Abstract
The Candida haemulonii complex (Candida haemulonii, Candida haemulonii var. vulnera, and Candida duobushaemulonii) comprises emerging opportunistic human fungal pathogens with recognized multidrug-resistance profiles. Little is known about the virulence markers produced by this fungal complex. However, it is recognized that Candida spp. express a large array of peptidases, which play multiple roles in different aspects of fungal-host interactions. In the present study, we have identified proteolytic enzymes in clinical isolates of the C. haemulonii complex using zymographic assays. Peptidases able to hydrolyze gelatin, casein, albumin, hemoglobin, and immunoglobulin G were detected in cell-free supernatants and cellular extracts taken from the three species forming the C. haemulonii complex. Overall, peptidases were preferentially evidenced at physiological pH and temperatures of 37–42 °C, with molar masses between 35 and 85 kDa. Peptidase profiles of C. haemulonii and C. haemulonii var. vulnera isolates were quite similar, contrasting to the peptidases produced by C. duobushaemulonii. Almost all peptidases were inhibited by phenylmethanesulfonyl fluoride (PMSF), thus classifying them as serine-type peptidases. Additionally, proteolytic cleavage of soluble azoalbumin was blocked by PMSF (65–95% inhibition depending on the fungal isolate). These unprecedented results have demonstrated the capability of the C. haemulonii complex to produce serine-type peptidases with an ability to cleave a broad spectrum of proteins, including key host components.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Candidiasis represents the most important opportunistic mycosis in hospital settings and is one of the main causes of human infectious diseases worldwide. Although Candida albicans remains the most notorious species responsible for candidiasis, non-albicans Candida species, with reduced susceptibility to antifungal agents commonly used in the medical practice, have represented, in recent years, a substantial part of the clinical isolates collected in hospitals around the globe (Chen et al. 2009; Nucci et al. 2010; Pfaller et al. 2010; Papon et al. 2013; Sardi et al. 2013; Caggiano et al. 2015; Sehnaz et al. 2015).
Candida haemulonii was originally described in 1962 as Torulopsis haemulonii and was isolated from the gut of a fish (Haemulon sciurus) (van and Kolipinski 1962). Lavarde et al. (1984) reported the first clinical isolate of C. haemulonii which was obtained from the blood of a patient with renal failure. Since then, various kinds of infections associated with this yeast have been described, mainly in immunocompromised patients (Gargeya et al. 1991; Rodero et al. 2002; Khan et al. 2007; Ruan et al. 2010; Crouzet et al. 2011; Cendejas-Bueno et al. 2012; Ramos et al. 2015). Due to the similarities in the physiological characteristics, isoenzyme profiles, and DNA re-association experiments, Lehmann et al. (1993) described, for the first time, the C. haemulonii complex. In that study, 25 isolates from different geographic origins and clinical sources were investigated, being described as two genetically distinct groups: C. haemulonii group I and C. haemulonii group II. Subsequently, Cendejas-Bueno et al. (2012) suggested the following new classification of this fungal complex, taking into consideration both molecular and phenotypic approaches: C. haemulonii (formerly known as C. haemulonii group I), C. duobushaemulonii (formerly known as C. haemulonii group II), and C. haemulonii var. vulnera (a variety of Candida haemulonii). Undoubtedly, among the most striking features of the C. haemulonii species complex are its resistance to amphotericin B, its resistance/low susceptibility to azoles (e.g., fluconazole, itraconazole, and voriconazole), and its limited susceptibility to echinocandins (e.g., caspofungin). Indeed, it is this multidrug-resistance profile that has hindered the treatment of patients with deep infections and has increased the frequency of clinical failures followed by death (Rodero et al. 2002; Khan et al. 2007; Kim et al. 2009; Crouzet et al. 2011; Cendejas-Bueno et al. 2012; Li et al. 2015; Ramos et al. 2015).
Despite the growing importance of C. haemulonii complex infections in medicine, its interactions with the infected host are still poorly understood. However, it is well-known that fungi have the ability to digest polymeric compounds found in their environment into smaller components, which are easily absorbed by cells and utilized as carbon and nitrogen sources (Mahon et al. 2009). This characteristic is reflected in the high levels of activity of the hydrolytic enzymes found in cell extracts and conditioned culture supernatants of many of these microorganisms (Mahon et al. 2009). Recently, our research group described the production of different classes of hydrolytic enzymes/activities by species forming the C. haemulonii complex, which included aspartic peptidase, caseinolytic and hemolytic activities, phytase, esterase, and phospholipase (Ramos et al. 2017a). For instance, in that work, aspartic peptidase activity was detected (using agar plate methodology) in all of the clinical isolates of the C. haemulonii complex. In general, C. duobushaemulonii isolates were classified as “excellent” producers (Pz values between 0.399 and 0.100), while C. haemulonii and C. haemulonii var. vulnera isolates were categorized as “good” producers (Pz values 0.699–0.400) (Ramos et al. 2017a).
Microbial peptidases play physiological and pathological roles, for example, during nutrition, growth, differentiation, signaling, colonization, invasion, intracellular survival, evasion, dissemination, and immunomodulation of host defenses (Santos 2011). In fungi, several studies have indicated that the aspartic-, serine-, and metallo-type endopeptidases, as well as aminopeptidases, carboxypeptidases, and dipeptidylpeptidases, perform crucial biological roles and may also have important virulence attributes (Segal 2006; Yike 2011; Bochenska et al. 2013; Silva et al. 2014). From our present study, involving nine Brazilian clinical isolates, we herein report that secreted and cell-associated serine-type peptidases of the C. haemulonii species complex are able to cleave different key host proteins. This work is an important contribution to the understanding of the virulence factors linked to this fungal complex.
Materials and methods
Chemicals
Reagents used in electrophoresis, buffer components, peptidase inhibitors (trans-epoxysuccinyl-l-leucylamido(4-guanidino)butane (E-64), phenylmethanesulfonyl fluoride (PMSF), 1,10-phenanthroline, and pepstatin A), gelatin, casein, bovine serum albumin (BSA), human serum albumin (HSA), hemoglobin (Hgb), human immunoglobulin G (IgG), azoalbumin, 2,3-bis (2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino) carbonyl]-2H-tetrazolium hydroxide (XTT), and propidium iodide were obtained from Sigma-Aldrich (St Louis, USA). All other reagents were of analytical grade.
Microorganisms and growth conditions
Three Brazilian clinical isolates of each member forming the C. haemulonii complex were used in all of the experiments performed in the present study: C. haemulonii (LIPCh2 from sole of the foot, LIPCh7 from toe nail, and LIPCh12 from blood), C. duobushaemulonii (LIPCh6 from toe nail, LIPCh8 from blood, and LIPCh10 from bronchoalveolar lavage), and C. haemulonii var. vulnera (LIPCh5 from toe nail, LIPCh9 from urine, and LIPCh11 from blood) (Ramos et al. 2015). The fungal isolates were previously identified by the automatized system VITEK® 2 and characterized by ITS1–5.8S-ITS2 gene sequencing (Ramos et al. 2015). The yeasts (inoculum of 106 cells per mL) were cultured on Sabouraud broth at 37 °C for 48 h in an orbital incubator shaker (200 rpm) (Ramos et al. 2017b). All of the clinical isolates studied herein have reached their exponential phase at 48 h of in vitro growth on Sabouraud liquid medium as previously published by our research group (Ramos et al. 2017b).
Procurement of culture supernatants and cellular extracts
The fungal cultures (400 mL) were harvested by centrifugation (4000g, 10 min, 4 °C), and the supernatants were filtered through a 0.22-μm membrane (Millipore, São Paulo, Brazil). The cell-free culture supernatants were concentrated 100-fold in an Amicon ultrafiltration system (Amicon®, Beverly, USA) using a 10-kDa membrane (Santos and Soares 2005). The same volume of Sabouraud medium was also concentrated and used as a control to check for possible peptidase activity. In parallel, the number of fungal cells was counted using a Neubauer chamber, and 108 fungi were washed three times with phosphate buffered saline (PBS; 10 mmol/L NaH2PO4, 10 mmol/L Na2HPO4, 150 mmol/L NaCl; pH 7.2), suspended in 2.5% Triton X-100, and distributed in Eppendorf tubes containing glass beads (500 μm). Cellular lysis was performed in a cell homogenizer (FastPrep) by 5 cycles of 30 s, alternating with an ice bath for 2 min. Then, the mixtures were centrifuged at 10,000g for 10 min at 4 °C, and the supernatants were collected. Protein concentration in both cellular and extracellular extracts was determined by the method described by Lowry et al. (1951) using BSA as the standard.
Viability tests
The ability of fungal cells to survive under growth conditions (37 °C for 48 h on Sabouraud medium) was assessed by measuring (i) mitochondrial dehydrogenase activity using a colorimetric assay that quantifies the metabolic reduction of XTT to a water-soluble, brown formazan product and (ii) propidium iodide uptake by damaged fungal cells due to a loss in cell membrane integrity. In these experiments, fungi were treated with either 0.4% paraformaldehyde or sodium azide (0.95 g/L) for 30 min in order to obtain non-viable cells to use as a negative control in the viability tests.
Zymographic assay
Peptidases were detected and characterized by electrophoresis on 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) containing 0.1% gelatin, casein, BSA, HSA, Hgb, or IgG as proteinaceous substrates incorporated into the gels (Heussen and Dowdle 1980). The gels were loaded with 10–45 μL of cell-free supernatants (varying from 400 to 1400 μg of proteins) and cellular extracts (varying from 40 to 140 μg of proteins), following electrophoresis at a constant potential of 200 V at 4 °C for 2 h. Subsequently, the gels were incubated in 2.5% Triton X-100 for 1 h at room temperature under constant agitation in order to permit the enzymes’ renaturation. The gels were incubated for 48 h under different conditions. The effect of pH on the peptidase activity was assessed by incubating the gels at 37 °C in 50 mmol/L sodium citrate (pH 5.0), 50 mmol/L sodium phosphate (pH 7.0), and 50 mmol/L glycine-NaOH (pH 9.0) buffers. The influence of temperature was assessed by incubating the gels at 28, 37, and 42 °C in 50 mmol/L sodium phosphate buffer (pH 7.0). The effect of peptidase inhibitors was also assessed by incubating the gels at 37 °C in 50 mmol/L sodium phosphate buffer (pH 7.0) in the absence (control) or in the presence of peptidase inhibitors able to block different peptidase classes: 10 μmol/L pepstatin A (aspartic peptidase inhibitor), 10 μmol/L E-64 (cysteine peptidase inhibitor), 10 mmol/L 1,10-phenanthroline (metallopeptidase inhibitor), and 10 mmol/L PMSF (serine peptidase inhibitor). The gels were then stained overnight with 0.2% Coomassie brilliant blue R-250 in methanol-acetic acid-water (50:10:40) and destained in methanol-acetic acid-water (5:10:85), to intensify the digestion halos. The peptidase activities in the gels were revealed by the presence of colorless zones indicative of protein digestion. The gels were dried, scanned, and digitally processed (Santos and Soares 2005). The molar mass of the peptidases was calculated by comparison with the mobility of Pierce™ Prestained Protein MW Marker (Thermo Fisher Scientific, São Paulo, Brazil).
Dendrogram analysis
Peptidase patterns observed in both cell-free culture supernatants and cellular extracts by zymographic assays with gelatin, casein, BSA, HSA, Hgb, and IgG were used to construct a dendrogram through the unweighted pair group method analysis (UPGMA). For these analyses, the PAST3 software package (Hammer et al. 2001) was applied. The results of peptidases’ fingerprinting were collected into a matrix indicating the presence or absence (scored as 1 or 0, respectively) of specific peptidase bands in each zymographic analysis.
Quantitative peptidase activity assay
The peptidase activity was quantified using the method described by Plantner (1991). Briefly, the cell-free culture supernatants and cellular extracts (400 μg and 100 μg of protein, respectively) were incubated with azoalbumin (1.6 mg/mL) in 10 mmol/L sodium phosphate (pH 7.0), for 2 h at 37 °C, in the absence (control) or in the presence of 10 mmol/L PMSF. Following incubation, the reaction was terminated by adding 40 μL of 50% trichloroacetic acid, and samples were kept on ice for 1 h. The precipitate formed in each reaction was then removed by centrifugation at 500g for 15 min, and the fluid phase (200 μL) was added to 15 μL of sodium hydroxide (10 mmol/L) in a 96-well plate. For the background of the tests, only azoalbumin and sodium phosphate buffer were incubated at 37 °C for 2 h. In this case, the reaction was stopped with trichloroacetic acid prior to the addition of the fungal samples. The absorbance was measured at 440 nm in a microplate reader (SpectraMax M3; Molecular Devices, Sunnyvale, USA), and the specific peptidase activity was expressed in arbitrary units (U) per milligram of protein, where 1 unit of enzymatic activity is equivalent to the variation in optical density of 0.001 nm per min at 440 nm.
Statistical analysis
All the experiments were performed in triplicate, in three independent experimental sets, and data were expressed as mean ± standard deviation (SD). Data were analyzed by Student’s t test using GraphPad Prism 5.0 software. P values less than or equal to 0.05 were considered statistically significant.
Results
Detection of peptidases in clinical isolates of the C. haemulonii complex
For all experimental sets, the fungal cells were obtained after 48 h of in vitro growth on Sabouraud medium, which correspond to cells at exponential growth phase. These cells were metabolically active as judged by the capability of their mitochondrial dehydrogenases in reducing the XTT salt to formazan and also by the lack of incorporation of a considerable amount of propidium iodide (< 10%) into the fungal cells (assessed using flow cytometry) (data not shown). Both of these viability assays attested that the fungi were viable along the experiments, suggesting that the extracellular contents were not arising from the lysis of damaged/dead cells.
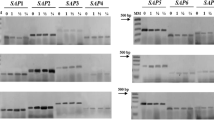
Peptidase profiles of both cell-free culture supernatants (Fig. 1a; Table 1) and cellular extracts (Fig. 1b; Table 1) from nine clinical isolates forming the C. haemulonii complex were initially evaluated by SDS-PAGE containing different protein substrates (gelatin, casein, BSA, HSA, IgG, or Hgb) copolymerized into the gel matrix. Overall, peptidase activity was detected in all the studied fungal isolates, except for the supernatant obtained from LIPCh11 in which no peptidase activity was detected regardless of the substrate evaluated. Two gelatinases, with apparent molar masses of 50 and 60 kDa, were detected in supernatants and cellular extracts of the isolates belonging to the C. haemulonii and C. haemulonii var. vulnera (Fig. 1a, b; Table 1). Furthermore, peptidases of 35 and 40 kDa, which were visualized in both extracellular and cellular samples, showed broad hydrolytic capabilities, degrading the remaining test proteinaceous substrates (casein, BSA, HSA, IgG, and Hgb). Besides that, a cell-associated 65-kDa peptidase, able to degrade only IgG, and a 70-kDa peptidase, able to degrade casein, BSA, and HSA, evidenced in both supernatants and cellular extracts, were also detected in the C. haemulonii and C. haemulonii var. vulnera isolates (Fig. 1a, b; Table 1). In contrast, a great variability on the production of either cell-associated or secreted peptidases was clearly evident in the clinical isolates of C. duobushaemulonii, showing a typical isolate-dependent peptidase profile (Fig. 1a, b; Table 1). Interestingly, a good correlation between the peptidase profiles of C. haemulonii and C. haemulonii var. vulnera was observed in supernatants and in cellular extracts. However, these profiles differed significantly from those found for C. duobushaemulonii in both of the evaluated samples (Fig. 1a, b; Table 1).
Peptidase profiles detected in culture supernatants and cell extracts from clinical isolates of the Candida haemulonii complex. a Analysis of the peptidase activity profiles in culture supernatants and in b cell extracts of C. haemulonii (isolates LIPCh2, LIPCh7, and LIPCh12), C. haemulonii var. vulnera (LIPCh5, LIPCh9, and LIPCh11), and C. duobushaemulonii (LIPCh6, LIPCh8, and LIPCh10) after electrophoresis on 12.5% SDS–PAGE with 0.1% gelatin, casein, BSA, HSA, Hgb, and IgG as substrates incorporated into the gel matrix. Gel strips containing the samples were incubated in 10 mmol/L sodium phosphate buffer, pH 7.0, for 48 h at 37 °C. The apparent molecular mass (MM) of the detected peptidases is expressed in kDa. The numbers on the top of each lane correspond to the code of the fungal isolates: 2, LIPCh2; 7, LIPCh7; 12, LIPCh12; 5, LIPCh5; 9, LIPCh9; 11, LIPCh11; 6, LIPCh6; 8, LIPCh8; and 10, LIPCh10
Subsequently, a similarity analysis (based on UPGMA) was applied using all peptidase bands detected in both cell-free culture supernatants and cellular extracts (visualized by the zymographic assay) as markers (Fig. 2). At about 34% of similarity, two major groups were clearly separated for the C. haemulonii species complex: one formed by C. haemulonii and C. haemulonii var. vulnera isolates and the other one formed by C. duobushaemulonii isolates (Fig. 2).
Effects of pHs, temperatures, and inhibitors on the peptidases produced by the C. haemulonii complex
Based on the zymographic profiles (Fig. 1) and dendrogram analysis (Fig. 2), the supernatants and cellular extracts, with different proteolytic bands on each test substrate, were selected to further investigate their biochemical properties.
pH
In general, most of the peptidases detected in the fungal supernatants (Fig. 3a) and in the cellular extracts (Fig. 3b) showed activity at all of the evaluated pH values, presenting the highest hydrolytic capability at pH 7.0, when gels were incubated at 37 °C for 48 h. However, some peptidases detected in the cellular extract of LIPCh8 with activity over BSA, HSA (40, 70, and 80 kDa), and casein (50 kDa) appeared to be most active at pH 5.0 (Fig. 3).
Effects of pH on the peptidase activities detected in culture supernatants and cell extracts from clinical isolates of the Candida haemulonii complex. In this set of experiments, the clinical isolates presenting different peptidase profiles were selected. a Analysis of the peptidase activity profiles in culture supernatants and in b cell extracts of the C. haemulonii complex after electrophoresis on 12.5% SDS–PAGE with 0.1% gelatin, casein, BSA, HSA, Hgb, and IgG as substrates incorporated into the gels. Gel strips containing the samples were incubated in 50 mmol/L sodium citrate buffer, pH 5.0, 50 mmol/L sodium phosphate, pH 7.0, and 50 mmol/L glycine-NaOH, pH 9.0, for 48 h at 37 °C. The apparent molecular mass (MM) of the detected peptidases is expressed in kDa. The numbers on the left margins of the pictures correspond to the code of the fungal isolates. 2, LIPCh2 (C. haemulonii); 8, LIPCh8; and 10, LIPCh10 (C. duobushaemulonii)
Temperature
The vast majority of the peptidases detected in the fungal supernatants (Fig. 4b) and in the cellular extracts (Fig. 4b) showed activity at all of the test temperatures, with the highest cleavages equally occurring at 37 and 42 °C, after 48 h at neutral pH (Fig. 4).
Effects of temperature on the peptidase activities in culture supernatants and in cell extracts from clinical isolates of the Candida haemulonii complex. In this set of experiments, the clinical isolates presenting different peptidase profiles were selected. a Analysis of the peptidase activity profiles in culture supernatants and in b cell extracts of the C. haemulonii complex after electrophoresis on 12.5% SDS–PAGE with 0.1% gelatin, casein, BSA, HSA, Hgb, and IgG as substrates incorporated into the gels. Gel strips containing the samples were incubated in 10 mmol/L sodium phosphate buffer, pH 7.0, for 48 h, at 28, 37, or 42 °C. The apparent molecular mass (MM) of the detected peptidases is expressed in kDa. The numbers on the left margins of the pictures correspond to the code of the fungal isolates. 2, LIPCh2 (C. haemulonii); 8, LIPCh8; and 10, LIPCh10 (C. duobushaemulonii)
Peptidase inhibitors
Overall, PMSF (at 10 mmol/L) partially or fully inhibited the enzymatic activities of all the peptidases detected in the fungal supernatants (Fig. 5a) and in the cellular extracts (Fig. 5b) of the C. haemulonii species complex when incubated for 48 h at pH 7.0 and 37 °C. Additionally, 1,10-phenanthroline (10 mmol/L) was also able to inhibit the enzymatic activity of some secreted and cell-associated peptidases with activity over BSA and IgG (Fig. 5a, b). Both pepstatin A (10 μmol/L) and E-64 (10 μmol/L) did not interfere with the peptidase activities observed in all the evaluated systems (Fig. 5).
Effect of inhibitors on the peptidase activities in culture supernatants and cell extracts from clinical isolates of the Candida haemulonii complex. In this set of experiments, the clinical isolates presenting different peptidase profiles were selected. a Analysis of the peptidase activity profiles in culture supernatants and in b cell extracts of the C. haemulonii complex after electrophoresis on 12.5% SDS–PAGE with 0.1% gelatin, casein, BSA, HSA, Hgb, and IgG as substrates incorporated into the gels. Gel strips containing the samples were incubated in 50 mmol/L sodium phosphate buffer, pH 7.0, for 48 h at 37 °C, in the absence (ctrl) or in the presence of 10 mmol/L PMSF, 10 mmol/L 1,10-phenanthroline (Phen), 10 μmol/L E-64, or 10 μmol/L pepstatin (Peps). The apparent molecular mass (MM) of the detected peptidases is expressed in kDa. The numbers on the left margins of the pictures correspond to the code of the fungal isolates. 2, LIPCh2 (C. haemulonii); 8, LIPCh8; and 10, LIPCh10 (C. duobushaemulonii)
Quantification of peptidase activity
To varying extents, peptidase activity was quantified in all of the analyzed samples (supernatants and cellular extracts) using azoalbumin as soluble substrate (Fig. 6). Regarding the clinical isolates of each member of this fungal complex, differences in peptidase activities were detected in supernatants (≈ 4–9 U/mg for C. haemulonii; 3–6 U/mg for C. haemulonii var. vulnera; and 6–9 U/mg for C. duobushaemulonii) (Fig. 6a) and in cellular extracts (≈ 7–16 U/mg for C. haemulonii; 11–15 U/mg for C. haemulonii var. vulnera; and 7–10 U/mg for C. duobushaemulonii) (Fig. 6b). However, no significant differences (P > 0.05) were detected considering the mean of peptidase activity among the species forming the C. haemulonii complex (Fig. 6a, b, insert). In parallel, PMSF (at 10 mmol/L) significantly inhibited the peptidase activity in all of the supernatants (≈ 79–87% for C. haemulonii; 75–84% for C. haemulonii var. vulnera; and 63–72% for C. duobushaemulonii) and in the cellular extracts (≈ 64–69% for C. haemulonii; 77–96% for C. haemulonii var. vulnera; and 75–80% for C. duobushaemulonii) from the C. haemulonii complex (Fig. 6a, b).
Peptidase activity measured in the culture supernatants and in cell extracts from clinical isolates of the Candida haemulonii complex. a Measurement of peptidase activity in culture supernatants and in b cell extracts of C. haemulonii (Ch; isolates LIPCh2, LIPCh7, and LIPCh12), C. haemulonii var. vulnera (Chv; LIPCh5, LIPCh9, and LIPCh11), and C. duobushaemulonii (Cd; LIPCh6, LIPCh8, and LIPCh10) using azoalbumin (1.6 mg/mL) as the protein substrate in 50 mmol/L sodium phosphate (pH 7.0), for 2 h at 37 °C, in the absence or in the presence of 10 mmol/L PMSF. The peptidase activity is expressed in arbitrary units (U) per milligram of protein, where one unit of activity is equivalent to the variation in optical density of 0.001 nm per min at 440 nm. The bars represent the mean ± SD from at least three independent experiments. The asterisks indicate significant difference in the peptidase activity considering the isolates treated (white bars) or not (black bars) with PMSF (P < 0.05). Inserts represent the mean of peptidase activity (in the absence of PMSF) among the isolates of each member of the C. haemulonii complex. The numbers on the bottom of each bar correspond to the code of the fungal isolates: 2, LIPCh2; 7, LIPCh7; 12, LIPCh12; 5, LIPCh5; 9, LIPCh9; 11, LIPCh11; 6, LIPCh6; 8, LIPCh8; and 10, LIPCh10
Discussion
The difficulties encountered in the treatment of infections caused by the C. haemulonii species complex have driven the search for new drugs and novel therapeutic targets. In this context, peptidases are involved in different interactive stages between fungi and hosts, being considered as potential virulence attributes (Hube 2000), and it is the fact which has prompted us to investigate the potential peptidases’ arsenal produced by the C. haemulonii complex.
Candida haemulonii and C. haemulonii var. vulnera isolates produced two secreted/cell-associated peptidases (50 and 60 kDa) that hydrolyze gelatin. Zymography also revealed peptidases in the supernatants and in the cellular extracts of C. haemulonii and C. haemulonii var. vulnera having different hydrolytic capacities, being able to cleave casein, BSA, HSA, IgG, and Hgb (35 and 40 kDa), casein, BSA, and HSA (70 kDa) and only IgG (65 kDa). Conversely, a wide variety of secreted and cell-associated peptidases with the ability to degrade all of the abovementioned substrates were detected in C. duobushaemulonii isolates. Gelatin and casein were used as substrates because they are easily hydrolyzed by various peptidases (Rao et al. 1998). Besides that, gelatin is a denatured form of collagen, itself a known constituent of the extracellular matrix of the connective tissue of animals. In Candida spp., it has been suggested that the ability of peptidases to degrade extracellular matrix proteins may be related to their role in removing host barriers during the in vivo infection process, providing nutrients for growth and favoring the penetration/colonization of host tissue (Hube 2000). The hydrolysis of Hgb (main protein in red blood cells) and albumin (a carrier protein found in high amounts in serum) by Candida spp. has been associated with the uptake of nutrients (iron and amino acids) during mammalian infection (Moors et al. 1992; Chaffin et al. 1998; Hube 2000; Santos and Soares 2005; de Melo et al. 2007; Noble 2013; Ramachandra et al. 2014). The IgG molecules are known to play important roles in humoral host defense mechanisms against infections caused by Candida spp.; however, their degradation by fungal peptidases could result in its immunological escape (Casadevall 1995; Santos et al. 2006).
The peptidase profiles observed in C. haemulonii and C. haemulonii var. vulnera were identical for either supernatants or cellular extracts. Contrarily, differences in the proteolytic profiles were clearly observed among the isolates of C. duobushaemulonii, as well as between them and the isolates of C. haemulonii and C. haemulonii var. vulnera. These data reflect the genetic similarity existing between the microbial members of the complex, being most striking between C. haemulonii and C. haemulonii var. vulnera (Cendejas-Bueno et al. 2012). Additionally, the quantitative evaluation of peptidase activities indicated differences between the isolates of each member of the C. haemulonii complex; however, the mean proteolytic activities were not different among the species forming this fungal complex.
PMSF blocked the activity of almost all of the peptidases produced by C. haemulonii complex, thus classifying them as serine-type peptidases. Fungal serine peptidases constitute an important group of intra- and extracellular peptidases with both regulatory and nutritional roles (Hube 2000; Monod et al. 2002), which present broad substrate specificity and hydrolytic activity at neutral and alkaline pH (de Souza et al. 2015). Corroborating these premises, the serine peptidases of the C. haemulonii complex showed activity across a wide range of pH and temperature. Moreover, the pH of maximum activity (7.0) is close to the pH range of the sole of the foot (pH 5.0–7.82) (Marshall et al. 1987), blood (pH 7.35–7.45) (de Melo et al. 2007), and bronchoalveolar lavage (pH 5.96–6.58) (Lozo Vukovac et al. 2014), sources from which the fungal isolates were recovered. Regarding temperature, the range of maximum peptidase activity observed (37–42 °C) suggests the effectiveness of these enzymes during the infection in an individual with normal body temperature (36.5–37 °C) or someone in feverish condition (> 37 °C).
Several studies have suggested the importance of serine peptidases as virulence factors in human pathogenic fungi (Jousson et al. 2004; Behnsen et al. 2010). Rodier et al. (1994) described two main cellular serine peptidases of 50 and 60 kDa found in C. albicans samples recovered from urine, feces, vagina, and mouth, which degraded gelatin at 37 °C and over a wide pH range (5.0–8.0). Ito et al. (2010) reported the presence of a 64-kDa, PMSF-sensitive serine peptidase in the cellular extract of C. glabrata and which had optimum enzymatic activity over casein at pH 7.0 and 37 °C. Secreted serine peptidases have also been studied in Candida genus. Our group reported, for the first time, the presence of extracellular acidic serine peptidases (30–58 kDa) in oral clinical isolates of C. albicans recovered from HIV-positive and healthy children (de Brito Costa et al. 2003). Santos and Soares (2005) identified a 50-kDa serine peptidase secreted by C. guilliermondii isolated from bronchoalveolar lavage which was able to hydrolyze relevant human proteins, including HSA, IgG, fibronectin, and laminin, at 37 °C and physiological pH. A 50-kDa serine peptidase was also identified in the supernatant of an isolate of C. albicans recovered from urine, which was active within a wide pH range (5.0–7.2) at 37 °C, and was able to hydrolyze human serum proteins and extracellular matrix components (Santos and Soares 2005). Extracellular serine peptidases possessing an ability to cleave key host proteins were also reported in clinical isolates of C. rugosa (50, 94, and 120 kDa), C. lipolytica (60 kDa) (de Melo et al. 2007), C. tropicalis and C. dubliniensis (44 to 104 kDa) (Portela et al. 2010), and C. parapsilosis (60 kDa) (Vermelho et al. 2010).
Collectively, our results reveal the capability of clinical isolates of the C. haemulonii species complex to produce serine peptidases that can hydrolyze a broad spectrum of protein substrates, some of which have great significance in the fungi-host interface (e.g., IgG, HSA, and Hgb). Moreover, these peptidases are active across a wide range of pH and temperature, which may explain the ability of the C. haemulonii species complex to colonize several sites of the human body. Future in vivo investigations are needed to determine the biological significance of these peptidases and, additionally, to explore the possibility of using these enzymes as new targets for the development of antifungal drugs based on peptidase inhibitors.
References
Behnsen J, Lessing F, Schindler S, Wartenberg D, Jacobsen ID, Thoen M, Zipfel PF, Brakhage AA (2010) Secreted Aspergillus fumigatus protease Alp1 degrades human complement proteins C3, C4, and C5. Infect Immun 78:3585–3594
Bochenska O, Rapala-Kozik M, Wolak N, Bras G, Kozik A, Dubin A, Aoki W, Ueda M, Mak P (2013) Secreted aspartic peptidases of Candida albicans liberate bactericidal hemocidins from human hemoglobin. Peptides 48:49–58
Caggiano G, Coretti C, Bartolomeo N, Lovero G, de Giglio O, Montagna MT (2015) Candida bloodstream infections in Italy: changing epidemiology during 16 years of surveillance. Biomed Res Int 2015:256580
Casadevall A (1995) Antibody immunity and invasive fungal infections. Infect Immun 63:4211–4218
Cendejas-Bueno E, Kolecka A, Alastruey-Izquierdo A, Theelen B, Groenewald M, Kostrzewa M, Cuenca-Estrella M, Gomez-Lopez A, Boekhout T (2012) Reclassification of the Candida haemulonii complex as Candida haemulonii (C. haemulonii group I), C. duobushaemulonii sp. nov. (C. haemulonii group II), and C. haemulonii var. vulnera var. nov.: three multiresistant human pathogenic yeasts. J Clin Microbiol 50:3641–3651
Chaffin WL, Lopez-Ribot JL, Casanova M, Gozalbo D, Martinez JP (1998) Cell wall and secreted proteins of Candida albicans: identification, function, and expression. Microbiol Mol Biol Rev 62:130–180
Chen SC, Marriott D, Playford EG, Nguyen Q, Ellis D, Meyer W, Sorrell TC, Slavin M (2009) Candidaemia with uncommon Candida species: predisposing factors, outcome, antifungal susceptibility, and implications for management. Clin Microbiol Infect 15:662–669
Crouzet J, Sotto A, Picard E, Lachaud L, Bourgeois N (2011) A case of Candida haemulonii osteitis: clinical features, biochemical characteristics, and antifungal resistance profile. Clin Microbiol Infect 17:1068–1070
Gargeya IB, Pruitt WR, Meyer SA, Ahearn DG (1991) Candida haemulonii from clinical specimens in the USA. J Med Vet Mycol 29:335–338
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron 4:1–9
Heussen C, Dowdle EB (1980) Electrophoretic analysis of plasminogen activators in polyacrylamide gels containing sodium dodecyl sulfate and copolymerized substrates. Anal Biochem 102:196–202
Ito M, Yamada T, Makimura K, Ishihara Y, Satoh K, Kikuchi K, Nomura Y, Ishii Y, Abe S (2010) Intracellular serine protease from Candida glabrata species detected and analyzed by zymography. Med Mycol 1:29–35
Jousson O, Lechenne B, Bontems O, Mignon B, Reichard U, Barblan J, Quadroni M, Monod M (2004) Secreted subtilisin gene family in Trichophyton rubrum. Gene 339:79–88
Khan ZU, Al-Sweih NA, Ahmad S, Al-Kazemi N, Khan S, Joseph L, Chandy R (2007) Outbreak of fungemia among neonates caused by Candida haemulonii resistant to amphotericin B, itraconazole, and fluconazole. J Clin Microbiol 45:2025–2027
Kim MN, Shin JH, Sung H, Lee K, Kim EC, Ryoo N, Lee JS, Jung SI, Park KH, Kee SJ, Kim SH, Shin MG, Suh SP, Ryang DW (2009) Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features. Clin Infect Dis 48:e57–e61
Lavarde V, Daniel F, Saez H, Arnold M, Faguer B (1984) Peritonite mycosique a Torulopsis haemulonii. Bull Soc Fr Mycol Med 13:173–176
Lehmann PF, Wu LC, Pruitt WR, Meyer SA, Ahearn DG (1993) Unrelatedness of groups of yeasts within the Candida haemulonii complex. J Clin Microbiol 31:1683–1687
Li W, Hu YA, Li FQ, Shi LN, Shao HF, Huang M, Wang Y, Han DD, Liao H, Ma CF, Zhang GY (2015) Distribution of yeast isolates from invasive infections and their in vitro susceptibility to antifungal agents: evidence from 299 cases in a 3-year (2010 to 2012) surveillance study. Mycopathologia 179:397–405
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275
Mahon CS, O’Donoghue AJ, Goetz DH, Murray PG, Craik CS, Tuohy MG (2009) Characterization of a multimeric, eukaryotic prolyl aminopeptidase: an inducible and highly specific intracellular peptidase from the non-pathogenic fungus Talaromyces emersonii. Microbiology 155:3673–3682
Melo AC, Dornelas-Ribeiro M, De Souza EP, Macrae A, Fracalanzza SE, Vermelho AB (2007) Peptidase profiles from non-albicans Candida spp. isolated from the blood of a patient with chronic myeloid leukemia and another with sickle cell disease. FEMS Yeast Res 7:1004–1012
Monod M, Capoccia S, Lechenne B, Zaugg C, Holdom M, Jousson O (2002) Secreted proteases from pathogenic fungi. Int J Med Microbiol 292:405–419
Moors MA, Stull TL, Blank KJ, Buckley HR, Mosser DM (1992) A role for complement receptor-like molecules in iron acquisition by Candida albicans. J Exp Med 175:1643–1651
Noble SM (2013) Candida albicans specializations for iron homeostasis: from commensalism to virulence. Curr Opin Microbiol 16:708–715
Nucci M, Queiroz-Telles F, Tobon AM, Restrepo A, Colombo AL (2010) Epidemiology of opportunistic fungal infections in Latin America. Clin Infect Dis 51:561–570
Papon N, Courdavault V, Clastre M, Bennett RJ (2013) Emerging and emerged pathogenic Candida species: beyond the Candida albicans paradigm. PLoS Pathog 9:e1003550
Pfaller MA, Diekema DJ, Gibbs DL, Newell VA, Ellis D, Tullio V, Rodloff A, Fu W, Ling TA (2010) Results from the ARTEMIS DISK Global Antifungal Surveillance Study, 1997 to 2007: a 10.5-year analysis of susceptibilities of Candida species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion. J Clin Microbiol 48:1366–1377
Plantner JJ (1991) A microassay for proteolytic activity. Anal Biochem 195:129–131
Portela MB, Kneipp LF, Ribeiro de Souza IP, Holandino C, Alviano CS, Meyer-Fernandes JR, de Araujo Soares RM (2010) Ectophosphatase activity in Candida albicans influences fungal adhesion: study between HIV-positive and HIV-negative isolates. Oral Dis 16:431–437
Ramachandra S, Linde J, Brock M, Guthke R, Hube B, Brunke S (2014) Regulatory networks controlling nitrogen sensing and uptake in Candida albicans. PLoS One 9:e92734
Ramos LS, Branquinha MH, Santos ALS (2017a) Different classes of hydrolytic enzymes produced by multidrug-resistant yeasts comprising the Candida haemulonii complex. Med Mycol 55:228–232
Ramos LS, Figueiredo-Carvalho MH, Barbedo LS, Ziccardi M, Chaves AL, Zancope-Oliveira RM, Pinto MR, Sgarbi DB, Dornelas-Ribeiro M, Branquinha MH, Santos ALS (2015) Candida haemulonii complex: species identification and antifungal susceptibility profiles of clinical isolates from Brazil. J Antimicrob Chemother 70:111–115
Ramos LS, Oliveira SSC, Souto XM, Branquinha MH, Santos ALS (2017b) Planktonic growth and biofilm formation profiles in Candida haemulonii species complex. Med Mycol 55:785–789
Rao MB, Tanksale AM, Ghatge MS, Deshpande VV (1998) Molecular and biotechnological aspects of microbial proteases. Microbiol Mol Biol Rev 62:597–635
Rodero L, Cuenca-Estrella M, Cordoba S, Cahn P, Davel G, Kaufman S, Guelfand L, Rodriguez-Tudela JL (2002) Transient fungemia caused by an amphotericin B-resistant isolate of Candida haemulonii. J Clin Microbiol 40:2266–2269
Rodier MH, Moudni BE, Ghazali M, Lacroix C, Jacquemin JL (1994) Electrophoretic detection of cytoplasmic serine proteinases (gelatinases) in Candida albicans. Exp Mycol 18:267–270
Ruan SY, Kuo YW, Huang CT, Hsiue HC, Hsueh PR (2010) Infections due to Candida haemulonii: species identification, antifungal susceptibility and outcomes. Int J Antimicrob Agents 35:85–88
Santos ALS (2011) Protease expression by microorganisms and its relevance to crucial physiological/pathological events. World J Biol Chem 2:48–58
Santos ALS, Carvalho IM, Silva BA, Portela MB, Alviano CS, Soares RMA (2006) Secretion of serine peptidase by a clinical strain of Candida albicans: influence of growth conditions and cleavage of human serum proteins and extracellular matrix components. FEMS Immunol Med Microbiol 46:209–220
Santos ALS, Soares RMA (2005) Candida guilliermondii isolated from HIV-infected human secretes a 50 kDa serine proteinase that cleaves a broad spectrum of proteinaceous substrates. FEMS Immunol Med Microbiol 43:13–20
Sardi JC, Scorzoni L, Bernardi T, Fusco-Almeida AM, Mendes Giannini MJ (2013) Candida species: current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options. J Med Microbiol 62:10–24
Segal B (2006) Molecular pathogenesis of fungal infections. In: Runge MS, Patterson C (eds) Principles of molecular medicine. Humana Press, pp 920–933
Sehnaz A, Sevtap AA, Dolunay G, Sibel A, Omrum U, Murat A (2015) Epidemiology of candidaemia in a tertiary care university hospital: 10-year experience with 381 candidaemia episodes between 2001 and 2010. Mycoses 58:498–505
Silva NC, Nery JM, Dias AL (2014) Aspartic proteinases of Candida spp.: role in pathogenicity and antifungal resistance. Mycoses 57:1–11
Souza PM, Bittencourt ML, Caprara CC, de Freitas M, de Almeida RP, Silveira D, Fonseca YM, Ferreira Filho EX, Pessoa Junior A, Magalhaes PO (2015) A biotechnology perspective of fungal proteases. Braz J Microbiol 46:337–346
van U, Kolipinski MC (1962) Torulopsis haemulonii nov. sp., a yeast from the Atlantic Ocean. Antonie Van Leeuwenhoek 28:78–80
Vermelho AB, Mazotto AM, de Melo AC, Vieira FH, Duarte TR, Macrae A, Nishikawa MM, da Silva Bon EP (2010) Identification of a Candida parapsilosis strain producing extracellular serine peptidase with keratinolytic activity. Mycopathologia 169:57–65
Yike I (2011) Fungal proteases and their pathophysiological effects. Mycopathologia 171:299–323
Acknowledgments
The authors would like to thank Dr. Malachy McCann (Chemistry Department, National University of Ireland Maynooth, Co. Kildare, Ireland) for the valuable critical English review and Dr. Diogo de Azevedo Jurelevicius (Instituto de Microbiologia Paulo de Góes, Universidade Federal do Rio de Janeiro, Brazil) for the help with the dendrogram analysis.
Funding
This study was supported by grants from Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Souto, X.M., Ramos, L.S., Branquinha, M.H. et al. Identification of cell-associated and secreted serine-type peptidases in multidrug-resistant emergent pathogens belonging to the Candida haemulonii complex. Folia Microbiol 64, 245–255 (2019). https://doi.org/10.1007/s12223-018-0651-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-018-0651-y