Abstract
Potato virus Y (PVY) is a common and devastating pathogen of potato worldwide and insecticide control alone for aphid vectors is not effective due to the rapid transmission of the virus by aphids following probing. Three Ry genes for extreme resistance to PVY have been identified and are used in potato breeding programs for the development of PVY-resistant varieties. Tightly linked molecular markers have been developed for all three genes and are used for marker-assisted selection (MAS). However, some varieties with known extreme resistance to PVY do not amplify any molecular markers for these resistance genes. Sante and its progeny Bistra are examples of two varieties with extreme resistance to PVY and yet have no amplification of Ry associated molecular markers. The source of PVY resistance in Bistra was mapped using SNP genotyping of 190 of its progeny which had been characterized for response to PVY infection using a N-Wilga strain. A major QTL was found on the proximal end of chromosome 11, in close proximity to Ryadg, one of the three known extreme resistance genes for PVY. However, no known markers which detect Ryadg amplify in Sante or Bistra which indicates they may possess a currently unidentified resistance gene, or the genetic linkages of molecular markers typically associated with the presence of Ryadg no longer exist due to recombination between the gene and its associated molecular marker primer annealing sites.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Potato virus Y (PVY) is one of the most common and destructive pathogens afflicting potato growers in the United States and worldwide. Infected plants produce a reduced harvest with lower quality tubers (Gray et al. 2010; Karasev and Gray 2013). PVY is non-persistently transmitted by a variety of aphids and insecticides alone are not effective at controlling the disease in the field due to the rapid transmission of the virus following aphid probing of the host plant (MacKenzie et al. 2014). To date, the best means of preventing PVY infection is by the cultivation of varieties having extreme resistance to PVY conferred by an Ry gene. Extreme resistance is characterized by the PVY inoculated plant showing no symptoms and when tested only a very low amount of virus can be detected (Barker 1996; Solomon-Blackburn and Barker 2001; Karasev and Gray 2013). Grech-Baran et al. (2019) showed that plants transformed with the functional Rysto gene showed none to negligible amounts of PVY RNA. Hypersensitive resistance is conferred by Ny genes and is PVY strain specific and is sensitive to temperature conditions (Karasev and Gray 2013). Extreme resistant examples include the varieties Payette Russet (Novy et al. 2017), Castle Russet (V. Sathuvalli, pers. communication), Mackinaw and Saginaw Chipper (D. Douches, pers. communication), Eva (evaluated as NY103) (Kasai et al. 2000; Plaisted et al. 2001), Upstate Abundance and Lady Liberty, (W. DeJong, pers. Communication), and Ciklamen (Heldak et al. 2007), with others in development. Extreme resistance includes resistance to multiple strains of PVY (Valkonen 1994; Barker 1996) and varieties Payette Russet, Eva, and Ciklamen have be shown to be resistant against multiple PVY strains and multiple isolates of these strains (Whitworth et al. 2021).
Extreme resistance to PVY is conferred by three currently known Ry genes. Ryadg originates from Solanum tuberosum ssp. andigena (Munoz et al. 1975) and has been mapped to an R-gene cluster at the proximal end of chromosome 11 (Hämäläinen et al. 1998; Hämäläinenet al. 1997). A variety of molecular markers have been developed to track this resistance source including RySC3 (Kasai et al. 2000) which is widely used, as well as M6 and M45 (Brigneti et al. 1997; Herrera et al. 2018) which are believed to flank the resistance gene (Herrera et al. 2018). Rysto originates from S. stoloniferum (Ross 1958) and has been mapped to the distal end of chromosome 12 (Flis et al. 2005; Song et al. 2005). It can be detected by the molecular marker set Yes3A (Song and Schwarzfischer 2008) and was identified and cloned in 2019 (Grech-Baran et al. 2019). The most recently reported resistance gene, Rychc, originates from S. chacoense (Asama et al. 1982) and has been identified and mapped to the distal end of Chromosome 9 (Hosaka et al. 2001; Li et al. 2022; Akai et al. 2023) and successfully cloned (Li et al. 2022), where it can be traced by the molecular marker set Ry186 (Mori et al. 2012). These markers allow marker assisted selection (MAS) to be employed during the breeding process and greatly improve the ability of breeders to integrate PVY resistance into new varieties. Many PVY resistant varieties amplify at least one of the markers mentioned above and can be conclusively identified as having specific resistance genes, with the recently developed multiplex PVY PCR assay able to assess previously unidentified material having extreme resistance genes quickly and efficiently (Elison et al. 2020).
While most varieties with extreme resistance to PVY can be easily identified via molecular markers, the Dutch variety Sante is reported to possess extreme resistance to multiple PVY strains (Gruden et al. 2000; Heldak et al. 2007) yet it has consistently failed to amplify any molecular marker developed in targeting an Ry gene (Cernák et al. 2008; Flis et al. 2005; Heldak et al. 2007; Whitworth et al. 2009). Sante has both S. tuberosum spp. andigena and S. stoloniferum in its background, several generations back (Hutten & Berloo 2001). Several prior studies have asserted that this is the result of a recombination event during the breeding of Sante which separated a hypothetical Rysto gene from its associated markers (Flis et al. 2005; Heldak et al. 2007; Hinrichs et al. 1998). This is a reasonable assertion given that a small number of such events can occur during every crossing attempt and that Sante has a source of S. stoloniferum and S. tuberosum ssp.andigena germplasm in its ancestry. However, there has never been any direct evidence to support this hypothesis and Sante contains other sources of wild germplasm in its ancestry that could have provided an alternative source of PVY resistance. There remains a possibility that Sante could represent a novel source of PVY resistance rather than a known source separated from its marker. Even if it does contain a known resistance source, markers do not currently exist which allow tracking of the resistance gene in Sante and its progenies.
While the source of resistance in Sante has yet to be determined, its extreme resistance to PVY and other useful traits has resulted in its use as a parent for numerous progenies, many of which also display extreme resistance to PVY without amplifying any of the molecular markers associated with any of the Ry genes. Among these are a collection of Slovenian varieties (Dolničar and Pilih 2012) used in the Aberdeen breeding program which have Sante as a parent and display extreme resistance to PVY in the field. Unfortunately, it is not possible to use MAS on the progeny of Sante or on its descendants (e.g. Slovenian variety Bistra) due to the lack of amplification of markers associated with the PVY-resistance gene.
A population was generated from a cross between Bistra and a PVY susceptible breeding clone from the USDA ARS Aberdeen breeding program to map the source of the PVY resistance in Bistra. By inoculating 190 individuals from this population with PVY and comparing the phenotypic results to single nucleotide polymorphism (SNP) data for the population, the resistance source in Bistra, and its descendants were localized to the proximal end of chromosome 11. This represents a potential new source of PVY resistance.
Materials and Methods
Population Development and Evaluation for PVY Resistance
The population designated A13300 was generated by a cross between the Slovenian variety Bistra, kindly provided by Peter Dolničar from the Agricultural Institute of Slovenia, and the USDA ARS Aberdeen breeding clone A98345-1 (Fig. 1). A98345-1 is susceptible to PVY and exhibits none of the existing genetic markers for PVY resistance. Bistra has been shown to be resistant to PVY in field trials (Table 1), but also does not amplify any existing genetic markers for PVY resistance (internal data). Sante is the female parent of Bistra and both show extreme resistance to PVY (Table 1). True potato seed from this family was germinated in seedling flats containing a 50/50 mix of vermiculite and perlite and soaked in a solution containing 1 teaspoon JR Peters Professional Blue® liquid fertilizer (16-16-16) and 5 ml Baicor® chelated iron (0-0-1) per 3.8 L. Flats were covered loosely with a layer of thin, clear plastic shaded by a layer of black cloth to restrict heat and sunlight and allowed to germinate. When plantlets were approximately 2.5 cm tall, 224 clones were individually planted and grown in a greenhouse environment. Plants were allowed to grow to maturity and senesce, after which all tubers were harvested. Thirty-two clones did not produce tubers or produced tubers which were too small to use and so were not carried forward. The remaining 192 clones were placed into cold storage for one month. Tubers were then removed from cold storage and placed at room temperature in a dark area until they began to sprout (roughly 4–6 weeks). As tubers developed sprouts, five tubers from each clone were planted and grown in a greenhouse. Both parents (Bistra and A98345-1) were also planted in the same manner at this time and were evaluated in the same manner as their progeny for the rest of the analysis.
After a period of 4–6 weeks depending on the growth rate of the clone, five plants of each breeding clone were hand inoculated with PVY. Inoculation was conducted using the isolate OR3 (GenBank KY848021) of the PVY N-Wilga strain on five replicate plants of each clone. N-Wilga was chosen for its prevalence in the field in the Pacific Northwest region of the U.S. (Tran et al. 2022). Inoculum was created by the addition of OR3 infected tobacco leaf tissue blended at a ratio of 1 g tissue to 3 ml phosphate buffer (0.1M K2HPO4, 0.025 M KH2PO4, pH 7.5). Carborundum (320 grit) was dusted onto three leaflets of each plant and a small piece of cheesecloth saturated in the inoculum was gently rubbed on each leaflet. Leaflets were then washed to remove excess carborundum. One week after the initial inoculation, all plants were re-inoculated using the same protocol. Three and five weeks after the first inoculation, plants were tested via ELISA using the method of Clark and Adams (Clark and Adams 1977) with monoclonal antibodies for PVYAll obtained from SASA (Edinburgh, Scotland). Plates included both positive and negative controls and an absorbance (A405) value greater than 0.1 was regarded as positive for PVY. Clones for which at least two plants tested positive were deemed to be susceptible to PVY. Clones for which zero plants tested positive for both ELISA tests were deemed to be resistant to PVY.
DNA Extraction from Leaf Tissue
Collection of leaf tissue was conducted from each clone during the first phase of greenhouse growth when each clone was represented by a single plant. Approximately 0.4 g of leaf tissue was collected and placed in a 2.0 ml Omni International reinforced microcentrifuge tube containing three 2.8 mm ceramic beads. DNA extraction was conducted using the DNeasy Plant Mini Kit (Qiagen) with modifications as follows: 500 ul Buffer AP 1, 3 ul RNase A, 150 ul Buffer P3, 650 ul sample applied to the QIAshredder mini spin column, 550 µl of the flow through recovered, 825 µl Buffer AW2, 50 µl Buffer AE for 1st elution, and another 25 µl for the 2nd elution. Sample concentrations were checked using a nanodrop spectrophotometer and stored at -20OC. Assessment for markers associated with PVY resistance (Table 2) was determined via PCR. PCRs were conducted according to the methods outlined in Elison et al. (2020) and Herrera et al. (2018)
SNP Genotyping
Samples from the 192 clones along with the two parents were evaluated using an Agilent Tapestation™ 4200 system to determine DNA concentration and quality. Due to the requirement to submit samples in 96 well plates, the two clones with the worst quality DNA were removed and the remaining 190 clones and two parents were prepared for SNP genotyping. These samples were genotyped with the SolCAP Infinium Potato SNP array Version 3 (21,027 SNPs) (Hamilton et al. 2011), and the Illumina iScan system at the Neogene SNP Genotypic Center (Lansing, MI). Basic DNA sample quality evaluation and obtainment of SNP theta scores were conducted through GenomeStudio software (Park et al. 2019; Staaf et al. 2008). The R-package, Clustercall version 1.5 was used to translate SNP theta values into autotetraploid marker genotypes (AAAA, AAAB, AABB, ABBB, and BBBB) and to filter out uninformative SNPs (Schmitz Carley et al. 2017). Any SNP markers that registered as no-calls in either or both parents and weren’t able be analyzed by MAPpoly were removed. Any SNPs with incorrect physical map positions after analysis were omitted to remove the phenomenon of immoderate extended linkage maps. Several arguments equipped in MAPpoly continuously filtered out unnecessary markers such as non-conforming and redundant markers.
The filter_segregation argument was used to perform a chi-squared (χ2) test, which matches expected genotype frequencies against observed frequency and calculates the associated p-value. Bonferroni correction was used to distinguish informative markers (p < 0.05). The make_seq_mappoly argument omitted markers which did not significantly meet the expected segregation ratios based on Mendelian inheritance. Markers were omitted which were either uninformative, co-segregating, or unaffiliated with any of the 12 linkage groups.
Construction of Linkage and QTL maps
Automated marker selection and linkage mapping processes were performed by the R-package MAPpoly v. 0.2.3 which can analyze ploidy levels up to eight when hidden Markov models (HMM) are applied (Mollinari and Garcia 2019; Mollinari et al. 2020; RCoreTeam 2020). After the translated SNP marker dataset was imported into MAPpoly, uninformative markers were first filtered out through the filter_missing, filter_segregation, make_seq_mappoly, and elim.redundant functions. As described in Park et al. (2021), MAPpoly was used to select markers for developing and refining 12 linkage groups through two-point analysis, Unweighted Pair Group Method with Arithmetic Mean (UPGMA) hierarchical clustering method, multidimensional scaling (MDS) (da Silva Pereira et al. 2020; Preedy and Hackett 2016; Sharma et al. 2013), and the potato reference genome PGSC Version 4.03 (http://solanaceae.plantbiology.msu.edu).
The 12 complete linkage groups were transferred to QTLpoly, an R-package designed for QTL analysis of polyploid organisms. QTLpoly was used to scrutinize and combine both the linkage groups and phenotype data to depict 12 QTL maps (da Silva Pereira et al. 2020). Among the various functions implemented in the software, the remim function, which runs a random-effect multiple interval mapping (REMIM) model, was used to fit various random-effect QTL by evaluating a single parameter per QTL. QTLpoly was used to conduct linear score statistics tests (Qu et al. 2013) at every position and compare its p-value to a prescribed critical value. A continuous pattern over the whole range of the unit interval was observed from the p-values as a result of weighted sums of the scores from the profiled likelihood (da Silva Pereira et al. 2020; Qu et al. 2013). The p-values were converted to LOP scores (LOP = – log10 (p-value)) by QTLpoly to visualize and compare newly detected QTLs in this study intuitively and to calculate support intervals of those QTLs. As suggested by the QTLpoly software developer, the QTLs with four or higher LOP scores were adopted as significant QTL peaks (da Silva Pereira et al. 2020). In addition, the QTLpoly (R-package) provides information on support intervals defined as the QTL peak adjacent to a zone with LOP higher than or equal to LOP – d, where d is a constant, which subtracts the highest LOP in that region (da Silva Pereira et al. 2020; Lander and Green 1987). The current study used approximately 95% support intervals by using LOP – 1.5 (da Silva Pereira et al. 2020). The heritability of the significant QTL was calculated by the fit model function equipped in QTLpoly. Each calculated QTL heritability indicated the ratio between the QTL variance component and total variance. The symbol “h2QTL” was used to indicate the heritability of each significant QTL (da Silva Pereira et al. 2020). It should be noted that the QTL heritability (h2QTL) should be distinguished from the general heritability representing how well a trait was inheritable from two parents to their progeny. After a significant QTL was localized, the closest SNPs to the QTL were reported together.
Analyses of Allele Effects
After localizing the significant QTL and the most adjacent SNPs to the QTL locations, allele effects at each SNP position were visualized, by QTLpoly, with bar graphs. The graphs reflect the contribution of each parent to the mean of the whole mapping population for the trait. For example, how much each homolog of the four homologs of the two parents adds to or subtracts from the mean given one of 190 observed genotypes (da Silva Pereira et al. 2020). They were also helpful to find the location of a target allele (e.g., resistant allele) among the four homologs of each parent (da Silva Pereira et al. 2020; Park et al. 2021).
Results
Marker Selection Process and Linkage Group Assembly
One hundred and ninety clones resulting from a cross between Bistra and breeding clone A98345-1 were inoculated with PVY and tested in the greenhouse. Of these, 84 (44%) were susceptible to PVY and 106 (56%) were resistant to PVY indicating the observed segregation pattern did not significantly deviate from a 1:1 ratio, χ2(1, N = 190) = 2.55, p = 0.11. As expected Bistra tested as a resistant clone and A98345-1 tested as a susceptible clone. The same 190 clones along with their parents were SNP genotyped with SolCAP Infinium Potato SNP array Version 3 (21,027 SNPs) to identify the location of the PVY resistance source in Bistra. After analysis using ClusterCall software 22 clones were removed due to missing values identified by the filter_missing function. This left 168 clones for final analysis.
After multiple steps for marker evaluation and filtering processes (Park et al. 2021), a total of 4659 informative SNPs were identified and used to assemble the 12 linkage maps, which represent the fundamental potato chromosome number. The selected SNPs were regularly distributed without significant gaps between them on each chromosome, resulting in dense linkage groups without a massive gap between contiguous SNPs (Fig. 2). The number of selected markers for each linkage group, their map lengths, the number of markers segregated in the two parents, and map coverage rates are given in Table 3. A high concordance (Supplemental Fig. 1) and map coverage rate (Table 3) were observed when comparing the complete linkage groups with the reference genome (PGSC version 4.03).
Identification of the Source of PVY Resistance
QTL maps with detailed information were generated including the chromosome number and location of significant QTLs as well as the LOP score (Fig. 3). Only one QTL for the PVY resistance phenotype (PVY_R_ch11_QTL) was observed at ~ 2.08 cM on chromosome 11 with the highest LOP score which was the maximum score reported by QTLpoly. QTLpoly also reported support intervals, h2QTL, and the closest SNP markers to the mapped QTL. The QTL heritability (h2QTL) of PVY_R_ch11_QTL was 50% and the closest SNP marker was solcap_snp_c2_13432. The support interval of PVY_R_ch11_QTL occupied 2.08–4.96 cM. Details of the range of the support interval and a linkage map are included in supplemental Fig. 2 and supplemental Table 1. Due to the qtl_effects function in QTLpoly, allele effects at PVY_R_ch11_QTL could be investigated (Fig. 4). This reflects how much each of the four homologs from the two parents adds to or subtracts from the mean effect given one of 168 observed genotypes (da Silva Pereira et al. 2020). The X-axis of Fig. 4 represents the four homologs of each parent with “a – d” as the four homologs of Bistra (resistant) and “e – h” as those of A98345-1 (susceptible). The Y-axis indicates the allele effect on each homolog which can be either positive or negative. As expected, a single allele from Bistra (homolog “a”) substantially contributed to a reduction in PVY infection with no other alleles from either parent meaningfully contributing to infection.
QTL maps for PVY resistance. X-axis shows all the positions of the 12 different linkage groups. Y-axis visualizes the height of the QTL. QTL with four or more LOP scores were selected as significant. The LOP score of the significant QTL at 2.08 cM on chromosome 11 was over 15.65, the maximum value provided by QTLpoly
Allele effects of PVY_R_ch11_QTL. The X-axis indicates four homologs of each parent. The “a” to “d” were four homologs of Bistra and “e” to “h” were those of A98345-1. The Y-axis indicated the amount of each allele effect. Positive and negative effects are associated with susceptibility and resistance, respectively. The strength of the effect is also correlated with the brightness of the bar, so the alleles for A98345-1 appear faint while those for Bistra appear bright, especially allele “a”
Testing of Ry adg Markers on Bistra
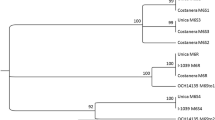
The resistant locus identified on Bistra is located in the same region as the Ryadg gene which is known to confer extreme resistance to PVY. To determine if the resistant source is the same as Ryadg, multiple sets of molecular markers which are known to amplify Ryadg, as well markers linked to Rysto and Rychc, were applied to both Bistra and its resistant parent Sante and several varieties possessing one of the three genes for extreme resistance (Table 4). The standard marker used to identify Ryadg is RySC3 (Kasai et al. 2000) which amplifies a region within an NBS-LRR protein at around 1,660,000 bp on chromosome 11. Two additional markers (M6 and M45) developed by Brigneti et al. (1997) and refined by Hererra et al. (2018) are also close markers for the presence of Ryadg and are believed to flank the Ryadg resistance gene. All of these markers are able to amplify in standard ADG + varieties such as Reiche and Costanera. All three of the Ryadg associated markers failed to amplify in standard susceptible varieties such as Yukon Gold and Desiree, as well as in PVY-resistant varieties Castle and Ursus (presence only of Rysto) and Sakai 35 (presence only of Rychc). In addition, all three markers failed to amplify in Sante and Bistra, despite the close proximity of the identified resistance source to the rough location of Ryadg (Table 4). In addition, an older marker, ADG2, was tested on many of the same varieties in Whitworth et al. (2009) and also failed to amplify in Sante.
Discussion
The mapping data is very clear that the source of PVY resistance in Bistra is located on the proximal end of chromosome 11 and that a single allele in Bistra is responsible for the resistance. The data is consistent with the hypothesis that extreme resistance to PVY in Bistra is conferred by a single dominant R-gene on chromosome 11.
Unfortunately, due to limitations in QTL mapping, it is impossible to determine the exact location of this gene, but it is likely contained within a known R-gene cluster (Hämäläinen et al. 1998) found on the proximal end of chromosome 11. The closest SNP marker to the resistance source (solcap_snp_c2_13432) is located at roughly bp 790,000 on the chromosome, but this is just the location of the marker, not the actual gene. Due to the close proximity between the closest SNP marker found in Bistra and the approximate location of Ryadg it is difficult to determine if these represent one or two resistance sources through SNP mapping alone, as the resolution is not precise enough to make distinctions on the order of below 1–2 Mb. However, analysis of molecular markers in the region suggests that these may be independent resistance sources. Every marker tested to assess the presence of Ryadg failed to amplify in Bistra or Sante, despite the fact that these markers are regarded as being very tightly linked to Ryadg. Unfortunately, no markers currently exist which are capable of tracking PVY resistance in Bistra or Sante, so it remains unknown if such hypothetical markers would amplify in clones such as Reiche or Costanera which are known to amplify the Ryadg marker.
If the resistance source in Bistra is Ryadg, with the same Solanum tuberosum ssp. andigena progenitor as with Reiche or Costanera, then multiple meiotic events would have needed to occur to separate the gene from its markers. Herrera et al. (2018) claims that the markers M45 and M6 flank either side of Ryadg which means that a single meiotic recombination would not separate the gene from both markers. Two events, one on each side are needed for this to occur. Given that both markers are estimated to be within 0–2 centimorgans of Ryadg, these events are moderately likely to occur individually (roughly 1 or 2 events per 100 progeny) but for them to occur together or sequentially is extremely unlikely (roughly 1 double event per 10,000 progeny). While it is possible that Bistra may represent examples of breakage of linkage between Ryadg and its associated molecular markers, it would be a highly unlikely occurrence.
Another possible explanation is that the gene for extreme PVY resistance identified in Bistra does represent Ryadg. If Bistra derived its resistance gene from a Solanum tuberosum ssp. andigena clone divergent from the progenitor donor of Ryadg for Reiche or Costanera, then flanking chromosome regions of Bistra may lack the necessary primer annealing sites necessary for successful marker amplification relative to Reiche or Costanera.
Regardless of the identity of the resistance source in Bistra, the presence of resistant material which cannot be tracked by any known molecular markers slows development in breeding programs attempting to incorporate PVY resistance for the genesis of new varieties. As such, the creation of molecular markers targeting this resistance source would be of great value to the breeding community, and the identification of the closest SNP to the resistance source allows for the development of new markers.
The PVY_R_ch11_QTL showed an extremely and exclusively high LOP score, which reached the software limit. Furthermore, its QTL heritability (h2QTL), representing the quantity of the QTL variance compared to total variance, was also extremely high (50%). Based on those outstanding scores of the PVY_R_ch11_QTL, it is likely that the resistance source in Bistra is an R-gene.
More research is needed with additional populations to develop markers for this gene. Rysto and Rychc genes have been cloned (Grech-Baran et al. 2019; Akai et al. 2023) and once Ryadg has been cloned then sequencing can be used to determine if Bistra derived PVY resistance is due to Ryadg. If this is the case, more robust markers will be need to be developed for Ryadg. If future research identifies a new Ry gene, then a new source of PVY resistance will be available to breeding programs.
References
Akai, K., K. Asano, C. Suzuki, E. Shimosaka, S. Tamiya, T. Suzuki, T. Takeuchi, and T. Ohki. 2023. De novo genome assembly of the partial homozygous dihaploid potato identified PVY resistance gene (Rychc) derived from Solanum chacoense. Breeding Science 73: 168–179.
Asama, K., and N. M. H I, T I. 1982. New potato variety Konafubuki. Bull Hokkaido Pref Agr Exp Stn 48: 75–84.
Barker, H. 1996. Inheritance of resistance to potato viruses Y and A in progeny obtained from potato cultivars containing gene ry: evidence for a new gene for extreme resistance to PVA. Theoretical and Applied Genetics 93: 710–716.
Brigneti, G., J. Garcia-Mas, and D. C. Baulcombe. 1997. Molecular mapping of the Potato virus Y resistance gene rysto in potato. Theoretical and Applied Genetics 94: 198–203.
Cernák, I., J. Taller, I. Wolf, E. Fehér, G. Babinszky, Z. Alföldi, G. Csanádi, and Z. Polgár. 2008. Analysis of the applicability of molecular markers linked to the PVY extreme resistance gene rysto, and the identification of new markers. Acta Biologica Hungarica 59: 195–203.
Clark, M. F., and A. N. Adams. 1977. Characteristics of the microplate method of enzyme linked immunosorbent assay for the detection of plant viruses. Journal of General Virology 34: 475–483.
G. da Silva Pereira, D. C. Gemenet, M. Mollinari, B. A. Olukolu, J. C. Wood, F. Diaz, V. Mosquera, W, J. Gruneberg, A. Khan, C. R. Buell, G. C. Yencho, Z-B. Zeng. 2020. Multiple QTL Mapping in Autopolyploids: A random-effect model approach with application in a hexaploid sweetpotato full-sib population. Genetics 215:579–595.
Dolničar, P., and K. Rudolf Pilih. 2012. Gene bank and potato breeding in Slovenia. Acta Agriculturae Slovenica 99: 377–386.
Elison, G. L., D. G. Hall, R. G. Novy, and J. L. Whitworth. 2020. Development and application of a multiplex marker assay to detect PVY resistance genes in Solanum tuberosum. American Journal of Potato Research 97: 289–296.
Flis, B., J. Hennig, D. Strzelczyk-Żyta, C. Gebhardt, and W. Marczewski. 2005. The Ry-f sto gene from Solanum stoloniferum for extreme resistant to Potato virus Y maps to potato chromosome XII and is diagnosed by PCR marker GP122 718 in PVY resistant potato cultivars. Molecular Breeding 15: 95–101.
Gray, S., S. De Boer, J. Lorenzen, A. Karasev, J. Whitworth, P. Nolte, R. Singh, A. Boucher, and H. Xu. 2010. Potato virus Y: an evolving concern for potato crops in the United States and Canada. Plant Disease 94: 1384–1397.
Grech-Baran, M., K. Witek, K. Szajko, A. I. Witek, K. Morgiewicz, I. Wasilewicz-Flis, H. Jakuczun, W. Marczewski, J. D. G. Jones, and J. Hennig. 2019. Extreme resistance to Potato virus Y in potato carrying the rysto gene is mediated by a TIR-NLR immune receptor. Plant Biotechnology Journal 18: 655–667.
Gruden, K., B. Strukelj, M. Ravnikar, and B. Herzog-Velikonja. 2000. A putative viral resistance-connected protein isolated from potato cultivar Sante resistant to PVYNTN infection. Phyton 40: 191–200.
Hämäläinen, J. H., K. N. Watanabe, J. P. T. Valkonen, A. Arihara, R. L. Plaisted, E. Pehu, L. Miller, and S. A. Slack. 1997. Mapping and marker-assisted selection for a gene for extreme resistance to potato virus Y. Theoretical and Applied Genetics 94: 192–197.
Hämäläinen, J. H., V. A. Sorri, K. N. Watanabe, C. Gebhardt, and J. P. T. Valkonen. 1998. Molecular examination of a chromosome region that controls resistance to potato Y and a potyviruses in potato. Theoretical and Applied Genetics 96: 1036–1043.
Hamilton, J. P., C. N. Hansey, B. R. Whitty, K. Stoffel, A. N. Massa, A. Van Deynze, W. S. De Jong, D. S. Douches, and C. R. Buell. 2011. Single nucleotide polymorphism discovery in elite north American potato germplasm. Bmc Genomics 12: 302.
Heldak, J., M. Bezo, V. Stefunova, and A. Gallikova. 2007. Selection of DNA markers for detection of extreme resistance to Potato virus Y in tetraploid potato (Solanum tuberosum L.) F1 progenies. Czech Journal of Genetics and Plant Breeding 43: 125–134.
Herrera, M. R., L. J. Vidalon, J. D. Montenegro, C. Riccio, F. Guzman, I. Bartolini, and M. Ghislain. 2018. Molecular and genetic characterization of the ry adg locus on chromosome XI from Andigena potatoes conferring extreme resistance to Potato virus Y. Theoretical and Applied Genetics 131: 1925–1938.
Hinrichs, J., S. Berger, and J. G. Shaw. 1998. A hypersensitive response-like mechanism is involved in resistance of potato plants bearing the rysto gene to the potyviruses Potato virus Y and Tobacco Etch virus. Journal of General Virology 79: 167–176.
Hosaka, K., Y. Hosaka, M. Mori, T. Maida, and H. Matsunaga. 2001. Detection of a simplex RAPD marker linked to resistance to Potato virus Y in a tetraploid potato. American Journal of Potato Research 78: 191–196.
Hutten, R. C. B., and R. van Berloo. 2001. An online potato pedigree database. URL: http://www.plantbreeding.wur.nl/PotatoPedigree/. Accessed 22 Feb. 2024.
Karasev, A. V., and S. M. Gray. 2013. Continuous and emerging challenges of Potato virus Y in potato. Annual Review of Phytopathology, pp 571–586.
Kasai, K., Y. Morikawa, V. A. Sorri, J. P. T. Valkonen, C. Gebhardt, and K. N. Watanabe. 2000. Development of SCAR markers to the PVY resistance gene RY(adg) based on a common feature of plant disease resistance genes. Genome 43: 1–8.
Lander, E. S., and P. Green. 1987. Construction of multilocus genetic linkage maps in humans. Proceedings of the National Academy of Sciences 84:2363–2367.
Li, G., J. Shao, Y. Wang, T. Liu, Y. Tong, S. Jansky, C. Xie, B. Song, and X. Cai. 2022. Rychc confers Extreme Resistance to Potato virus Y in Potato. Cells 11: 2577.
MacKenzie, T. D. B., M. S. Fageria, X. Nie, and M. Singh. 2014. Effects of crop management practices on current-season spread of Potato virus Y. Plant Disease 98: 213–222.
Mollinari, M., and A. A. F. Garcia. 2019. Linkage analysis and haplotype phasing in experimental autopolyploid populations with high ploidy level using hidden markov models. G3 Genes|Genomes|Genetics 9: 3297–3314.
Mollinari, M., B. A. Olukolu, GdS Pereira, A. Khan, D. Gemenet, G. C. Yencho, and Z-B. Zeng. 2020. Unraveling the hexaploid sweetpotato inheritance using ultra-dense multilocus mapping. G3 Genes|Genomes|Genetics 10: 281–292.
Mori, K., N. Mukojima, T. Nakao, S. Tamiya, Y. Sakamoto, N. Sohbaru, K. Hayashi, H. Watanuki, K. Nara, K. Yamazaki, T. Ishii, and K. Hosaka. 2012. Germplasm release: Saikai 35, a male and female fertile breeding line carrying Solanum Phureja-derived cytoplasm and potato cyst nematode resistance (H1) and potato virus Y resistance (Rychc) genes. American Journal of Potato Research 89: 63–72.
Munoz, F. J., R. L. Plaisted, and H. D. Thurston. 1975. Resistance to Potato virus Y in Solanum tuberosum spp. andigena. American Potato Journal 52: 107–115.
Novy, R. G., J. L. Whitworth, J. C. Stark, B. L. Schneider, N. R. Knowles, M. J. Pavek, L. O. Knowles, B. A. Charlton, V. Sathuvalli, S. Yilma, C. R. Brown, M. Thornton, T. L. Brandt, and N. Olsen. 2017. Payette Russet: a dual-purpose potato cultivar with cold-sweetening resistance, low acrylamide formation, and resistance to late blight and Potato virus Y. American Journal of Potato Research 94: 38–53.
Park, J., C. A. Hackett, L-M. Dandurand, X. Wang, and W. S. De Jong. 2019. QTL for resistance to Globodera Rostochiensis pathotype Ro2 and G. Pallida pathotype Pa2/3 in autotetraploid potato. American Journal of Potato Research 96: 552–563.
Park, J., A. N. Massa, D. Douches, J. Coombs, D. Akdemir, G. C. Yencho, J. L. Whitworth, and R. G. Novy. 2021. Linkage and QTL mapping for tuber shape and specific gravity in a tetraploid mapping population of potato representing the russet market class. BMC Plant Biology 21: 507.
Plaisted, R. L., D. E. Halseth, B. B. Brodie, S. A. Slack, J. B. Sieczka, B. J. Christ, K. M. Paddock, and M. W. Peck. 2001. Eva: a midseason golden nematode- and virus-resistant variety for use as tablestock or chipstock. American Journal of Potato Research 78: 65–68.
Preedy, K. F., and C. A. Hackett. 2016. A rapid marker ordering approach for high-density genetic linkage maps in experimental autotetraploid populations using multidimensional scaling. Theoretical and Applied Genetics 129: 2117–2132.
Qu, L., T. Guennel, and L. Marshall Scott. 2013. Linear score tests for variance components in linear mixed models and applications to genetic association studies. Biometrics 69: 883–892.
RCoreTeam. 2020. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing.
Ross, H. 1958. Inheritance of extreme resistance to Potato virus Y in Solanum stoloniferum and its hybrids with Solanum tuberosum. Proc Third Conf Potato Virus Diseases:204–211.
Schmitz Carley, C. A., J. J. Coombs, D. S. Douches, P. C. Bethke, J. P. Palta, R. G. Novy, and J. B. Endelman. 2017. Automated tetraploid genotype calling by hierarchical clustering. Theoretical and Applied Genetics 130: 717–726.
Sharma, S. K., D. Bolser, J. de Boer, M. Sønderkær, W. Amoros, M. F. Carboni, J. M. D’Ambrosio, G. de la Cruz, A. Di Genova, D. S. Douches, M. Eguiluz, X. Guo, F. Guzman, C. A. Hackett, J. P. Hamilton, G. Li, Y. Li, R. Lozano, A. Maass, D. Marshall, D. Martinez, K. McLean, N. Mejía, L. Milne, S. Munive, I. Nagy, O. Ponce, M. Ramirez, R. Simon, S. J. Thomson, Y. Torres, R. Waugh, Z. Zhang, S. Huang, R. G. F. Visser, C. W. B. Bachem, B. Sagredo, S. E. Feingold, G. Orjeda, R. E. Veilleux, M. Bonierbale, J. M. E. Jacobs, D. Milbourne, D. M. A. Martin, and G. J. Bryan. 2013. Construction of reference chromosome-scale pseudomolecules for potato: Integrating the potato genome with genetic and physical maps. G3 Genes|Genomes|Genetics 3:2031–2047.
Solomon-Blackburn, R. M., and H. Barker. 2001. A review of host major-gene resistance to potato viruses X, Y, A and V in potato: genes, genetics and mapped locations. Heredity 86: 8–16.
Song, Y. S., and A. Schwarzfischer. 2008. Development of STS markers for selection of extreme resistance (rysto) to PVY and maternal pedigree analysis of extremely resistant cultivars. American Journal of Potato Research 85: 159–170.
Song, Y. S., L. Hepting, G. Schweizer, L. Hartl, G. Wenzel, and A. Schwarzfischer. 2005. Mapping of extreme resistance to PVY (Rysto) on chromosome XII using anther-culture-derived primary dihaploid potato lines. Theoretical and Applied Genetics 111: 879–887.
Staaf, J., J. Vallon-Christersson, D. Lindgren, G. Juliusson, R. Rosenquist, M. Höglund, Å Borg, and M. Ringnér. 2008. Normalization of Illumina Infinium whole-genome SNP data improves copy number estimates and allelic intensity ratios. Bmc Bioinformatics 9: 409.
Tran, L. T., K. J. Green, M. Rodriguez-Rodriguez, G. E. Orellana, C. N. Funke, O. V. Nikolaeva, A. Quintero-Ferrer, M. Chikh-Ali, L. Woodell, N. Olsen, and A. V. Karasev. 2022. Prevalence of recombinant strains of Potato Virus Y in seed potato planted in Idaho and Washington States between 2011 and 2021. Plant Disease 106: 810–817.
Valkonen, J. P. T. 1994. Natural genes and mechanisms for resistance to viruses in cultivated and wild potato species (Solanum spp). Plant Breeding 112: 1–16.
Whitworth, J. L., R. G. Novy, D. G. Hall, J. M. Crosslin, and C. R. Brown. 2009. Characterization of broad spectrum Potato Virus Y Resistance in a Solanum tuberosum ssp. andigena-derived population and select breeding clones using molecular markers, grafting, and field inoculations. American Journal of Potato Research 86: 286–296.
Whitworth, J. L., S. M. Gray, J. T. Ingram, and D. G. Hall. 2021. Foliar and tuber symptoms of U.S. Potato varieties to multiple strains and isolates of Potato virus Y. American Journal of Potato Research 98: 93–103.
Acknowledgements
The authors would like to thank Darren Hall for his expert help in inoculations and technical support. The authors would like to thank Peter Dolničar for his generosity in providing Slovenian material which was instrumental for this work. This work was partially funded by USDA-NIFA Specialty Crop Research Initiative award no. 2020-51181-32136.
Funding
This work was partially funded by USDA-NIFA Specialty Crop Research Initiative award no. 2020-51181-32136.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Elison, G.L., Park, J., Novy, R.G. et al. A Potential New Source of Extreme Resistance to Potato Virus Y in the Potato Variety Bistra. Am. J. Potato Res. 101, 248–256 (2024). https://doi.org/10.1007/s12230-024-09954-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12230-024-09954-6