Abstract
Combined techniques like fluorescence, circular dichroism (CD), and Fourier transform infrared (FT-IR) were applied to study the interaction between ifosfamide-loaded superparamagnetic iron oxide nanoparticles (ISION) and human serum albumin (HSA) at physiological pH. The fluorescence results showed the existance of a static quenching mode for HSA-ISION interaction. The binding constant at 298 K was in the order of 105 M−1, showing high affinity occurred between ISION and HSA. The thermodynamic parameters obtained at three different temperatures (288, 298, and 308 K) indicated that the dominant binding forces are the electrostatic forces, for the values of the enthalpy change (ΔH°) and entropy change (ΔS°) were found to be −1.31 × 104 J · mol−1 and 55.4 J · mol−1 · K−1, respectively. Fluorescence resonance energy transfer (FRET) analysis revealed that the average binding distance (r) between Trp of HSA and ISION was 5.25 nm. The results of CD and FT-IR spectra demonstrated that the secondary structure of HSA was altered in the presence of ISION.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ifosfamide is a nitrogen mustard alkylating agent used in the treatment of a variety of cancers, including testicular cancer, breast cancer, nlymphoma, soft tissue sarcoma, bone cancer, and lung cancer [1–3]. Because of its nonspecific interaction with normal tissues, ifosfamide leads to significant normal tissue toxicity and limits dosages of it is far below the tumor sites required to destroy most malignant lesions [4].
In order to solve the problem, drug targeting and combined treatments are being investigated, for instance, molecular coupling of the anticancer drug to specific molecules like low-density lipoproteins, a drug-loaded poly(lactic-co-glycolic acid)-Au half-shell nanoparticles for tumor-specific delivery applied in heat treatment, and an aqueous suspension of magnetic nanoparticles retained in tumor by application of an external magnetic field [5]. Among all the types of targeted drug deliverers, nano-Fe3O4 has shown great prospect due to its biocompatibility and unique magnetic properties [6]. Moreover, methods of ifosfamide load to iron oxide nanoparticle and its cytotoxicity has been reported.
HSA is the most abundant protein in plasma, and is able to bind and transport various endogenous and exogenous substances such as fatty acids, drugs, dyes and metal ions. The biodistribution, free concentration and the metabolism of varieties of drugs can be significantly influenced by drug-protein interactions in the circulation system [7, 8]. As a result, it is imperatively important to study the interaction of different drugs with the protein. HSA is composed of three structurally homologous domains (I-III) with each two sub-domains in each. It has one Trp residue which possess intrinsic fluorescence: Trp-214, which is located in the hydrophobic binding cavity of sub-domain IIA [9]. The binding sites of HSA for different ligands may be in the three domains, and the most important regions of drug binding sites are usually located in hydrophobic pockets of sub-domains IIA and IIIA which are called Sudlow’s sites I and II, respectively. As a result, HSA is often chosen as a model protein in the related pharmacodynamic research by virtue of its physiological and pharmacological importance [10–12].
Spectroscopy, like fluorescence, FT-IR, UV-vis and CD, are powerful tools to study the binding of ligands to proteins due to their sensitivity, availability and non-destructive measurement. However, so far the binding profile of ISION with HSA has never been investigated. The aim of this study was to analyze the fluorescence quenching mechanism, binding properties and conformational changes of HSA induced by binding of ISION.
Material and Methods
Material
FeSO4 · 7H2O, FeCl3 · 6H2O, acetone, 25 % aqueous ammonia, and 35 % hydrochloric acid were bought from Sinopharm Chemical Reagent Co., Ltd. (Beijing, China). HSA and dodecanebenzenesulfonic acid sodium salt are all analytical reagent grade and were obtained from Aladdin (Shanghai, China). Ultrapure water of 18 M Ω cm−1 was used throughout the experiments.
Apparatus and Methods
Iron oxide nanoparticles were synthesized using protocols described previously. Briefly, 1.39 g of FeSO4 · 7H2O and 2.70 g of FeCl3 · 6H2O were added to a 250 mL beaker with stirring at 600 rpm. The pH of mixture was firstly 1.5 (stay for 30 min) and then was adjusted fast to 9.5 by adding ammonia. After 30 min 9.5 mL of oleic acid was added at 70 °C within 25 min with stirring. Then ifosfamide was loaded according to the methods described by Liang [13]. Transmission electron microscope (TEM) images were captured on a JEOL 2100F microscope (JEOL, Tokyo, Japan). Size distribution of the as-prepared nanoparticles was measured by Mastersizer 2000 laser particle size analyzer (Malvern, England). Then, the drug-loaded nanoparticle was chosen to interact with HSA, as shown in Fig. 1
For fluorescence measurement, an F-7000 spectrophotometer (Hitachi, Japan) is used for the fluorescence measurement with emission and excitation slits of 5 nm and a scan speed of 1200 nm min-1. Each spectrum was the average of three runs with scanning voltage of lamp set to 650 V. Inner filter effects (IFE) are considered for the related calculations using the following equation [14]:
where CFp and CFs are the correction factors for the absorption of emission and excitation radiation, respectively. They all depend on the total absorbance of the sample at λem and λex, where the system absorbs with values of Aem and Aex, respectively. Fobs is the observed fluorescence.
The fluorescence quenching mechanism is usually divided into static or dynamic as described by the well known Stern-Volmer formula [15]:
where F 0 and F are the fluorescence intensities in the absence and presence of quencher, respectively. [Q] is the concentration of the quencher. K q is the biomolecular quenching rate constant and τ 0 is the average lifetime of the biomolecule without quencher, which is 10-8 s. K SV is the Stern-Volmer quenching constant.
If the quenching is a static one, the number of substantive binding number n and the binding constant K A is usually calculated using [16]:
The free-energy change (ΔG ○), enthalpy change (ΔH ○) and the entropy change (ΔS ○) for the interaction between ISION and HSA were determined based on the van’t Hoff equation [17]:
and
According to the Förster's theory of nonradiative energy transfer, the efficiency of energy transfer, E, is calculated using the following formula [18]:
where F and F 0 are the fluorescence intensity of protein in the absence and presence of equimolar concentration of acceptor (ligand), r is the distance from the the ligand to Trp of HSA, and R 0 is the Förster critical distance when 50 % of the excitation energy is transferred and determined by:
where k 2 is the factor describing the relative orientation of the transition dipoles of the donor and acceptor, N is the refractive index of medium, Φ is the quantum yield of donor in the absence of acceptor and overlap integral J is the extent of overlap between the fluorescence emission spectrum of the donor and the absorption spectrum of the acceptor. J is described by:
where F(λ) is the fluorescence intensity of the fluorescent donor (HSA here) at the studied wavelengths; ε(λ) is the molar absorption coefficient of the acceptor (the drug) at the studied wavelengths.
Synchronous fluorescence was performed on the same equipment by adjusting the excitation and emission wavelength intervals (Δλ) at 15 or 60 nm. The experimental condition is the same with the fluorescence measurement.
CD spectra were acquired on a MOS-450 (Bio-Logic, France) spectropolarimeter at 298 K within 190-240 nm. The spectra were recorded with nitrogen protection by using 1 cm cell length with a scan rate of 200 nm min-1. Each spectrum is accumulated from three runs.
The FT-IR spectra of HSA in presence and absence of ISION at 298 K were obtained in the range of 1500-1700 cm-1. The protien concentration was made stable at 50 μM whereas that of ISION was 50 μM in presence of buffer solution.
Results and Discussion
ISION Characterization
The ISION nanoparticles were characterized by TEM and the images were shown in Fig. 2A. It can be seen that the size of the spherical particles is between 10 and 40 nm. The size distribution of the ISION particles was also confirmed with dynamic light scattering (DLS) technique. As was shown in Fig. 2B, the results are in line with the TEM observations such and over 90 % of the particles are in the range of 15-40 nm.
Fluorescence Quenching of HSA by ISION and Quenching Mode Determination
Fluorescence quenching is the result of the quantum yield decrease of fluorophore fluorescence induced by various molecular interactions with a quencher, such as excited state reactions, energy transfer, static and dynamic quenching [19, 20]. Fluorescence spectroscopy is a powerful tool to detect the interaction of a drug with a biomacromolecule as it has high sensitivity and availability in physiological environments. For biomacromolecules, the fluorescence detection gives information of binding details like the binding mode, binding constants, binding sites and protein-ligand distances.
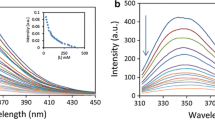
The fluorescence intensity of HSA is mainly from the Trp, Tyr and Phe residues. In fact, the intrinsic fluorescence of HSA is almost contributed by Trp alone (more than 95 %), because Phe has very low quantum yield and the fluorescence of Tyr is almost totally quenched if it is ionized or located near to an amino group, which is confirmed and supported by Sulkowska through experimental results [21]. As a result, alterations of fluorescence intensity of the protein are attributed by Trp residue when ligand bound to HSA. As was shown in Fig. 3, the fluorescence intensity of HSA decreased with gradual addition of ISION, demonstrating micronenvironmental alterations of Trp residue.
To make clear of the possible quenching mechanisms, the fluorescence quenching data at three different temperatures (288, 298 and 308 K) were recorded and the Stern-Volmer equation (Eq. (1)) was used for calculations.
Fig. 4 shows the Stern-Volmer equation of the system at three different temperatures. Besides, the related thermodynamic constants were listed in Table 1. The maximum scattering collision quenching constant of various quencher is known to be 2×1010 M-1 s-1 for dynamic quenching. The K q values got from the experimental data suggest that the mechanisms of all quenching here were the static one, for K q at all temperatures is less than 2×1010 M-1 s-1. It should be noted that KSV increased followed by decreasing temperatures, indicating the occurrence of static quenching mechanism [22]. Moreover, ISION shows the largest K SV at 288 K and highest sensitivity to the change of the temperature, and at 308 K, ISION shows the lowest quenching efficiency towards HSA.
In static quenching mode, the binding constant K A and the number binding sites n of the association between ISION and HSA can be calculated by plotting Log(F0/F-1) vs Log [ISION] (Fig. 5) and the results are summarized in Table 2. From Table 2, it can be seen that the value of n at experimental temperatures was all about 1, demonstrating the existence of a single binding site for ISION in HSA molecule. The binding constants KA at 298 K are in the order of 105, showing strong binding force between ISION and HSA.
Thermodynamic Parameters and the Nature of Binding Forces
Through calculating the fluorescence quenching data using van’t Hoff equation, thermodynamic parameters were got and listed in Table 3. The binding force of the system and be determined through these thermodynamic parameters. Ross and Subramanian [23] have made the thermodynamic law of judging the primary binding forces: if ΔH○ > 0 and ΔS○ > 0, a hydrophobic interaction plays the main role; if ΔH○ < 0 and ΔS○ < 0, it suggest that hydrogen bond and van de Waals force are the dominating force; if ΔH○ < 0 and ΔS○ > 0, this implies that electrostatic interactions are the driving forces. Besides, ΔG○ at different temperatures were also obtained according to Van’t Hoff equation. Negative value of ΔG○ at all three temperatures indicates the binding is spontaneous, and the spontaneous process is driven by both enthalpy and entropy as evidenced by negative ΔH○ and positive ΔS○. The dominating force is obviously an electrostatic interaction from the calculated data. The ifosfmide molecule carries many charges. The O atom in P=O structure processes more electrons, showing negative charge properties. The electrophilic effect of the phosphoryl group may reduce the intensity of the electron cloud of N atom, causing it to be positively-charged. Thus these charged atoms may interact with opposite-charged amino acid residues through electrostatic forces. Besides, it could be speculated that unfolding of the protein lead to different driven force during different binding process. The influence to such denaturation effect caused by loading drug differences could be explored in the future work.
Conformational Study by CD Spectroscopy
CD spectroscopy is a sensitive and effective technique to monitor the protein structural changes, particularly secondary structure. In quest of the possible effects of ISION binding on the secondary structure of HSA, CD studies in the presence of different ISION concentrations were carried out. The CD spectra of HSA exhibit two negative bands in the ultraviolet region at 208 and 220 nm, which is specific for α-helical structures of protein, which is assigned to n→π* transfer for the peptide bond of α-helix . Changes in intensity of the two peaks in the presence of the drug may be used to examine alterations in α-helical content of the protein. Fig. 6 shows the CD spectra of HSA with various amouts of ISION. As can be seen from Fig. 6, the binding of ISION to HSA caused decreasing intensity at all wavelengths of far-UV CD without apparent shifts for all the peaks, demonstrating that binding of the drug brings slight decrement in the α-helix and increment of β-turn structure of HSA, and this may be due to the complexation of ISION and HSA. Therefore it is obvious that ISION binding causes structural changes of HSA, with loss of helical structure stability and conformational destruction [24-26]. The secondary structural contents were calculated utilzing the CDpro program and were shown in Table 4. The results exhibit decrement of α-helical structure from 57.12 % to 51.61 % and 46.79 % and increment of β-turn structure from 10.92 % to 11.88 % and 14.53 % at molar ration of HSA: ISION of 1:0, 1:1 and 1:2, respectively (Table 4).
Fluorescent Resonant Energy Transfer
Fluorescence resonance energy transfer (FRET) is a useful and sensitive method for measuring the distance between the donor and acceptor in vitro. Spectroscopic technique can be applied to monitor the proximity and relative angular orientation of the fluorescence donor and acceptor [27]. Energy transfer may occur through a direct electrodynamics interaction between the excited fluorescence donors and accepters, which will happen when the following conditions are satisfied: First of all, the donor can produce fluorescence light; second, the donor's fluorescence emission spectrum and the acceptor's UV-vis absorbance spectrum overlap; third, the distance between the donor and the acceptor is less than 8 nm.
For this, equimolar concentrations of ISION and HSA were employed, and from the above conditions and the formulas, J, E, R0 and r can be calculated. Supplementary Data Fig. S1 shows the overlap of the fluorescence spectrum of HSA with the absorption spectrum of ISION. The value of J was found to be 2.53×10-14 cm3 M-1. Using the values of K 2 = 2/3, N = 1.336 and Φ = 0.15, the values of R 0, r and E were found to be 4.63, 5.25 nm and 0.031, respectively. As demonstrated above, the distance between donor and acceptor must be less than 8 nm. This criterion is satisfied in this case and thus quenching of Trp fluorescence in the presence of ISION is attributed to energy transfer. Static quenching mechanism was again confirmed based on higher value of r compared to the value of R0. The short distance between the bound ISION and Trp residue in the present study suggests that ISION as a whole did interact with HSA.
Synchronous Fluorescence and Three-Dimensional Fluorescence Spectroscopy
In quest of the possible effect of ISION binding on the tertiary structure of HSA, synchronous fluorescence spectra were obtained in the absence and presence of ISION, which are displayed in Fig. 7. We got characteristic information for the Tyr or Trp residues by setting the scanning interval Δλ at 15 or 60 nm, respectively, where the shifts in the maximum wavelength reveals an polar microenvironmental changes around these residues. A weak red shift (from 279 to 281 nm for Trp) can be observed in Fig. 7 when Δλ = 60 nm; meanwhile the fluorescence intensity of HSA decreased stepwisely in the presence of different concentrations of ISION. However, when Δλ was set at 15 nm both fluorescence intensity and peak position did not change significantly. This phenomenon implied that the binding of ISION to HSA was mainly located closely to the Trp residue [28]. Considering the minor red shifts in the emission peaks, it can be safely concluded that the Trp microenvironment undergoes a slight change in the presence of ISION.
Increasing use of three-dimensional fluorescence spectra as a fluorescence analysis technique in recent years has proved it a proper tool for detecting conformational changes in HSA in presence of ISION. Fig. 8 shows the three-dimensional fluorescence spectrum of HSA (A), and ISION-HSA (B) respectively. Peak a shows the Rayleigh scattering peak where emission wavelength equals to excitation wavelength, whereas the strong peak b exhibits the spectral characteristic of fluorophore residues (Trp and Tyr) and peak c is associated with the polypeptide backbone structure of HSA [29]. The 3D fluorescence contour spectra of the protein show contour maximums at about λex/λem = 280/340 nm due to π–π* transition of aromatic amino acids in HSA. In Fig. 8A, the fluorescence peak of HSA can be seen at λex/λem=277/337 nm and after addition of the drug, this peak moved to λex/λem = 280/340 nm as shown in Fig. 8B. The Stokes shift and fluorescence quenching of the fluorescence characteristic peaks elucidates that the complexation formed between ISION and HSA induced some microenvironmental and conformational changes in the protein.
FT-IR Spectra
From FT-IR results we got additional evidence for ISION-HSA interaction. Amide bands vibrations of the peptide moieties can be evidenced by the differences in FTIR spectra of a protein. Of all the amide modes of the peptide groups, the single most widely used one in studies of protein secondary structures is the amide I. The amides I and II peak take place in the region of 1600-1700 cm-1 and 1500-1600 cm-1, respectively. Compared with amide II, amide I band is more sensitive to the protein secondary structural alterations [30]. Therefore, the amide I band is more meaningful for secondary structural studies.
From Fig. 9, the peak position of amide I was moved from 1646.3 to 1651.0 cm-1 and amide II from 1543.1 to 1543.8 cm-1 in the FT-IR spectrum of HSA upon binding to ISION. This phenomenon demonstrates that the ISION interacted with HSA and the secondary structure as well as the conformation of HSA was changed thereby.
Conclusions
In this study, ISION was synthesized and the interaction between HSA and ISION were investigated using spectroscopy like fluorescence, CD and FT-IR. Steady state spectroscopy gave the information that ISION could substabtially quench the intrinsic fluorescence of HSA through static quenching mode. In this binding process, electrostatic interaction plays an important role in stabilizing the HSA–ISION complex and van’t Hoff calculation results proved it to be spontaneous. The binding of ISION to HSA caused the conformational perturbation of HSA, which was confirmed by synchronous, CD, FT-IR and 3D fluorescence spectroscopy. Above all, the research on the interaction between biophysically important protein HSA and ISION may help understand the role that ifosfamide-loaded nanoparticle contributes to their application as a potential formulation in the pharmaceutical research and development in the future.
References
Loehrer PJ, Lauer R, Roth BJ, Williams SD, Kalasinski LA, Einhorn LH. Salvage therapy in recurrent germ cell cancer: ifosfamide and cisplatin plus either vinblastine or etoposide. Ann Intern Med. 1988;109(7):540–6.
Loehrer P, Gonin R, Nichols CR, Weathers T, Einhorn LH. Vinblastine plus ifosfamide plus cisplatin as initial salvage therapy in recurrent germ cell tumor. J Clin Oncol. 1998;16(7):2500–4.
Grier HE, Krailo MD, Tarbell NJ, Link MP, Fryer CJ, Pritchard DJ, et al. Addition of ifosfamide and etoposide to standard chemotherapy for Ewing’s sarcoma and primitive neuroectodermal tumor of bone. N Engl J Med. 2003;348(8):694–701.
Cullen M, Billingham L, Woodroffe C, Chetiyawardana A, Gower N, Joshi R, et al. Mitomycin, ifosfamide, and cisplatin in unresectable non–small-cell lung cancer: effects on survival and quality of life. J Clin Oncol. 1999;17(10):3188–94.
Alexiou C, Schmid RJ, Jurgons R, Kremer M, Wanner G, Bergemann C, et al. Targeting cancer cells: magnetic nanoparticles as drug carriers. Eur Biophys J. 2006;35(5):446–50.
Dobson J. Magnetic nanoparticles for drug delivery. Drug Dev Res. 2006;67(1):55–60.
Nowak I, Shaw LM. Mycophenolic acid binding to human serum albumin: characterization and relation to pharmacodynamics. Clin Chem. 1995;41(7):1011–7.
Rich RL, Day YS, Morton TA, Myszka DG. High-resolution and high-throughput protocols for measuring drug/human serum albumin interactions using BIACORE. Anal Biochem. 2001;296(2):197–207.
Frostell-Karlsson Å, Remaeus A, Roos H, Andersson K, Borg P, Hämäläinen M, et al. Biosensor analysis of the interaction between immobilized human serum albumin and drug compounds for prediction of human serum albumin binding levels. J Med Chem. 2000;43(10):1986–92.
He XM, Carter DC. Atomic structure and chemistry of human serum albumin. 1992.
Zhao XC, Liu RT. Recent progress and perspectives on the toxicity of carbon nanotubes at organism, organ, cell, and biomacromolecule levels. Environ Int. 2012;40:244–55.
Chuang VTG, Kragh-Hansen U, Otagiri M. Pharmaceutical strategies utilizing recombinant human serum albumin. Pharm Res. 2002;19(5):569–77.
Liang S, Liu Y, Xiang J, Qin M, Yu H, Yan G. Fabrication of a new fluorescent polymeric nanoparticle containing naphthalimide and investigation on its interaction with bovine serum albumin. Colloids Surf B: Biointerfaces. 2014;116:206–10.
Gu Q, Kenny JE. Improvement of inner filter effect correction based on determination of effective geometric parameters using a conventional fluorimeter. Anal Chem. 2009;81(1):420–6.
He W, Li Y, Xue C, Hu Z, Chen X, Sheng F. Effect of Chinese medicine alpinetin on the structure of human serum albumin. Bioorg Med Chem. 2005;13(5):1837–45.
Matei I, Hillebrand M. Interaction of kaempferol with human serum albumin: a fluorescence and circular dichroism study. J Pharm Biomed Anal. 2010;51(3):768–73.
Zhao XC, Liu RT, Chi ZX, Teng Y, Qin PF. New insights into the behavior of bovine serum albumin adsorbed onto carbon nanotubes: comprehensive spectroscopic studies. J Phys Chem B. 2010;114(16):5625–31.
Wang S, Makhina EN, Masia R, Hyrc KL, Formanack ML, Nichols CG. Domain organization of the ATP-sensitive potassium channel complex examined by fluorescence resonance energy transfer. J Biol Chem. 2013;288(6):4378–88.
Charbonneau D, Beauregard M, Tajmir-Riahi HA. Structural analysis of human serum albumin complexes with cationic lipids. J Phys Chem B. 2009;113(6):1777–84.
Kanakis C, Tarantilis P, Polissiou M, Diamantoglou S, Tajmir-Riahi H. Antioxidant flavonoids bind human serum albumin. J Mol Struct. 2006;798(1):69–74.
Sułkowska A. Interaction of drugs with bovine and human serum albumin. J Mol Struct. 2002;614(1):227–32.
Zhao X, Liu R, Teng Y, Liu X. The interaction between Ag(+) and bovine serum albumin: a spectroscopic investigation. Sci Total Environ. 2011;409(5):892–7.
Ross PD, Subramanian S. Thermodynamics of protein association reactions: forces contributing to stability. Biochemistry. 1981;20(11):3096–102.
Zhao X, Sheng F, Zheng J, Liu R. Composition and stability of anthocyanins from purple Solanum tuberosum and their protective influence on Cr(VI) targeted to bovine serum albumin. J Agric Food Chem. 2011;59(14):7902–9.
Zhang Y, Qi ZD, Zheng D, Li CH, Liu Y. Interactions of chromium (III) and chromium (VI) with bovine serum albumin studied by UV spectroscopy, circular dichroism, and fluorimetry. Biol Trace Elem Res. 2009;130(2):172–84.
Bourassa P-, Bariyanga J, Tajmir-Riahi H-A. Binding Sites of Resveratrol, Genistein and Curcumin with Milk α-and β-Caseins. J Phys Chem B. 2013.
Forster T. Modern Quantum Chemistry III. In: Sinanoglu OE, editor. New York: Academic; 1965. p. 93–137.
Hu YJ, Liu Y, Xiao XH. Investigation of the interaction between berberine and human serum albumin. Biomacromolecules. 2009;10(3):517–21.
Zhang YZ, Li HR, Dai J, Chen WJ, Zhang J, Liu Y. Spectroscopic studies on the binding of cobalt(II) 1,10-phenanthroline complex to bovine serum albumin. Biol Trace Elem Res. 2010;135(1–3):136–52.
Purcell M, Neault JF, Tajmir-Riahi HA. Interaction of taxol with human serum albumin. Biochim Biophys Acta Protein Struct Mol Enzymol. 2000;1478(1):61–8.
Acknowledgments
The authors gratefully thank the Scientific Research Program of Yantai City (2008162) and Shandong Provincial Natural Science Foundation (ZR2013HL004) for their financial support.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Fig. S1
(DOCX 60 kb)
Rights and permissions
About this article
Cite this article
Kong, L., Hu, J., Qin, D. et al. Interaction of Ifosfamide-Loaded Superparamagnetic Iron Oxide Nanoparticles with Human Serum Albumin—A Biophysical Study. J Pharm Innov 10, 13–20 (2015). https://doi.org/10.1007/s12247-014-9199-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12247-014-9199-9