Abstract
KlUpc2p, a transcription factor belonging to the fungal binuclear cluster family, is an important regulator of ergosterol biosynthesis and azole drug resistance in Kluyveromyces lactis. In this work, we show that the absence of KlUpc2p generates Rag− phenotype and modulates the K. lactis susceptibility to oxidants and calcofuor white. The KlUPC2 deletion leads to increased expression of KlMGA2 gene, encoding an important regulator of hypoxic and lipid biosynthetic genes in K. lactis and also KlHOG1 gene. The absence of KlUpc2p does not lead to statistically significant changes in glycerol, corroborating the expression of KlGPD1 gene, encoding NAD+-dependent glycerol-3-phosphate dehydrogenase, that is similar in both the deletion mutant and the parental wild-type strain. Increased sensitivity of Klupc2 mutant cells to brefeldin A accompanied with significant increase in KlARF2 gene expression point to the involvement of KlUpc2p in intracellular signaling. Our observations highlight the connections between ergosterol and fatty acid metabolism to modulate membrane properties and point to the possible involvement of KlUpc2p in K. lactis oxidative stress response.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fungi belonging to Saccharomycotina clade contain a specific family of transcriptional regulators (Zn2Cys6), many of which are essential for adaptation to various types of stresses (MacPherson et al. 2006). The main transcription factors involved in Saccharomyces cerevisiae ergosterol biosynthesis regulation, Upc2p, Ecm22p, and Sut1p, belong to this zinc cluster family. In the absence of ergosterol Upc2p and Ecm22p bind to the sterol response element (SRE) in the promoters of ergosterol (ERG) genes to activate their transcription (Vik and Rine 2001). Under anaerobic conditions, Upc2p also plays a role in the uptake of sterols by regulating the expression of genes encoding cell wall mannoproteins (DAN1/TIR) and ABC transporters (AUS1, PDR11) (Wilcox et al. 2002). Along with the SRE binding proteins, Upc2p and Ecm22p, the heme-binding protein Hap1p and the repressors Rox1p and Mot3p coordinate ergosterol gene expression in S. cerevisiae (Davies and Rine 2006; reviewed in Jorda and Puig 2020).
Kluyveromyces lactis, an ascomycetous yeast species closely related to S. cerevisiae, received considerable attention due to its distinct metabolic and physiological properties (Breunig et al. 2000; Rodicio and Heinisch 2013). In aerobiosis, S. cerevisiae primarily uses fermentative metabolism, whereas in K. lactis, respiratory metabolism is predominant. In S. cerevisiae, where respiration and mitochondrial functions are dispensable, glucose is consumed via glycolysis. On the contrary, in the respiratory K. lactis yeast, glucose is metabolized through the pentose phosphate pathway and the preferences between respiration and fermentation depend on oxygen availability (Kiers et al. 1998). K. lactis belongs to petite-negative yeast species that are not able to survive without mitochondrial DNA and proteosynthesis. Petite mutants can be obtained in specific K. lactis nuclear background only (Chen and Clark-Walker 1999). Wild-type K. lactis strains are able to grow on glucose in the presence of antimycin A, an inhibitor of mitochondrial respiration, although many K. lactis mutants (natural or induced) fail to grow in such conditions. Impaired growth on glucose medium supplemented with antimycin A is known as Rag− phenotype (resistance to antimycin A on glucose) and is usually ascribed to defective glycolysis and/or its regulation (reviewed in Breunig et al. 2000). However, Rag− phenotype in K. lactis was also observed as a result of KlHSL1 (the K. lactis homologue of a gene encoding a serine-threonine protein kinase regulating the morphogenesis checkpoint in S. cerevisiae) and KlMGA2 (transcriptional activator involved in various kinds of stresses) gene deletions (Cialfi et al. 2011; Micolonghi et al. 2012). The fact, that low oxygen induction of KlMGA2 requires the plasma membrane glucose sensor Rag4 in K. lactis, suggests that low oxygen response is dependent on glucose availability in this yeast (Rolland et al. 2006; Micolonghi et al. 2012). Thus, the coordination of the oxygen-response pathways at the transcription factor level maintains metabolic homeostasis in the cell, primarily the requirement for acetyl-CoA in lipid biosynthesis (Burr and Espenshade 2018).
K. lactis has a lower redundancy of genes compared to S. cerevisiae because it did not undergo a whole genome duplication (WGD) (Bussereau et al. 2006; González-Siso et al. 2009). In S. cerevisiae, three ARF (ADP-ribosylation factors) GTPases have been identified, and two of them, the paralogs ScARF1 and ScARF2, regulate the formation of coated vesicles in intracellular trafficking within the Golgi. K. lactis contains only one ortholog of ScARF1 and ScARF2, the KlARF2. The activity of ARFs is regulated by small G-proteins that are essential for a variety of cellular processes (Labbaoui et al. 2017).
The absence of an uptake system for ergosterol precludes the growth of K. lactis in strictly anoxic conditions (Snoek and Steensma 2006). K. lactis contains only one orthologue of S. cerevisiae Upc2p and Ecm22p (Bussereau et al. 2006). In the previous work, we showed that K. lactis cells lacking KlUPC2 exhibit hypersusceptibility to azole drugs and are not able to activate expression of ERG genes in the presence of azoles. KlUpc2p was shown to be important for the appropriate regulation of plasma membrane ion transporters KlEna1p (Na+-ATPase) and KlPma1p (H+-ATPase) (Toth Hervay et al. 2020). In this work, we further extend these observations and show that KlUpc2p is required not only for optimal growth on glucose and modulation of membrane properties but also for oxidative stress response.
Materials and methods
Strains and culture conditions
The following yeast strains were used: K. lactis CBS2359ku80 (MATα ku80::loxP; Kooistra et al. 2004) and its isogenic upc2Δ deletion mutant (Toth Hervay et al. 2020). For cultivation standard YPD medium (1% yeast extract and 2% peptone supplemented with carbon sources as needed, typically 2% glucose) was used. The solid media contained 2% Bacto agar.
Drug susceptibility assays
Overnight yeast cultures grown in YPD were diluted to a density of 1.0 × 108 cells/mL. Cell suspensions from tenfold dilutions were spotted onto agar plates containing the indicated concentrations of drugs in 5 μl aliquots. Growth at 28 °C was scored after 2 days. The drug concentrations used were as follows: CW, 14, 18, and 20 µg/mL; menadione, 10, 20, and 30 µg/mL; brefeldin A, 20 and 30 µg/mL; and H2O2, 5 and 6 mmol/L.
Quantitative real-time PCR
The levels of gene transcripts were assessed by quantitative real-time PCR as described previously (Konecna et al. 2016). For statistical analyses, the Student’s t-test was used and the level of statistical significance was set to P < 0.05. The primers used are listed in Table 1.
Glycerol assay
Cells grown in liquid YPD with or without 5 µg/mL of calcofluor white were processed as described previously (Konecna et al. 2018). Glycerol concentration in supernatants gained from 3 × 108 cells was estimated using the Free Glycerol Reagent Kit (Sigma- Aldrich). For statistical analyses, the Student’s t-test was used and the level of statistical significance was set to P < 0.05.
Results
KlUpc2p absence generates Rag − phenotype
The Klupc2Δ mutant showed slightly reduced growth in almost all conditions tested compared to its parental strain. However, growth of the Klupc2Δ mutant was further reduced when cultures were grown on YPD plates containing the respiratory inhibitor antimycin A (Fig. 1A). Inspired by the previous findings of Cialfi et al. (2011) and Micolonghi et al. (2012), we tested whether the addition of unsaturated fatty acids or ergosterol to the growth medium could restore the growth of Klupc2Δ mutant on glucose in the presence of antimycin A. As Fig. 1A shows, the presence of Tween 80, but not ergosterol, restored the growth of Klupc2Δ cells in glucose media containing antimycin A. This observation indicates a functional correlation between the defective growth and unsaturated fatty acid biosynthesis mediated by KlUpc2p. The KlUPC2 deletion leads to a distinct resistance of mutant cells to menadione (Fig. 1B). This observation corroborates the finding of Becerra et al. (2004), who observed a mild increase in mRNAs transcribed from K. lactis genes participating in the K. lactis rag2/Klpgi1 mutant (lacking phosphoglucose isomerase) oxidative stress defence.
Susceptibility of Kluyveromyces lactis cells to AA (supplemented with Tween 80 or ergosterol) (A); CW and menadione (B). Spotting assays were performed with a tenfold dilution of overnight culture grown on YPD medium containing the indicated compounds. The plates were incubated for 2 days at 28 °C. AA antimycin A, CW calcofluor white
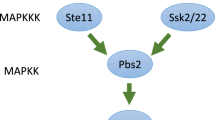
KlUPC2 gene deletion results in increased KlHOG1 expression
Figure 1B shows increased tolerance of mutant cells to calcofluor white as the result of KlUPC2 gene deletion. Foregoing research showed that the antifungal effect of calcofluor white is either the result of its binding to the cell wall chitin or depends on a functional HOG pathway (García-Rodriguez 2000; Roncero 2002). However, difference in the chitin content between the Klupc2Δ deletion mutant and the parental wild-type strain was not detected (data not shown). To find out whether the KlUPC2 gene deletion could influence the KlHOG1 transcription, the KlHOG1 expression was assayed in the mutant strain. Figure 2A shows an increase of mRNA transcribed from the KlHOG1 in the Klupc2Δ mutant cells compared with that in the parental strain. Following incubation of cells in the medium containing calcofluor white before the RNA isolation, the transcript levels of KlHOG1 gene further increased in both the mutant and wild-type strains (Fig. 2A). The Hog1p activation could lead to increased expression of GPD1 (glycerol-3-phosphate dehydrogenase) resulting in stimulation of de novo glycerol synthesis. Therefore, we determined the glycerol content in the parental strain and the Klupc2Δ mutant. As shown in Fig. 2B, the changes in glycerol content in the Klupc2Δ mutant were not statistically relevant as compared to that in the parental wild-type strain and this pattern was basically unaffected by the presence of calcofluor white. This finding was corroborated by estimating the levels of GPD1 gene transcripts in the Klupc2Δ mutant and parental cells, that were not significantly different (Fig. S1)
KlHOG1 gene expression (A) and glycerol content (B) in Klupc2Δ mutant and the corresponding wild-type strain. The gene transcript level in parental wild-type strain was set as 1. The values were normalized to the β-actin mRNA level. Glycerol concentration was determined as described in “Materials and methods.” Data represent the mean of three independent experiments (performed in triplicate). Error bars indicate the SDs. For statistical analyses, the Student’s t-test was used and the level of statistical significance was set to P < 0.05
KlUPC2 gene deletion results in increased KlMGA2 expression
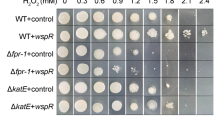
The cellular processes needed to combat redox stresses in yeast are different for different oxidants or reductants. The ability of Klupc2Δ mutant cells to grow in the presence of hydrogen peroxide and diamide was reduced compared to that of the parental wild-type strain (Fig. 3A). To analyze the possible relationship between KlUpc2p and KlMga2p, we determined the transcript levels of KlMGA2 gene in both the parental wild-type strain and the Klupc2Δ mutant. Figure 3B shows an increased expression of KlMGA2 gene in the Klupc2Δ mutant as compared to the parental wild-type strain. The transcript level of KlMGA2 gene in Klupc2Δ mutant cells, following the incubation of cells in the presence of hydrogen peroxide (1 mmol/L, for 1 h), was even higher (Fig. 3B). This result demonstrates the possibility that KlUpc2p prevents the induction of KlMGA2 expression in the presence of hydrogen peroxide.
KlUPC2 gene deletion results in increased susceptibility of Klupc2Δ mutant cells to hydrogen peroxide and diamide (A) and increased KlMGA2 gene expression (B) that is even higher in the presence of hydrogen peroxide, compared with that in the corresponding wild-type strain. The gene transcript level in parental wild-type strain was set as 1. The values were normalized to the β-actin mRNA level. For statistical analyses, the Student’s t-test was used and the level of statistical significance was set to P < 0.05
KlUpc2p controls vesicle trafficking within the Golgi
A possible explanation of Klupc2Δ drug sensitivity could be the defect in the ergosterol biosynthesis or troubles in transporting this lipid to the cell membrane. Figure 4A demonstrates increased susceptibility of Klupc2Δ cells to brefeldin A, an inhibitor of the guanine nucleotide exchange protein, necessary for Arf2p activation. Therefore, we decided to examine the KlARF2 expression in the parental wild-type strain and Klupc2Δ mutant cells. As Fig. 4B shows, the KlUPC2 gene deletion results in almost six fold increase in KlARF2 expression relative to that in the parental wild-type strain. The up-regulation of KlARF2 expression in the absence of KlUpc2p could be considered as a cellular strategy to compensate for its decrease in activity.
KlUPC2 gene deletion leads to increased susceptibility of mutant strain to brefeldin A (A) and marked increase in the KlARF2 gene expression (B). The gene transcript level in parental wild-type strain was set as 1. The values were normalized to the β-actin mRNA level. For statistical analyses, the Student’s t-test was used and the level of statistical significance was set to P < 0.05
Discussion
In this study, we further analyzed the impact of the KlUPC2 deletion in K. lactis. UPC2 gene in Saccharomycotina yeast species encodes the Zn2Cys6 transciption factor, member of the fungus specific family, that functions as the major regulator of ergosterol biosynthesis. Other factors beyond the paralogues Upc2p/Ecm22p, the heme-binding protein Hap1p, Mga2p, and the repressors Rox1p and Mot3p, coordinate ergosterol genes expression in S. cerevisiae (Davies and Rine 2006; reviewed in Jorda and Puig 2020). Recent reports point to a link between Pdr1p, the main transcription factor involved in the yeast S. cerevisiae and C. glabrata multidrug resistance, and Upc2p (Vu et al. 2019; Moye-Rowley 2020). Recently, we have shown that the KlUPC2 deletion increased the cell’s susceptibility to azole treatment. Reduced ergosterol content in the Klupc2Δ cells modulates their plasma membrane properties — the membrane is hyperpolarized and its fluidity decreased. Normal ergosterol homeostasis could be crucial for the proper regulation of plasma membrane ion transporters — the Ena1p Na+-ATPase or the Pma1p H+-ATPase in K. lactis (Toth Hervay et al. 2020). Here, we extend the published observations showing that deletion of KlUPC2 leads to a specific K. lactis Rag− phenotype, suggesting the link between ergosterol and carbohydrate metabolism in this yeast species. Coordination of carbohydrate response with lipid metabolism was also observed in C. parapsilosis, where sterol and carbohydrate biosynthesis are both regulated by Upc2p (Guida et al. 2011). We show that the presence of Tween 80 (a source of unsaturated fatty acids) in the growth medium suppresses the Rag− phenotype of the Klupc2Δ cells indicating a link between Upc2p and unsaturated fatty acid metabolism. Rag− phenotype in K. lactis was also generated as a result of KlMGA2 gene deletion (Micolonghi et al. 2012). KlMGA2 encodes the hypoxia responsive and lipid biosynthesis transcription factor involved also in glucose metabolism, cellular fitness, and light-stress response (Ottaviano et al. 2015; Santomartino et al. 2019; Camponeschi et al. 2021). The phenotypes generated by both the KlUPC2 and KlMGA2 gene deletions are restored by the addition of unsaturated fatty acids. In our previous paper, we showed decreased membrane fluidity of Klupc2Δ mutant. Lower fluidity of the membrane indicates lower Unsaturation Index (UI), due to a reduced content of unsaturated carbon–carbon bonds in fatty acid molecules. The decrease in the unsaturated fatty acid content can thus be compensated by the addition of Tween 80 to the growth media. The increased KlMGA2 expression observed as a result of KlUPC2 gene deletion could amend the KlupcΔ cells with the unsaturated fatty acids. Mga2p is constitutively expressed as inactive form bound to ER. Its activation requires appropriate stimuli (e.g., hypoxia), to induce ubiquitylation and the consequent proteolytic activation by the proteasome. Activation releases the transcriptional activator from the ER; the soluble Mga2p enters the nucleus where it activates gene expression through the stress responsive elements (STRE) in the promoters of regulated genes. Santomartino et al. (2019) showed stronger protection against ROS in Klmga2Δ cells. The increased expression of KlMGA2 gene observed in our Klupc2Δ mutant in the presence of hydrogen peroxide could be the result of inability to produce ergosterol in the Klupc2Δ mutant cells that is converted to an oxidative stress signal.
Along with the common antioxidant systems, one of the major factors contributing to the detoxification of ROS and the tolerance to oxidative stress in K. lactis is considered to be the ability to redirect the flux of metabolites from glycolysis to the pentose phosphate pathway providing the NADPH by the glucose-6-phosphate dehydrogenase. Such production of the reducing metabolite for the cellular redox systems is conserved between yeast and animals, underlining its importance in the process of oxidative stress adaptation (Grant 2008). The activation of stress signaling pathways as a result of KlUPC2 gene deletion is also in line with the increased KlHOG1 expression. However, we did not observed statistically relevant changes in glycerol content between the wild-type and the Klupc2Δ mutant strain. This is in compliance with the detected transcription level of KlGPD1 (glycerol-3-phosphate dehydrogenase). We propose that KlUPC2 gene deletion leads to activation of Hog pathway that is not functional. The observed increased sensitivity of the Klupc2Δ mutant to brefeldin A together with increased expression of KlARF2 gene points to the possible defect in membrane/protein trafficking in the Klupc2Δ mutant cells. Brefeldin A blocks Arf2p activation catalyzed by guanine nucleotide-exchange proteins. Arf2p, localized in the Golgi, controls intra-Golgi and Golgi-to-ER transport of cargo proteins by the formation of COPI carriers (Suda et al. 2018). Such trafficking could allow for changes in plasma membrane lipid composition, and strengthens the barrier function during stress condition (Babst 2020). K. lactis does not import exogenous sterols and relies mostly on the biosynthesis of endogenous ergosterol that has to be transported from ER to the plasma membrane (Snoek and Steensma 2006).
Promoter sequence analysis of the KlMGA2, KlHOG1, KlGPD1, and KlARF2 genes led to the identification of putative Stress response element (STRE), Yap1 recognition element (YRE) and Stb5p binding sites in all studied genes (Table S1). As both KlYap1p and KlStb5p are involved in oxidative stress response, it further points to the KlUpc2p involvement in oxidative stress response. Regulation of gene expression in response to the levels of oxygen involves several transcription factors. Similar to S. cerevisiae, the transcription factor KlHap1p mediates the induction of genes involved in respiration, lipid metabolism, and oxidative stress response. However, it seems to have lower number of target genes, some of which respond only under hypoxic conditions (Bao et al. 2008). Genes encoding enzymes of lipid metabolism are also expressed under hypoxic conditions in a KlMGA2-dependent manner (Micolonghi et al. 2012). Unlike in S. cerevisiae, KlRox1p is not involved in the hypoxic response in K. lactis, but rather contributes to metal ion resistance (Torres et al. 2012). Due to the crosstalk of different environmental signals, many aspects of regulation by these transcription factors are still unknown, but the coordination of the oxygen- responsive pathways at the transcriptional level efficiently maintains the requirement for acetyl-CoA in lipid biosynthesis (Burr and Espenshade 2018).
In our Klupc2Δ mutant, varying concentrations and types of sterols and a lack of ergosterol contributed to changes in the plasma membrane structure resulting in a decrease in plasma membrane fluidity. The disturbance in sterol content could be compensated by changes in the proportions of unsaturated fatty acids. Such plasticity in make-up of plasma membrane might increase the ability of cells to withstand of stress. We cannot exclude that the altered cellular sterol composition affects intracellular signaling and trafficking pathways, including the localization and activity of membrane proteins (Suchodolski and Krasowska 2019). As shown in Figs. 2A and S1, the KlUPC2 gene deletion leads to changes in the KlHOG1 but not in the KlGPD1 gene expression. This is in line with the work of Tanigawa et al. (2012), who observed the activation of HOG pathway as a result of the inhibition of sterol biosynthesis. Mojardín et al. (2018) proposed that KlGpd1p is part of a signaling pathway that governs transcriptional regulation to maintain the redox balance under changing environmental conditions. However, the exact molecular mechanism how the signal generated by the redox state is converted into the proper transcriptional response in the nucleus remains to be elucidated.
Altogether, Upc2p is a transcription regulator taking part in K. lactis ergosterol biosynthesis, carbon metabolism, intracellular signaling/trafficking pathways, and oxidative stress response.
References
Babst M (2020) Regulation of nutrient transporters by metabolic and environmental stresses. Curr Opin Cell Biol 65:35–41
Bao WG, Guiard B, Fang ZA et al (2008) Oxygen-dependent transcriptional regulator Hap1p limits glucose uptake by repressing the expression of the major glucose transporter gene RAG1 in Kluyveromyces lactis. Eukaryot Cell 7(11):1895–1905
Becerra M, Tarrio N, González-Siso MI et al (2004) Genome-wide analysis of Kluyveromyces lactis in wild-type and rag2 mtant strains. Genome 47:970–978
Breunig KD, Bolotin-Fukuhara M, Bianchi MM et al (2000) Regulation of primary carbon metabolism in Kluyveromyces lactis. Enzyme Microb Technol 26:771–780
Burr R, Espenshade PJ (2018) Oxygen-responsive transcriptional regulation of lipid homeostasis in fungi: Implications for anti-fungal drug development. Semin Cell Dev Biol 81:110–120
Bussereau F, Casaregola S, Lafay JF et al (2006) The Kluyveromyces lactis repertoire of transcriptional regulators. FEMS Yeast Res 6(3):325–335
Camponeschi I, Montanari A, Beccaccioli M et al (2021) Light-stress response mediated by the transcription factor KlMga2 in the yeast Kluyveromyces lactis. Front Microbiol 14(12):705012
Chen XJ, Clark-Walker GD (1999) α- and β-subunits of F1-ATPase are required for survival of petite mutants in Saccharomyces cerevisiae. Mol Gen Genet 262:898–908
Cialfi S, Uccelletti D, Carducci A et al (2011) KlHsl1 is a component of glycerol response pathways in the milk yeast Kluyveromyces lactis. Microbiology 157:1509–1518
Davies BSJ, Rine J (2006) A role for sterol levels in oxygen sensing in Saccharomyces cerevisiae. Genetics 174(1):191–201
García-Rodriguez LJ, Duran A, Roncero C (2000) Calcofluor antifungal action depends on chitin and a functional high-osmolarity glycerol response (HOG) pathway: evidence for a physiological role of the Saccharomyces cerevisiae HOG pathway under noninducing conditions. J Bacteriol 182(9):2428–2437
González-Siso MI, Garcia-Leiro A, Tarrio N et al (2009) Sugar metabolism, redox balance and oxidative stress response in the respiratory yeast Kluyveromyces lactis. Microb Cell Factories 8:1–17
Grant CM (2008) Metabolic reconfiguration is a regulated response to oxidative stress. J Biol 7:1–4
Guida A, Lindstadt C, Maguire SL et al (2011) Using RNA-seq to determine the transcriptional landscape and the hypoxic response of the pathogenic yeast Candida parapsilosis. BMC Genom 12:628
Jorda T, Puig S (2020) Regulation of ergosterol biosynthesis in Saccharomyces cerevisiae. Genes 11(7):795
Kiers J, Zeeman AM, Luttik M et al (1998) Regulation of alcoholic fermentation in batch and chemostat cultures of Kluyveromyces lactis CBS 2359. Yeast 14:459–469
Konecna A, Toth Hervay N, Bencova A et al (2018) Erg6 gene is essential for stress adaptation in Kluyveromyces lactis. FEMS Microbiol Lett. https://doi.org/10.1093/femsle/fny265
Konecna A, Toth Hervay N, Valachovic M et al (2016) ERG6 gene deletion modifies Kluyveromyces lactis susceptibility to various growth inhibitors. Yeast 33(12):621–632
Kooistra R, Hooykaas PJ, Steensma HY (2004) Efficient gene targeting in Kluyveromyces lactis. Yeast 21:781–792
Labbaoui H, Bogliolo S, Ghugtyal V et al (2017) Role of Arf GTPases in fungal morphogenesis and virulence. PLoS Pathog 13(2):e1006205
MacPherson S, Larochelle M, Turcotte B et al (2006) A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol Mol Biol Rev 70(3):583–604
Micolonghi C, Ottaviano D, Silvio ED et al (2012) A dual signalling pathway for the hypoxic expression of lipid genes, dependent on the glucose sensor Rag4, is revealed by the analysis of the KlMGA2 gene in Kluyveromyces lactis. Microbiology 158:1734–1744
Mojardín L, Vega M, Moreno F et al (2018) Lack of the NAD+-dependent glycerol 3-phosphate dehydrogenase impairs the function of transcription factors Sip4 and Cat8 required for ethanol utilization in Kluyveromyces lactis. Fungal Genet Biol 111:16–29
Moye-Rowley WS (2020) Linkage between genes involved in azole resistance and ergosterol biosynthesis. PLoS Pathog 16(9):e1008819
Ottaviano D, Montanari A, DeAngelis L et al (2015) Unsaturated fatty acids-dependent linkage between respiration and fermentation revealed by deletion of hypoxic regulatory KlMGA2 gene in the facultative anaerobe-respiratory yeast Kluyveromyces lactis. FEMS Yeast Res 15:fov028. https://doi.org/10.1093/femsyr/fov028
Rodicio R, Heinisch JJ (2013) Yeast on the milky way: genetics, physiology and biotechnology of Kluyveromyces lactis. Yeast 30:165–177
Rolland S, Hnatova M, Lamaire M et al (2006) Connection between the Rag4 glucose sensor and the KlRgt1 repressor in Kluyveromyces lactis. Genetics 174:617–626
Roncero C (2002) The genetic complexity of chitin synthesis in fungi. Curr Genet 41:367–378
Santomartino R, Camponeschi I, Polo G et al (2019) The hypoxic transcription factor KlMga2 mediates the response to oxidative stress and influences longevity in the yeast Kluyveromyces lactis. FEMS Yeast Res 19(3):foz020. https://doi.org/10.1093/femsyr/foz020
Snoek IS, Steensma HY (2006) Why does Kluyveromyces lactis not grow under anaerobic conditions? Comparison of essential anaerobic genes of Saccharomyces cerevisiae with the Kluyveromyces lactis genome. FEMS Yeast Res 6:393–403
Suchodolski J, Krasowska A (2019) Plasma membrane potential of Candida albicans measured by Di-4-ANEPPS fluorescence depends on growth phase and regulatory factors. Microorganisms 7(4):110
Suda Y, Kurokawa K, Nakano A (2018) Regulation of ER-Golgi transport dynamics by GTPases in budding yeast. Front Cell Dev Biol 5:122
Tanigawa M, Kihara A, Terashima M et al (2012) Sphingolipids regulate the yeast high osmolarity glycerol response pathway. Mol Cell Biol 32:2861–2870
Torres AM, Maceiras ML, Belmonte ER et al (2012) KlRox1p contributes to yeast resistance to metals and is necessary for KlYCF1 expression in the presence of cadmium. Gene 497(1):27–37
Toth Hervay N, Bencova A, Valachovic M et al (2020) UPC2 gene deletion modifies sterol homeostasis and susceptibility to metabolic inhibitors in Kluyveromyces lactis. Yeast 37:647–657
Vik A, Rine J (2001) Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol Cell Biol 21:6395–6405
Vu BG, Thomas GH, Moye-Rowley WS (2019) Evidence that ergosterol biosynthesis modulates activity of the Pdr1 transcription factor in Candida glabrata. Mbio 10(3):e00934-e1019
Wilcox LJ, Balderes DA, Wharton B et al (2002) Transcriptional profiling identifies two membrs of the ATP-binding cassette transporter superfamily required for sterol uptake in yeast. J Biol Chem 277:32466–32472
Funding
The authors received financial support from the Slovak Grant Agency of Science (VEGA 1/0697/18, VEGA 1/0388/22), The Slovak Research and Developmental Agency (APVV-19–0094), and Comenius University (UK/62/2021).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12223_2022_968_MOESM1_ESM.jpg
Supplementary file1 (JPG 622 kb). Fig. S1 KlGPD1 gene expression in wild-type and corresponding Klupc2Δ mutant strain. For statistical analyses, the Student’s t-test was used and the level of statistical significance was set to P < 0.05
Rights and permissions
About this article
Cite this article
Betinova, V., Toth Hervay, N., Elias, D. et al. The UPC2 gene in Kluyveromyces lactis stress adaptation. Folia Microbiol 67, 641–647 (2022). https://doi.org/10.1007/s12223-022-00968-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-022-00968-3