential to better understand the development of BC [28].
Many studies have indicated that the AURKA T91A (rs2273535) (Phe21Ile) polymorphism is a general low-penetrance susceptibility gene in breast [29], colorectal [30], and esophageal cancer [31]. However, results from these studies remain inconsistent. Consequently, Tang et al. made an association between the rs2273535 polymorphism and cancer susceptibility in a meta-analysis study of 27 published studies involving 19,267 multiple cancer cases and 29,359 controls from Caucasians and Asians [32]. The results of this study demonstrate that rs2273535 is associated with an increased risk of BC and esophageal cancer. Additionally, the stratified analysis performed regarding ethnicity demonstrates that rs2273535 is associated with cancer risk in Asians [32]. The studies of the AURKA T91A (rs2273535) (Phe21Ile) polymorphism in an affected Asian population with BC were conducted in China [33, 34] and Taiwan [35], whereas the studies in Caucasians were carried out in Germany [36, 37], Iceland [38], the UK [39], and the USA [40, 41]. In consequence, this is the first study conducted in a female Latin American mestizo population from a high altitude (2800 masl). It is noteworthy that this retrospective research presented a limited number of cases. However, it gives us relevant information about BC risk, histopathological characteristics, and its association with the AURKA T91A (rs2273535) (Phe21Ile) polymorphism.
In our study, we found that the AURKA Phe/Ile and Ile/Ile genotypes present a significantly higher risk factor in affected females with ORs of 2.6 (P = 0.004) and 3.8 (P = 0.002), respectively. While the combination of the Phe/Ile + Ile/Ile genotypes generated an OR of 2.9 (P = 0.001). Hence, there is an association between rs2273535 and BC in the mestizo population living at high altitude.
Concerning the SBR tumor grade, the highest percentage of the population presented SBRII (45%); however, SBRIII was presented in the 52.6% of the rare Ile/Ile homozygous individuals (P = 0.017), suggesting that there is a selective growth of cells with Ile/Ile genotype in tumorgenesis. This would determine a bad diagnosis for the carriers of this genotype in advanced stages.
It has been established that the Ile rare allele is more frequently found than Phe in patients with BC. Additionally, heterozygous patients for the Phe/Ile genotype have more aneuploidy cells than the normal Phe/Phe heterozygous patients [42], suggesting that the rare Ile allele is related to events that lead to tumor progression. On the other hand, cells with the Ile/Ile genotype have a deficient bond to the ubiquitin-conjugating enzyme E2 (UBE2N), suggesting that the substitution Phe31 > Ile inhibits the protein degradation of the Aurora A (Ile31), in late mitosis; and consequently, its oncogenic effect is provided [42].
The hormone receptor status indicates that the 26.7% of ER-, the 21.8% of PR+, and the 21.9% of the HER2- individuals have Ile/Ile genotype. Despite the fact that these receptors are the most important prognosis factors for BC, the P value of 0.561, 0.972, and 0.875, respectively, shows there is no statistically significant differences among the hormone receptors with the genotypes of the rs2273535 variant [43], obtaining results similar to the North American population [44]. On the other hand, there are other risk factors for the development of BC, such as early menarche, late menopause, and replacement hormone therapy [45]. The histopathological classification determined that the 79.5% of the affected individuals had invasive ductal carcinoma, and in connection with this subtype, the 28.6% presented Ile/Ile genotype (P = 0.398). The combined immunohistotype classification determined that the 47.4% of the individuals were ER+ RP+ HER2- (P = 0.875) and in connection with this subtype, the 22.2% presented Ile/Ile genotype; whereas the 3.2% of the individuals were ER- PR+ HER2- (P = 0.153), and in connection with this subtype, the 66.7% presented Ile/Ile genotype.
The logistic regression analysis showed that the Ile/Ile genotype was associated with SBRIII tumor grade with an OR of 12.0 (95% CI = 1.1–28.8; P = 0.040), and with the cell growth factor Ki-67 when ≥20 with an OR of 16.5 (95% CI = 2.7–101.3; P = 0.002), in comparison with the normal Phe/Phe homozygous genotype. On the other hand, there was no evidence of significant associations when the ER and PR status were analyzed separately as a whole (P > 0.05), no relation was found among the polymorphism and the overexpression of the HER2/neu receptor (P = 0.638), and the presence of ganglio metastasis (P = 0.679).
Our present study clarifies that the AURKA gene and its protein could act as potential predictive biomarker in BC. In conclusion, this study as well as our previous genetic studies on MTHFR in breast cancer [25], AR and MTHFR in prostate cancer [46, 47], EGFR in lung cancer [48], GPX-1 in bladder cancer [49], and hRAD5 in chronic myelogenous leukemia are important contributions in order to integrate pharmacogenetics and pharmacogenomics in clinical practice in Latin America [50–52].
Change history
05 September 2017
An erratum to this article has been published.
References
Cancer Research UK. Worldwide cancer statistics. In: Cancer statistics. 2014. www.cancerresearchuk.org/cancer info/cancerstats/world. Accessed 7 Nov 2014
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61:69–90
World Health Organization. International Agency for Research on Cancer. GLOBOCAN 2012: Estimated cancer incidence, mortality and prevalence worldwide in 2012. 2012. http://globocan.iarc.fr/ Default.aspx. Accessed 7 Nov 2016
Kumar V, Abbas A, Aster J. (2012) Robbins basic pathology. 9th ed. Philadelphia: Saunders
National Cancer Institute at the National Institutes of Health (2014) Histopathologic classification of breast cancer. In: Breast cancer treatment. http://www.cancer.gov/cancertopics/pdq/treatment/breast/healthprofessional/page2. Accessed 7 Nov 2014
Kumar R, Sharma A, Tiwari R (2012) Application of microarray in breast cancer: an overview. J Pharm Bioallied Sci 4:21–26
Banerji S, Cibulskis K, Rangel-Escareno C, Brown K, Carter S, Frederick A, Lawrance M, Sivachenko A, Sougnez C, Zou L, Cortes M, Fernandez-Lopez J, Peng S, Ardlie K, Auclair D, Bautista-Piña V, Duke F, Francis J, Jung J, Maffuz-Aziz A, Onofrio R, Parkin M, Pho N, Quintanar-Jurado V, Ramos A, Rebollar-Vega R, Rodriguez-Cuevas S, Romero-Cordoba S, Schumacher S, Stransky N, Thompson K, Uribe-Figueroa L, Baselga J, Beroukhim R, Polyak K, Sgroi D, Richardson A, Jimenez-Sanchez G, Lander E, Gabriel S, Garraway L, Golub T, Melendez-Zajgla J, Toker A, Getz G, Hidalgo-Miranda A, Meyerson M (2012) Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 486:405–409
Eisenstein M (2015) Startups use short-read data to expand long-read sequencing market. Nat Biotechnol 33:433–435
Wong KM, Hudson TJ, McPherson JD (2011) Unraveling the genetics of cancer: genome sequencing and beyond. Annu Rev Genomics Hum Genet 12:407–430
Meyerson M, Gabriel S, Getz G (2010) Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet 11:685–696
Eifert C, Powers RS (2012) From cancer genomes to oncogenic drivers, tumour dependencies and therapeutic targets. Nat Rev Cancer 12:572–578
Lovly CM, McDonald NT, Chen H, Ortiz-Cuaran S, Heukamp LC, Yan Y, Florin A, Ozretić L, Lim D, Wang L, Chen Z, Chen X, Lu P, Paik PK, Shen R, Jin H, Buettner R, Ansén S, Perner S, Brockmann M, Bos M, Wolf J, Gardizi M, Wright GM, Solomon B, Rusell PA, Rogers TM, Suehara Y, Red-Brewer M, Tieu R, de Stanchina E, Wang Q, Zhao Z, Johonson DH, Horn L, Wong KK, Thomas RK, Ladanyi M, Pao W (2014) Rationale for co-targeting IGF-1R and ALK in ALK fusion-positive lung cancer. Nat Med 20:1027–1034
Xia J, Jia P, Hutchinson KE, Dahlman KB, Johnson D, Sosman J, Pao W, Zhao Z (2014) A meta-analysis of somatic mutations from next generation sequencing of 241 melanomas: a road map for the study of genes with potential clinical relevance. Mol Cancer Ther 13:1918–1928
Jia P, Jin H, Meador CB, Xia J, Ohashi K, Liu L, Pirazzoli V, Dahmal KB, Politi K, Michor F, Zhao Z, Pao W (2013) Next-generation sequencing of paired tyrosine kinase inhibitor-sensitive and –resistant EGFR mutant lung cancer cell lines identifies spectrum of DNA changes associated with drug resistance. Genome Res 23:1434–1445
Dahlman KB, Xia J, Hutchinson K, Ng C, Hucks D, Jia P, Atefi M, Su Z, Branch S, Lyle PL, Hicks DJ, Bozon V, Glaspy JA, Rosen N, Solit DB, Netterville JL, Vnencak-Jones c, Sosman JA, Ribas A, Zhao Z, Pao W (2012) BRAF(L597) mutations in melanoma are associated with sensitivity to MEK inhibitors. Cancer Discov 2:791–797
Cheng F, Zhao J, Zhao Z (2016) Advances in computational approaches for prioritizing driver mutations and significantly mutated genes in cancer genomes. Brief Bioinform 17:642–656
Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW (2013) Cancer genome landscapes. Science 339:1546–1558
Khan J, Ezan F, Crémet J, Fautrel A, Gilot D, Lambert M, Benaud C, Troadec MB, Prigent C (2011) Overexpression of active aurora-C kinase results in cell transformation and tumour formation. PLoS One 6:e26512
Lim SK, Gopalan G (2007) Aurora-a kinase interacting protein 1 (AURKAIP1) promotes aurora-a degradation through an alternative ubiquitin-independent pathway. Biochem J 403:119–127
Fu J, Bian M, Jiang Q, Zhang C (2007) Roles of aurora kinases in mitosis and tumorigenesis. Mol Cancer Res 5:1–10
Sun T, Miao X, Wang J, Tan W, Zhou Y, Yu C, Lin D (2004) Functional Phe31Ile polymorphism in aurora a and risk of breast carcinoma. Carcinogenesis 25:2225–2230
Xu J, Wu X, Zhou WH, Liu AW, Wu JB, Deng JY, Yue CF, Yang SB, Wang J, Yuan ZY, Liu Q (2013) Aurora-a identifies early recurrence and poor prognosis and promises a potential therapeutic target in triple negative breast cancer. PLoS One 8:e56919
Mendiola M, Barriuso J, Mariño-Enríquez A, Redondo A, Domínguez-Cáceres A, Hernández-Cortés G, Pérez-Fernández E, Sánchez-Navarro I, Vera JA, Suárez A, Espinosa E, González-Barón M, Palacios J, Hardisson D (2009) Aurora kinases as prognostic biomarkers in ovarian carcinoma. Hum Pathol 40:631–638
Dai ZJ, Kang HF, Wang XJ, Shao YP, Lin S, Zhao Y, Ren HT, Min WL, Wang M, Liu XX (2014) Association between genetic polymorphisms in AURKA (rs2273535 and rs1047972) and breast cancer risk: a meta-analysis involving 37,221 subjects. Cancer Cell Int 14:91
López-Cortés A, Echeverría C, Oña-Cisneros F, Sánchez ME, Herrera C, Cabrera-Andrade A, Rosales F, Ortiz M, Paz-y-Miño C (2015) Breast cancer risk associated with gene expression and genotype polymorphisms of thefolate-metabolizing MTHFR gene: a case-control study in a high altitude Ecuadorian mestizo population. Tumor Biol 36:6451–6461
Rodriguez S, Gaunt TR, Day IN (2009) Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am J Epidemiol 169:505–514
Engle LJ, Simpson CL, Landers JE (2006) Using high-throughput SNP technologies to study cancer. Oncogene 25:1594–1601
Fejerman L, Ahmadiyeh N, Hu D, Huntsman S, Beckman KB, Caswell JL, Tsung K, John EM, Torres-Mejia G, Carvajal-Carmona L, Echeverry MM, Tuazon AM, Ramirez C, COLUMBUS Consortium, Gognoux CR, Eng C, Gonzalez-Burchard E, Henderson B, Le Marchard L, Kooperberg C, Hou L, Agalliu I, Kraft P, Lindström S, Perez-Stable EJ, Haiman CA, Ziv E (2014) Genome-wide association study of breast cancer in Latinas identifies novel protective variants on 6q25. Nat Commun 5:5260
Ruan Y, Song AP, Wang H, Xie YT, Han JY, Sajdik C, Tian XX, Fang WG (2011) Genetic polymorphisms in AURKA and BRCA1 are associated with breast cancer susceptibility in a Chinese Han population. J Pathol 225:535–543
Webb EL, Rudd MF, Houlston RS (2006) Case-control, kin-cohort and meta-analyses provide no support for STK15 F31I as a low penetrance colorectal cancer allele. Br J Cancer 95:1047–1049
Miao X, Sun T, Wang Y, Zhang X, Tan W, Lin D (2004) Functional STK15 Phe31Ile polymorphism is associated with the occurrence and advanced disease status of esophageal squamous cell carcinoma. Cancer Res 64:2680–2683
Tang W, Qiu H, Ding H, Sun B, Wang L, Yin J, Gu H (2013) Association between the STK15 F31I polymorphism and cancer susceptibility: a meta-analysis involving 43,626 subjects. PLoS One 8:e82790
Sun T, Miao X, Wang J, Tan W, Zhou Y, Yu C, Lin D (2004) Functional Phe31Ile polymorphism in aurora a and risk of breast carcinoma. Carcinogenesis 25:2225–2230
Dai Q, Cai QY, Shu XO, Ewart-Toland A, Wen WQ, Balmain A, Gao YT, Zheng W (2004) Synergistic effects of STK15 gene polymorphisms and endogenous estrogen exposure in the risk of breast cancer. Cancer Epidemiol Biomark Prev 13:2065–2070
Lo YL, Yu JC, Chen ST, Yang HC, Fann CS, Mau YC, Shen CY (2005) Breast cancer risk associated with genotypic polymorphism of the mitosis-regulating gene aurora-STK15/BTAK. Int J Cancer 115:276–283
MARIE-GENICA Consortium on Genetic Susceptibility for Menopausal Hormone Therapy Related Breast Cancer Risk (2010) Polymorphisms in the BRCA1 and ABCB1 genes modulate menopausal hormone therapy associated breast cancer risk in postmenopausal women. Breast Cancer Res Treat 120:727–736
Tchatchou S, Wirtenberger M, Hemminki K, Sutter C, Meindl A, Wappenschmidt B, Kiechle M, Bugert P, Schmutzler RK, Bartram CR, Burweinkel B (2007) Aurora kinases a and B and familial breast cancer risk. Cancer Lett 247:266–272
Vidarsdottir L, Bodvarsdottir SK, Hilmarsdottir H, Tryggvadottir L, Eyfjord JE (2007) Breast cancer risk associated with AURKA 91T -->a polymorphism in relation to BRCA mutations. Cancer Lett 250:206–212
Fletcher O, Johnson N, Palles C, dos Santos Silva I, McCormack V, Whittaker J, Ashworth A, Peto J (2006) Inconsistent association between the STK15 F31I genetic polymorphism and breast cancer risk. J Natl Cancer Inst 98:1014–1018
Cox DG, Hankinson SE, Hunter DJ (2006) Polymorphisms of the AURKA (STK15/aurora kinase) gene and breast cancer risk (United States). Cancer Causes Control 17:81–83
Egan KM, Newcomb PA, Ambrosone CB, Trentham-Dietz A, Titus-Ernstoff L, Hampton JM, Kimura MT, Nagase H (2004) STK15 polymorphism and breast cancer risk in a population-based study. Carcinogenesis 25:2149–2153
Royce ME, Xia W, Sahin AA, Katayama H, Johnston DA, Hortobagyi G, Hung MC (2004) STK15/ aurora-a expression in primary breast tumors is correlated with nuclear grade but not with prognosis. Cancer 100:12–19
Loman N, Johannson O, Kristoffersson U, Olsson H, Borg A (2001) Family history of breast and ovarian cancers and BRCA1 and BRCA2 mutations in a population-based series of early-onset breast cancer. J Natl Cancer Inst 93:1215–1223
Lindström L, Karlsson E, Wilking UM, Johansson U, Hartman J, Lidbrink EK, Hatschek T, Skoog L, Bergh J (2012) Clinically used breast cancer markers such as estrogen receptor, progesterone receptor and human epidermal growth factor receptor 2 are unstable throughout tumor progression. J Clin Oncol 30:2601–2608
Colditz G, Rosner BA, Chen WY, Holmes MD, Hankinson SE (2004) Risk factors for breast cancer according to estrogen and progesterone receptor status. J Natl Cancer Inst 96:218–228
López-Cortés A, Jaramillo-Koupermann G, Muñoz MJ, Cabrera A, Echeverría C, Rosales F, Vivar N, Paz-y-Miño C (2013) Genetic polymorphisms in MTHFR (C677T, A1298C), MTR (A2756G) and MTRR (A66G) genes associated with pathological characteristics of prostate cancer in the Ecuadorian population. Am J Med Sci 346:447–454
Paz-y-Miño C, Robles P, Salazar C, Leone PE, García Cárdenas JM, Naranjo M, López-Cortés A (2016) Positive association of the androgen receptor CAG repeat length polymorphism with the risk of prostate cancer. Mol Med Rep 14:1791–1798
Paz-y-Miño C, López-Cortés A, Muñoz MJ, Cabrera A, Castro B, Sánchez ME (2010) Incidence of the L858R and G719S mutations of the epidermal growth factor receptor oncogene in an Ecuadorian population with lung cancer. Cancer Genet Cytogenet 196:201–203
Paz-y-Miño C, Muñoz MJ, López-Cortés A, Cabrera A, Palacios A, Castro B, Paz-y-Miño N, Sánchez ME (2010) Frequency of polymorphisms pro198leu in GPX-1 gene and ile58thr in MnSOD gene in the altitude Ecuadorian population with bladder cancer. Oncol Res 18:395–400
Paz-y-Miño C, López-Cortés A, Muñoz MJ, Castro B, Cabrera A, Sánchez ME (2010) Relationship of an hRAD54 gene polymorphism (2290 C/T) in an Ecuadorian population with chronic myelogenous leukemia. Genet Mol Biol 33:646–649
Quiñones LA, Lavanderos MA, Cayun JP, Garcia-Martin E, Agundez JA, Caceres DD, Roco AM, Morales JE, Herrera L, Encina G, Isaza CA, Redal MA, Larovere L, Soria NW, Eslava-Schmalbach J, Castaneda-Hernandez G, López-Cortés A, Magno LA, Lopez M, Chiurillo MA, Rodeiro I, Castro de Guerra D, Teran E, Estevez-Carrizo F, Lares Assef I (2014) Perception of the usefulness of drug/gene pairs and barriers for pharmacogenomics in Latin America. Curr Drug Metab 15:202–208
López-Cortés A, Guerrero S, Redal M, Alvarado A, Quiñones L (2017) State of Art of Cancer Pharmacogenomics in Latin American Populations. Int J Mol Sci 18(6):639
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
None.
Rights and permissions
About this article
Cite this article
López-Cortés, A., Cabrera-Andrade, A., Oña-Cisneros, F. et al. Breast Cancer Risk Associated with Genotype Polymorphisms of the Aurora Kinase a Gene (AURKA): a Case-Control Study in a High Altitude Ecuadorian Mestizo Population. Pathol. Oncol. Res. 24, 457–465 (2018). https://doi.org/10.1007/s12253-017-0267-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12253-017-0267-6